Introduction
Systemic lupus erythematosus (SLE) is an autoimmune
disease involving multiple organs and is characterized by chronic
inflammation with autoantibody production. The pathogenesis of SLE
is complex and involves numerous immune cells and cytokines. B
cells are known to have an essential role in autoantibody
production in autoimmune disease (1). In addition, T cells have been
recognized as a crucial component in the pathogenicity of SLE
through their communication with B cells, which helps drive
autoantibody production (2). In
recent years, the importance of innate immunity in SLE pathogenesis
has become increasingly evident, particularly regarding the role of
Toll-like receptor (TLR) signaling pathways (3,4).
Furthermore, the abnormal expression of certain cytokines and their
receptors leads to immune hyperactivity in SLE; thus, cytokines
such as interferon (IFN)-α, interleukin (IL)-1, IL-6, and tumor
necrosis factor (TNF) are considered to be therapeutic targets for
SLE (5).
Previous studies have determined that several
cytokine genes are regulated by CCAAT/enhancer-binding protein β
[C/EBP β, also known as nuclear factor for IL 6, cysteine rich
protein 2, IL-6DBP, liver-enriched activating protein (LAP), NF-M,
α1 acid glycoprotein/enhancer-binding protein, or ApC/EBP), which
is a member of the C/EBP family of transcription factors (6). Three C/EBP β isoforms have been
identified: Liver-enriched activating protein* (LAP*, or C/EBP β1),
liver-enriched activating protein (LAP, or C/EBP β2), and
liver-enriched inhibitory protein (LIP, or C/EBP β3) (7). C/EBP β is expressed in immune cells,
specifically in myelomonocytic cells and macrophages (8–10). C/EBP
β has important roles in adipocyte differentiation, breast cancer
and the initiation of liver regeneration (11–13). In
myelomonocytic cells and monocytes/macrophages, C/EBP β is
regulated by a variety of differentiation- and
proliferation-inducing agents, including cytokines and inflammatory
substances (10), which mostly
increase C/EBP β expression. In murine J774.2
macrophage-like cells, C/EBP β mRNA and protein
expression was induced by TNF, IL-1 and IFN γ (14). Furthermore, the expression of
C/EBP β mRNA increased markedly during
differentiation to a macrophage lineage in M1 mouse myeloid
leukemia cells, U937 human histiocytic leukemia cells, HL-60
promyelocytic leukemia cells and human peripheral monocytes
(15). Vascular endothelial growth
factor (VEGF) reduced the expression levels of the inhibitory
isoform of C/EBP β (LIP) in the THP-1 cultured human monocytic
leukemia cell line (16). C/EBP β
contributes to the regulation of certain inflammatory cytokines,
such as TNF, IL-1β, IL-6, IL-10 and IL-12, which have important
roles in SLE pathogenesis (10,17–19), and
the serum levels of IL-1, IL-6, TNF-α, IFN-γ and VEGF are
significantly elevated in SLE patients (20–22).
Furthermore, IL-10 shows a positive correlation with C-reactive
protein and a negative correlation with complement component C3 in
SLE (20).
Single-nucleotide polymorphisms of TNF-α-induced
protein 3 (TNFAIP3) confer susceptibility to SLE (23). The TNFAIP3 protein, also known as
A20, is a potent anti-inflammatory signaling molecule, and TNFAIP3
downregulation and dysfunction are associated with inflammation in
SLE (23). C/EBP β was reported to
bind to the promoter of TNFAIP3 following lipopolysaccharide
(LPS) stimulation in RAW264.7 cells (24). However, the levels of C/EBP β and its
target gene products were increased in mice with knocked-out
TNFAIP3-interacting protein 1 (TNIP1), which also confers
susceptibility to SLE (25,26).
Although there is a known association between SLE
disease and the regulators and target gene products of C/EBP β, the
expression level of C/EBP β in immune cells from SLE
patients is unknown. Furthermore, the association between the
expression of C/EBP β and the expression of
TNFAIP3 and TNIP1 in SLE is unclear. Therefore, the
present study compared the expression of C/EBP β mRNA
in peripheral blood mononuclear cells (PBMCs) from patients with
SLE and healthy controls, and analyzed the association of
C/EBP β with TNFAIP3 and TNIP1 in order
to elucidate the role of C/EBP β expression in the
pathogenesis of SLE.
Materials and methods
Human subjects
A total of 20 patients with SLE who were diagnosed
according to the criteria of the American College of Rheumatology
(27) were enrolled in this study.
In addition, 20 gender- and age-matched healthy controls without
any rheumatological conditions were recruited. Individual disease
activity was quantified using the SLE disease activity index
(SLEDAI) score (28). All of the
blood samples collected from the patients with SLE and healthy
controls were used with informed consent and approval from the
Ethics Committee of Southwestern Hospital (Chongqing, China). The
study was performed in accordance with the 1964 Declaration of
Helsinki and its later amendments.
For all of the patients with SLE, routine blood and
urine tests were conducted using a hematology analyzer (Shenzhen
Mindray Bio-Medical Electronics Co., Ltd., Shenzhen, China) and a
urine sediment analyzer (Dirui Industrial Co., Ltd., Changchun,
China). The serum levels of complement components C3 and C4 were
detected using immunoturbidimetric assays (IMMAGE 800; Beckman
Coulter, Inc., Fullerton, CA, USA) according to the manufacturer's
instructions. Autoantibodies, including anti-nuclear (ANA),
anti-dsDNA, anti-Smith (anti-Sm) and anti-nuclear ribonuclear
protein (anti-nRNP) autoantibodies were detected using a EUROLINE
test (EUROIMMUN AG, Luebeck, Germany) according to the
manufacturer's instructions.
PBMC preparation and RNA
extraction
PBMCs were separated by density gradient
centrifugation, 1,200 rpm/min 15 min, from peripheral blood
anticoagulated with sodium citrate (Tianjin Haoyang Co., Ltd.,
Tianjin, China). Total RNA was extracted from 5×105
PBMCs using RNAiso Plus (Takara Biotechnology Co., Ltd., Dalian,
China) according to the manufacturer's instructions, and then
quantified by photometrical measurement.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For each sample, 1 µg RNA was reverse transcribed to
cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser
(Perfect Real Time) with gDNA Eraser (Takara Biotechnology Co.,
Ltd.). The quantity of C/EBP β, TNIP1 and
TNFAIP3 cDNA was then evaluated by qPCR. The expression
levels of β-actin mRNA were also determined and served as an
internal control. qPCR was performed using a SYBR Green I Real-Time
PCR kit (Takara Biotechnology Co., Ltd.) on a Stratagene Mx3000p
Real-Time PCR system (Agilent Technologies GmbH, Waldbronn,
Germany). The PCR thermal cycling conditions were as follows: 95°C
for 2 min; 50 cycles of 95°C for 30 sec, 62°C for 30 sec and 72°C
for 20 sec; and 1 cycle of 95°C for 1 min, 55°C for 30 sec and 95°C
for 30 sec. The primers used were as follows: C/EBP β
forward, 5′-CACAGCGACGACTGCAAGATCC-3′ and reverse,
5′-CTTGAACAAGTTCCGCAGGGTG-3′; TNIP1 forward,
5′-CAGAATGAGTTGCTGAAACA-3′ and reverse, 5′-TCTCCTCATCTTTGAATGCT-3′;
TNFAIP3 forward, 5′-GTGTATTTTGGGACTCCAGA-3′ and reverse,
5′-ACTTCTGGCAGTATCCTTCA-3′; and β-actin forward,
5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse, 5′-GGGCACGAAGGCTCATCATT-3′
(Sangon Biotech Co., Ltd., Shanghai, China). A melting-curve
analysis was performed to ensure specificity of the PCR products,
and all of the PCR products were subjected to electrophoresis in an
agarose gel to confirm the presence of a single band of the
expected size. The expression levels of C/EBP β,
TNIP1, and TNFAIP3 were normalized to β-actin and
determined using the comparative (2−ΔΔCt) method. The
experiment was repeated three times.
Statistical analysis
The relative expression of C/EBP β,
TNIP1, and TNFAIP3 mRNA for each sample is presented
as a mean ± standard deviation. The difference in
C/EBP β mRNA expression levels between the subject
groups was analyzed with the Mann-Whitney test. Correlation
analyses were conducted using Spearman's rank test. All analyses
were performed using GraphPad Prism software, version 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical characteristics of patients
with SLE
The demographic characteristics, clinical
manifestations, and laboratory measurements of the patients with
SLE are presented in Table I. The
demographic characteristics were not significantly different
between the SLE group and the healthy control group. Arthritis,
vasculitis and rash were present in 3, 2 and 4 of the 20 patients
with SLE, respectively. The mean SLEDAI score was 4.70 (range,
0–12). Anti-ANA, anti-dsDNA, anti-Sm and anti-nRNP autoantibodies
were detected in 20, 6, 7 and 10 of the 20 patients with SLE,
respectively. Thrombocytopenia and proteinuria were detected in 4
and 3 of the 20 patients with SLE, respectively. The mean level of
complement components C3 (0.66 g/l) and C4 (0.13 g/l) in patients
with SLE was lower than the controls (C3 normal range, 0.79–1.32
g/l; C4 normal range, 0.16–0.38 g/l).
 | Table I.Clinical characteristics of patients
with SLE. |
Table I.
Clinical characteristics of patients
with SLE.
| Parameter | SLE patients
(n=20) | Healthy controls
(n=20) |
|---|
| Demographic
characteristics |
|
|
| Female,
n (%) | 18 (90) | 18 (90) |
| Male, n
(%) | 2 (10) | 2 (10) |
| Age,
years | 33.20 (18–54) | 32.45 (20–52) |
| Clinical
features |
|
|
|
Arthritis, n (%) | 3 (15) | – |
|
Vasculitis, n (%) | 2 (10) | – |
| Rash, n
(%) | 4 (20) | – |
| SLEDAI,
points | 4.70 (0–12) | – |
| Laboratory
measurements |
|
|
| ANA, n
(%) | 20 (100) | – |
|
Anti-dsDNA, n (%) | 6 (30) | – |
|
Anti-Sm, n (%) | 6 (30) | – |
|
Anti-nRNP, n (%) | 10 (50) |
|
| C3,
g/l | 0.66
(0.38–1.36) | – |
| C4,
g/l | 0.13
(0.04–0.42) | – |
|
Thrombocytopenia, n (%) | 4 (20) | – |
|
Proteinuria, n (%) | 3 (15) | – |
C/EBP β mRNA expression was elevated
in PBMCs from patients with SLE
The expression of C/EBP β mRNA in
PBMCs was examined in 20 patients with SLE and 20 gender- and
age-matched healthy controls using RT-qPCR. The relative mRNA
expression levels of C/EBP β were significantly
higher in PBMCs from patients with SLE (1.1340), as compared with
the PBMCs from the healthy controls (0.6256; P=0.0179; Fig. 1).
C/EBP β mRNA expression was elevated
in SLE patients that were positive for anti-Sm or anti-nRNP
antibodies, or had a high ANA titer
The relative mRNA expression levels of
C/EBP β were higher in PBMCs from SLE patients with
high ANA titer (≤1:160; 1.254), as compared with patients with low
ANA titer (≥1:80; 0.4567; P=0.0262). In addition, the relative
expression levels of C/EBP β mRNA were significantly
higher in PBMCs from SLE patients positive for anti-Sm (1.657) or
anti-nRNP (1.550) antibodies, as compared with patients negative
for anti-Sm (0.8530; P=0.0324) or anti-nRNP (0.7185; P=0.0039)
antibodies (Fig. 2).
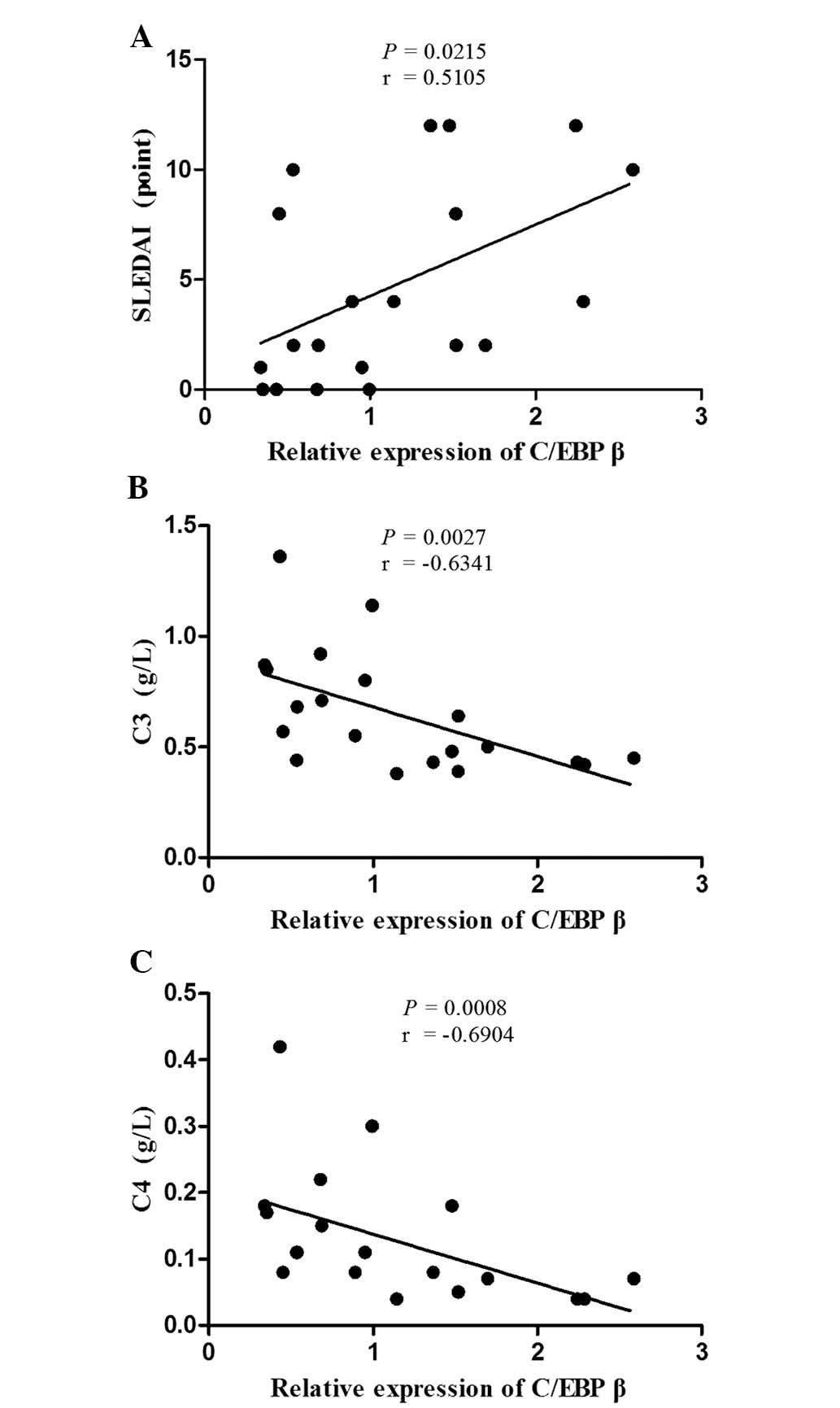
C/EBP β mRNA expression correlates
with disease activity in patients with SLE
The association between C/EBP β mRNA
expression and the demographic characteristics, clinical
manifestations and laboratory parameters was analyzed. As shown in
Fig. 3, C/EBP β
expression was positively correlated with the SLEDAI score
(r=0.5105; P=0.0215) in the patients with SLE. Furthermore,
C/EBP β expression was negatively correlated with the
serum levels of complement components C3 (r=−0.6341; P=0.0027) and
C4 (r=−0.6904; P=0.0008). No statistically significant association
was observed between C/EBP β mRNA expression levels
and the other characteristics, clinical manifestations or
laboratory parameters in patients with SLE.
C/EBP β mRNA expression is positively
correlated with TNIP1 and TNFAIP3 expression in patients with
SLE
The expression levels of TNIP1 and
TNFAIP3 mRNA were determined in the PBMCs from the 20
patients with SLE using RT-qPCR, and the association between
C/EBP β expression and TNIP1 and
TNFAIP3 expression was examined. As shown in Fig. 4, C/EBP β expression was
positively correlated with TNIP1 expression (r=0.5865;
P=0.0086) and TNFAIP3 expression (r=0.4692; P=0.0369) in the
patients with SLE.
Discussion
The results of the present study demonstrated the
upregulation of C/EBP β expression in patients with
SLE, specifically in patients with anti-Sm and anti-nRNP
antibodies, or high ANA titer, which is specificity with SLE or
common in SLE (29). A positive
correlation was observed between C/EBP β expression
and disease activity (SLEDAI score), and a negative correlation
between C/EBP β expression and the serum levels of
complement components C3 and C4, which are considered to be part of
the disease activity index. The data also demonstrated a positive
correlation between C/EBP β and
TNFAIP3/TNIP1 expression in patients with SLE. These
results suggest that increased expression of C/EBP β
and interactions between C/EBP β and
TNIP1/TNFAIP3 may be involved in the pathogenesis of
SLE.
SLE is a chronic inflammatory disease associated
with the dysfunction of numerous immune cells and cytokines. The
TLR signaling pathway has a crucial role as a trigger of
inflammation in SLE (30). TLR
activation (recognition of CpG-DNA by TLR9 or recognition of LPS by
TLR4) leads to the recruitment of TLR signaling complexes involving
TNF receptor-associated factor (TRAF)3 and TRAF6 via myeloid
differentiation marker 88 (31–33). The
signaling complexes activate C/EBP β through the mitogen-activated
protein kinase (MAPK) p38 (34,35). As
mentioned above, the majority of regulators and target gene
products of C/EBP β are abnormally expressed in patients with SLE,
and this aberrant expression contributes to the pathogenesis of
SLE. Therefore, C/EBP β is thought to be a potential pivotal
component of the inflammatory signaling pathway in SLE.
TNFAIP3 and TNIP1 were identified as
SLE susceptibility loci in a genome-wide association study
(36). TNFAIP3 is an inhibitor of
inflammation, and the deubiquitination of TNFAIP3 in T cells
negatively regulates NF-κB, which is crucial for inflammatory and
immune responses (37). Furthermore,
NF-κB and p38 were observed to regulate the transcription of
TNFAIP3 via C/EBP β in activated macrophages (24). TNFAIP3 mRNA expression was
shown to be reduced in patients with SLE (38). TNIP1 knockout mice showed an
increased expression of C/EBP β without changes to
NF-κB or MAPK, and developed an inflammatory disease with
characteristics similar to human SLE (26). In the present study, the positive
correlation between C/EBP β and
TNIP1/TNFAIP3 mRNA expression in patients with SLE
suggests that the interaction between C/EBP β and
TNIP1/TNFAIP3 may be involved in the pathogenesis of
SLE, and may indirectly demonstrate that C/EBP β could regulate
TNFAIP3 mRNA expression. However, TNFAIP3 and TNIP1
mRNA expression was observed to be reduced in patients with SLE
(38,39). Apparent discrepancies between the
TNFAIP3 and TNIP1 mRNA expression reported in the current study,
and those of earlier studies may be due to the dysfunction of
TNFAIP3 and TNIP1 single nucleotide polymorphisms (SNP) in
different regions (39,40). The National Center of Biotechnology
Information (http://www.ncbi.nlm.nih.gov/) website database states
that the SNP loci are in introns, exons, promoters, enhancers and
other regions. Some polymorphisms may cause reduced expression or
activity of anti-inflammatory A20, predisposing individuals to
develop SLE (41). Therefore, in SLE
the association between TNFAIP3 and TNIP1 SNP
mutations and expression and the function of its production
required further study. Future research we focus on investigating
the association between C/EBP β and
TNIP1/TNFAIP3 in SLE.
In recent years, the development of targeted
therapies for SLE has become an important research focus, due to
the fact that the available first-line drugs show poor efficacy and
lead to certain adverse reactions in patients with SLE. With the
exception of belimumab, which is an anti-B-cell-activating factor
(anti-BAFF) antibody, the efficacy and safety of the majority of
immune cell-targeted therapies for SLE, such as rituximab,
ofatumumab, ocrelizumab and veltuzumab (anti-CD20); epratuzumab
(anti-CD22); and atacicept (anti-BAFF and a proliferation-inducing
ligand) are not as good as expected (42). Therefore, a clear understanding of
the pathogenesis of SLE may significantly improve the targeted
therapy of SLE. An important step will be defining the function and
regulation of C/EBP β, a pivotal component of the SLE
inflammatory signaling pathway. Additional potential SLE
therapeutic targets include TNF and TNFAIP3, which
are closely associated with C/EBP β (43,44).
However, as C/EBP β is also involved in adipocyte
differentiation, breast cancer and liver regeneration (11–13), the
potential side effects of changes in C/EBP β
expression resulting from targeted therapy must be assessed.
In conclusion, the present study demonstrated that
C/EBP β mRNA expression was upregulated and
positively correlated with disease activity and the expression of
TNIP1 and TNFAIP3 mRNA in patients with SLE. These
results suggest that increased C/EBP β expression and
an interaction between C/EBP β and
TNIP1/TNFAIP3 may contribute to the inflammation
pathogenesis of SLE. These data also indicate that the influence of
C/EBP β should be considered during the development
of targeted therapies for SLE.
Acknowledgments
The authors are grateful to Professor Bing Ni
(Institute of Immunology, PLA, Third Military Medical University;
Chongqing, China) for suggestions on the experimental methods and
research. This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81201232 and
81472883).
References
|
1
|
Yanaba K, Bouaziz JD, Matsushita T, Magro
CM, St Clair EW and Tedder TF: B-lymphocyte contributions to human
autoimmune disease. Immunol Rev. 223:284–299. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mak A and Kow N: The pathology of T cells
in systemic lupus erythematosus. J Immunol Res. 2014:4190292014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Celhar T, Magalhães R and Fairhurst AM:
TLR7 and TLR9 in SLE: When sensing self goes wrong. Immunol Res.
53:58–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kunz M: Lupus erythematosus. Part I:
Epidemiology, genetics and immunology. J Dtsch Dermatol Ges.
11:709–719; quiz 720, 709–720; quiz 721. 2013.[(In English,
German)]. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davis LS, Hutcheson J and Mohan C: The
role of cytokines in the pathogenesis and treatment of systemic
lupus erythematosus. J Interferon Cytokine Res. 31:781–789. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ramji DP and Foka P:
CCAAT/enhancer-binding proteins: Structure, function and
regulation. Biochem J. 365:561–575. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sears RC and Sealy L: Multiple forms of
C/EBP beta bind the EFII enhancer sequence in the Rous sarcoma
virus long terminal repeat. Mol Cell Biol. 14:4855–4871. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamanaka R, Lekstrom-Himes J, Barlow C,
Wynshaw-Boris A and Xanthopoulos KG: CCAAT/enhancer binding
proteins are critical components of the transcriptional regulation
of hematopoiesis (Review). Int J Mol Med. 1:213–221.
1998.PubMed/NCBI
|
|
9
|
Li-Weber M, Salgame P, Hu C, Davydov IV
and Krammer PH: Differential interaction of nuclear factors with
the PRE-I enhancer element of the human IL-4 promoter in different
T cell subsets. J Immunol. 158:1194–1200. 1997.PubMed/NCBI
|
|
10
|
Huber R, Pietsch D, Panterodt T and Brand
K: Regulation of C/EBPβ and resulting functions in cells of the
monocytic lineage. Cell Signal. 24:1287–1296. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lane MD, Tang QQ and Jiang MS: Role of the
CCAAT enhancer binding proteins (C/EBPs) in adipocyte
differentiation. Biochem Biophys Res Commun. 266:677–683. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zahnow CA: CCAAT/enhancer-binding protein
beta: Its role in breast cancer and associations with receptor
tyrosine kinases. Expert Rev Mol Med. 11:e122009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fausto N: Liver regeneration. J Hepatol.
32(1): Suppl. S19–S31. 2000. View Article : Google Scholar
|
|
14
|
Tengku-Muhammad TS, Hughes TR, Ranki H,
Cryer A and Ramji DP: Differential regulation of macrophage
CCAAT-enhancer binding protein isoforms by lipopolysaccharide and
cytokines. Cytokine. 12:1430–1436. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Natsuka S, Akira S, Nishio Y, Hashimoto S,
Sugita T, Isshiki H and Kishimoto T: Macrophage
differentiation-specific expression of NF-IL6, a transcription
factor for interleukin-6. Blood. 79:460–466. 1992.PubMed/NCBI
|
|
16
|
Nolan A, Weiden MD, Thurston G and Gold
JA: Vascular endothelial growth factor blockade reduces plasma
cytokines in a murine model of polymicrobial sepsis. Inflammation.
28:271–278. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng H, Wang W, Zhou M, Li R, Pan HF and
Ye DQ: Role of interleukin-10 and interleukin-10 receptor in
systemic lupus erythematosus. Clin Rheumatol. 32:1255–1266. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yap DY and Lai KN: The role of cytokines
in the pathogenesis of systemic lupus erythematosus-from bench to
bedside. Nephrology (Carlton). 18:243–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fojtíková M, Cerná M and Pavelka K: A
review of the effects of prolactin hormone and cytokine on the
development and pathogenesis of autoimmune diseases. Vnitr Lek.
56:402–413. 2010.(In Czech). PubMed/NCBI
|
|
20
|
Cigni A, Pileri PV, Faedda R, Gallo P,
Sini A, Satta AE, Marras R, Carta E, Argiolas D, Rum I and Masala
A: Interleukin 1, interleukin 6, interleukin 10, and tumor Necrosis
factor α in active and quiescent systemic lupus erythematosus. J
Investig Med. 62:825–829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lyn-Cook BD, Xie C, Oates J, Treadwell E,
Word B, Hammons G and Wiley K: Increased expression of Toll-like
receptors (TLRs) 7 and 9 and other cytokines in systemic lupus
erythematosus (SLE) patients: Ethnic differences and potential new
targets for therapeutic drugs. Mol Immunol. 61:38–43. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou L, Lu G, Shen L, Wang L and Wang M:
Serum levels of three angiogenic factors in systemic lupus
erythematosus and their clinical significance. Biomed Res Int.
2014:6271262014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma A and Malynn BA: A20: Linking a complex
regulator of ubiquitylation to immunity and human disease. Nat Rev
Immunol. 12:774–785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai TY, Wu SD, Tsai MH, Chuang EY, Chuang
LL, Hsu LC and Lai LC: Transcription of Tnfaip3 is regulated by
NF-κB and p38 via C/EBPβ in activated macrophages. PLoS One.
8:e731532013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gateva V, Sandling JK, Hom G, Taylor KE,
Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et
al: A large-scale replication study identifies TNIP1, PRDM1, JAZF1,
UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus.
Nat Genet. 41:1228–1233. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou J, Wu R, High AA, Slaughter CA,
Finkelstein D, Rehg JE, Redecke V and Häcker H: A20-binding
inhibitor of NF-κB (ABIN1) controls Toll-like receptor-mediated
CCAAT/enhancer-binding protein β activation and protects from
inflammatory disease. Proc Natl Acad Sci USA. 108:E998–E1006. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan EM, Cohen AS, Fries JF, Masi AT,
McShane DJ, Rothfield NF, Schaller JG, Talal N and Winchester RJ:
The 1982 revised criteria for the classification of systemic lupus
erythematosus. Arthritis Rheum. 25:1271–1277. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gladman DD, Ibanez D and Urowitz MB:
Systemic lupus erythematosus disease activity index 2000. J
Rheumatol. 29:288–291. 2002.PubMed/NCBI
|
|
29
|
Wrone DA and Kwon NJ: Andrews' diseases of
the skin: Clinical dermatology. Archives of Dermatology. 137:518.
2001.
|
|
30
|
O'Neill LA, Bryant CE and Doyle SL:
Therapeutic targeting of Toll-like receptors for infectious and
inflammatory diseases and cancer. Pharmacol Rev. 61:177–197. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kawai T, Adachi O, Ogawa T, Takeda K and
Akira S: Unresponsiveness of MyD88-deficient mice to endotoxin.
Immunity. 11:115–122. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Häcker H, Vabulas RM, Takeuchi O, Hoshino
K, Akira S and Wagner H: Immune cell activation by bacterial
CpG-DNA through myeloid differentiation marker 88 and tumor
necrosis factor receptor-associated factor (TRAF)6. J Exp Med.
192:595–600. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Häcker H, Redecke V, Blagoev B,
Kratchmarova I, Hsu LC, Wang GG, Kamps MP, Raz E, Wagner H, Häcker
G, et al: Specificity in Toll-like receptor signalling through
distinct effector functions of TRAF3 and TRAF6. Nature.
439:204–207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Horie R, Ishida T, Maruyama-Nagai M, Ito
K, Watanabe M, Yoneyama A, Higashihara M, Kimura S and Watanabe T:
TRAF activation of C/EBPbeta (NF-IL6) via p38 MAPK induces HIV-1
gene expression in monocytes/macrophages. Microbes Infect.
9:721–728. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takeuchi O and Akira S: Pattern
recognition receptors and inflammation. Cell. 140:805–820. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu
Z, Xu JH, Cai ZM, Huang W, Zhao GP, et al: Genome-wide association
study in a Chinese Han population identifies nine new
susceptibility loci for systemic lupus erythematosus. Nat Genet.
41:1234–1237. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Düwel M, Welteke V, Oeckinghaus A, Baens
M, Kloo B, Ferch U, Darnay BG, Ruland J, Marynen P and Krappmann D:
A20 negatively regulates T cell receptor signaling to NF-kappaB by
cleaving Malt1 ubiquitin chains. J Immunol. 182:7718–7728. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li D, Wang L, Fan Y, Song L, Guo C, Zhu F,
Zhang L and Shi Y: Down-regulation of A20 mRNA expression in
peripheral blood mononuclear cells from patients with systemic
lupus erythematosus. J Clin Immunol. 32:1287–1291. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang DM, Cheng LQ, Zhai ZF, Feng L, Zhong
BY, You Y, Zhang N, Song ZQ, Yang XC, Chen FR and Hao F:
Single-nucleotide polymorphism and haplotypes of TNIP1 associated
with systemic lupus erythematosus in a Chinese Han population. J
Rheumatol. 40:1535–1544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mele A, Cervantes JR, Chien V, Friedman D
and Ferran C: Single nucleotide polymorphisms at the TNFAIP3/A20
locus and susceptibility/resistance to inflammatory and autoimmune
diseases. Adv Exp Med Biol. 809:163–183. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Musone SL, Taylor KE, Lu TT, Nititham J,
Ferreira RC, Ortmann W, Shifrin N, Petri MA, Kamboh MI, Manzi S, et
al: Multiple polymorphisms in the TNFAIP3 region are independently
associated with systemic lupus erythematosus. Nat Genet.
40:1062–1064. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yildirim-Toruner C and Diamond B: Current
and novel therapeutics in the treatment of systemic lupus
erythematosus. J Allergy Clin Immunol. 127:303–312; quiz 313–314.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sriskantharajah S and Ley SC: Cell
biology. Turning off inflammation signaling. Science.
327:1093–1094. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Aringer M and Smolen JS: TNF inhibition in
SLE: Where do we stand? Lupus. 18:5–8. 2009. View Article : Google Scholar : PubMed/NCBI
|