Introduction
Promoter methylation is important in epigenetics and
always leads to transcriptional silencing of tumor suppressor genes
in acute myeloid leukemia (AML) (1).
Although current chemotherapy regimens result in complete remission
in many cases, there is no standard and efficient therapy for
refractory AML (2). As aberrant DNA
methylation is common in AML, clinical trials using
epigenetically-targeted therapies have yielded particularly
promising results in the treatment of hematopoietic malignancies
(3). Several demethylating agents,
including azacytidine and decitabine, have been demonstrated to
improve AML prognosis (4).
Adenomatous polyposis col 2 (APC2) is a tumor
suppressor gene, encoding a protein that controls the stability and
nuclear export of β-catenin, which is a Wnt signaling pathway
transcriptional coactivator (5). Wnt
pathway inhibitors are methylated at a high frequency in AML
patients (6). The cytochrome P450
family 1 subfamily B polypeptide 1 (CYP1B1) gene, which is a
candidate target gene in numerous types of cancers, encodes a
member of the cytochrome P450 enzyme superfamily. Furthermore,
cytochrome P450 enzymes are involved in drug metabolism and the
synthesis of cholesterol, steroids and other lipids (7). A previous study has revealed high
CYP1B1 expression in human myeloid leukemia cell lines
(8) and another study identified a
significant incidence of methylation in the patients with acute
leukemia (9).
Alterations in the promoter methylation status,
which is considered to be an indicator of a molecular abnormality,
can be used to predict the chemotherapeutic outcomes of multiple
regimens towards individualized therapy. The aim of the present
study was to investigate changes in the methylation status in bone
marrow mononuclear cells during chemotherapy and to assess their
potential prognostic value in Han Chinese AML patients.
Materials and methods
Patient samples
Bone marrow specimens and associated
clinicopathological information documented prior to and following
chemotherapy were collected from 30 AML patients treated at the
Department of Hematology and Oncology at Yuyao People's Hospital
(Ningbo, China). There were 13 male and 17 female patients, with a
mean age of 47.8±15.4 years (range, 19–76 years). AML was diagnosed
in accordance with the revised French-American-British
classification, which included classification into subtypes M0-7
(10). In total, 13 different
chemotherapy regimens were chosen according to the status of the
patients. Among them only 6 patients were treated with one kind of
drug, including one male of subtype M5 and four females (two of
subtype M3, and one each of subtypes M4 and M6) who were treated
with cytarabine (Ara-c), and one female M4 patient who was treated
with idarubicin (IDA). The remaining 24 patients were treated with
multi-drug chemotherapy regimens: HAA [homo-harringtonine (HHT) +
cytarabine (Ara-C) + aclacinomycin (ACLA)]; IA (IDA + Ara-c); AAG
[Ara-C + ACLA + granulocyte colony-stimulating factor (G-CSF)];
ATRA combined with arsenic trioxide (AS2O3);
all trans–retinoic acid (ATRA) combined with
AS2O3 and daunorubicin (DNR); HA (HHT +
Ara-c); IA + HAG; AA (Ara-c + ACLA); HAG (HHT + Ara-c + G-CSF); HAG
+ IDA; and ATRA + As2O3 + HA. The clinical
parameters of the patients with AML are summarized in Table I. The Ethics Committee at Yuyao
People's Hospital approved the study. Written informed consent was
obtained from all patients after the possible consequences of
participating in the study had been explained.
 | Table I.Clinical parameters at baseline and
following chemotherapy in patients with acute myeloid leukemia. |
Table I.
Clinical parameters at baseline and
following chemotherapy in patients with acute myeloid leukemia.
| ID | Gender | Age (years) | Subtype | Regimen | Remission | APC2
methylation level before and after treatment | CYP1B1
methylation level before and after treatment |
|---|
| 1 | Male | 55 | M1 | HHT + Ara-C +
ACLA | Yes | HM to HM | HM to HM |
| 2 | Male | 49 | M1 | IDA + Ara-C | No | HM to HM | HM to HM |
| 3 | Male | 76 | M2 | Ara-C + ACLA +
G-CSF | No | HM to HM | HM to U |
| 4 | Male | 66 | M2 | IDA + Ara-C | Yes | HM to HM | HM to HM |
| 5 | Male | 23 | M3 | ATRA +
As2O3 | Yes | HM to HM | HM to HM |
| 6 | Male | 40 | M3 |
As2O3+ DNR +
ATRA | No | HM to HM | U to HM |
| 7 | Male | 59 | M3 | HHT + Ara-C | Yes | HM to HM | U to HM |
| 8 | Male | 67 | M4 | IDA + Ara-C + ACLA +
G-CSF + HHT | Yes | HM to HM | U to HM |
| 9 | Male | 34 | M4 | HHT + Ara-C | Yes | HM to HM | HM to HM |
| 10 | Male | 68 | M5 | Ara-C | Yes | HM to HM | HM to HM |
| 11 | Male | 59 | M5 | Ara-C + ACLA | Yes | HM to HM | HM to HM |
| 12 | Male | 48 | M5 | HHT + Ara-C +
ACLA | No | HM to HM | HM to HM |
| 13 | Male | 52 | M6 | HHT + Ara-C +
G-CSF | Yes | HM to HM | HM to HM |
| 14 | Female | 59 | M1 | Ara-C + ACLA +
G-CSF | No | HM to HM | U to HM |
| 15 | Female | 66 | M2 | Ara-C + ACLA +
G-CSF | Yes | HM to HM | HM to HM |
| 16 | Female | 56 | M2 | Ara-C + ACLA +
G-CSF | Yes | HM to HM | HM to HM |
| 17 | Female | 48 | M2 | HHT + Ara-C +
ACLA | Yes | HM to HM | HM to HM |
| 18 | Female | 50 | M2 | HHT + Ara-C + G-CSF
+ IDA | Yes | HM to HM | HM to HM |
| 19 | Female | 19 | M2 | HHT + Ara-C +
ACLA | Yes | HM to HM | HM to HM |
| 20 | Female | 53 | M2 | HHT + Ara-C | Yes | HM to HM | HM to HM |
| 21 | Female | 51 | M3 | ATRA +
As2O3+ HHT + Ara-C | Yes | HM to HM | HM to HM |
| 22 | Female | 42 | M3 | IDA + Ara-C | No | HM to HM | HM to U |
| 23 | Female | 30 | M3 | Ara-C | Yes | HM to HM | U to HM |
| 24 | Female | 31 | M3 | Ara-C | Yes | HM to HM | HM to HM |
| 25 | Female | 30 | M4 | IDA | Yes | HM to HM | HM to HM |
| 26 | Female | 30 | M4 | IDA + Ara-C | No | HM to HM | HM to HM |
| 27 | Female | 19 | M4 | HHT + Ara-C +
ACLA | Yes | HM to HM | HM to HM |
| 28 | Female | 42 | M4 | Ara-C | Yes | HM to HM | HM to HM |
| 29 | Female | 64 | M6 | HHT + Ara-C | No | HM to HM | HM to HM |
| 30 | Female | 50 | M6 | Ara-C | Yes | HM to HM | HM to HM |
DNA extraction and bisulfite DNA
modification
Genomic DNA was isolated from bone marrow nucleated
cells using a nucleic acid extraction analyzer (Lab-Aid 820; Zeesan
Biotech, Xiamen, China). The DNA concentration of each specimen was
measured via a NanoDrop 1000 spectrophotometer (Thermo Fisher
Scientific, Inc., Wilmington, NC, USA). All DNA samples were
modified using the reagents provided in the ZYMO EZ DNA
Methylation-Gold kit (Zymo Research Corp., Irvine, CA, USA).
Following bisulfite treatment, the converted DNA samples were
stored at −20°C.
Methylation-specific polymerase chain
reaction (MSP PCR)
Modified DNA samples were subjected to MSP using
APC and CYP1B1 MSP primers (11). Two pairs of primers were synthesized
by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China)
according to the sequences listed in Table II. Methylated primers were used to
amplify methylated regions and unmethylated primers to amplify
unmethylated regions. Each PCR reaction contained 1.5 µl sodium
bisulfite modified DNA, 0.5 µl each primer, 10 µl Zymo TaqTM Premix
(Zymo Research, Orange, CA, USA) and 7.5 µl DNAase/RNAase-free
water in a final reaction volume of 20 µl. DNA amplification was
performed using a Veriti® PCR machine (Applied
Biosystems, Thermo Fisher Scientific, Inc.). Thermocycling
conditions were as follows: Initial denaturation step at 95°C for
10 min followed by 35 cycles of amplification, each cycle included
a denaturation step at 94°C for 30 sec, an annealing step with a
primer-specific temperature for 45 sec and an elongation step at
72°C for 1 min. The final extension step was performed at 72°C for
7 min. The methylation status of each sample was determined using
one or two independent experiments. Water blank was used as a
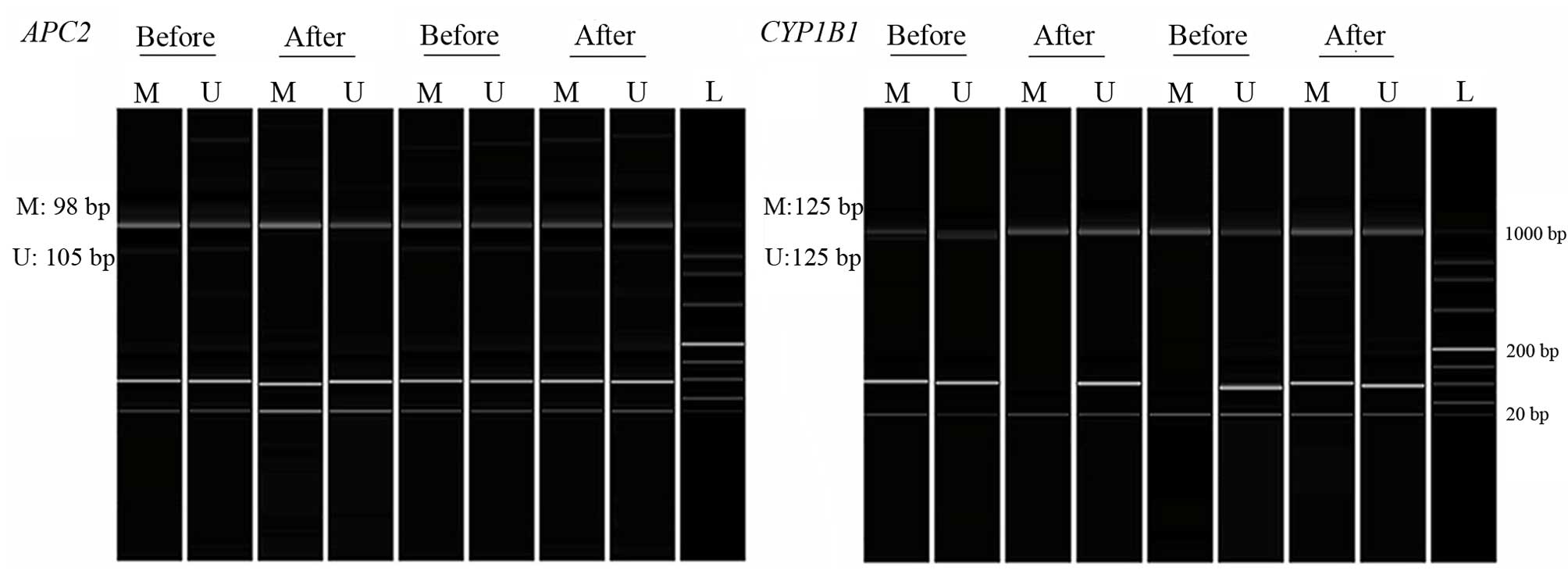
negative control. PCR products were analyzed using a Qsep100 DNA
Analyzer (Bioptic Inc., Taiwan, China). Samples were considered as
methylated or unmethylated according to the presence of clearly
visible peaks by the Q-analyzer software (Fig. 1). The sequences and details of the
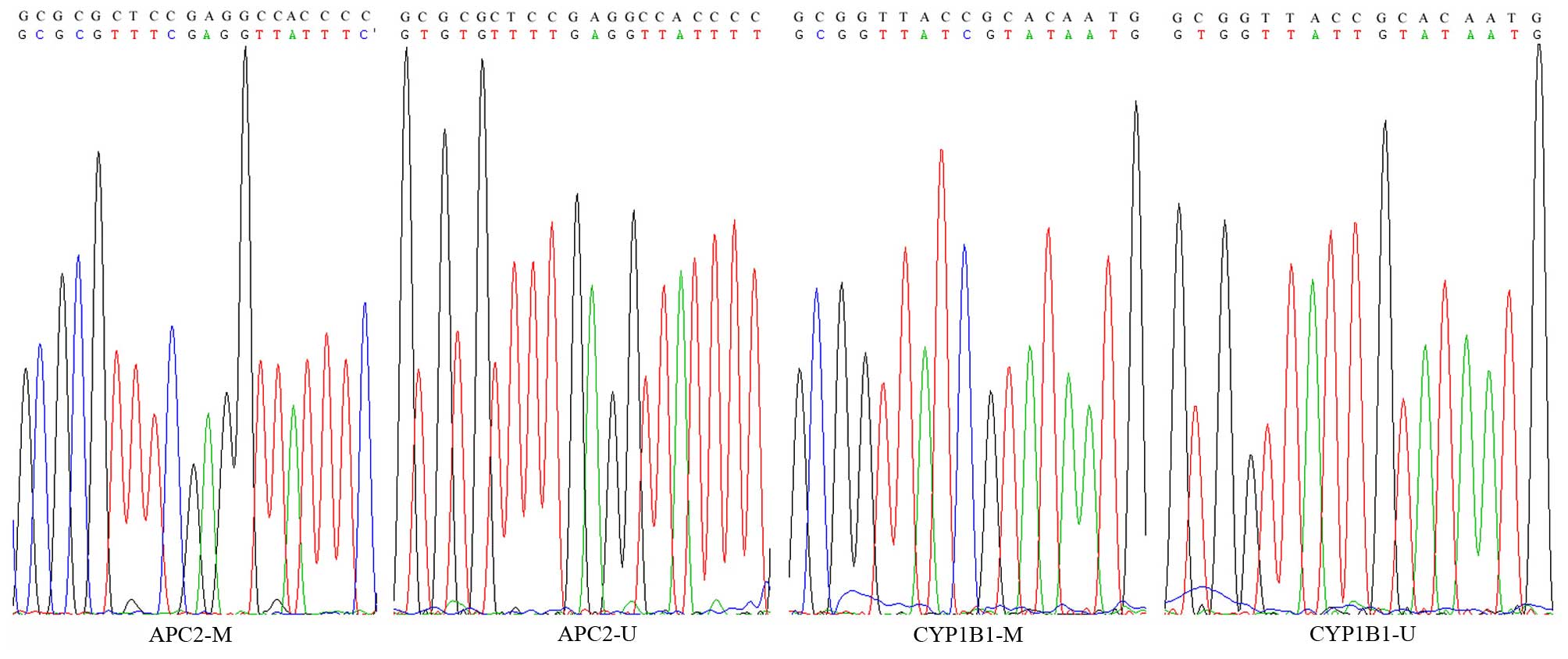
methylated and unmethylated primers are provided in Table II. DNA samples were randomly
sequenced using the Applied Biosystems 3730 DNA Analyzer (Thermo
Fisher Scientific, Inc.) to confirm complete bisulfite conversion
(Fig. 2).
 | Table II.Primers and PCR amplification
conditions. |
Table II.
Primers and PCR amplification
conditions.
| Gene | Primer set | Primer sequence
(5′-3′) | Amplified fragment
length (bp)/Annealing Temperature (°C) |
|---|
| APC2 | MF |
GTCGTTTGTTTAGGTTCGGATC | 98/60 |
|
| MR |
GACCCGAAATAACCTCGAAACG |
|
|
| UF |
TGGTAGTGTTGTTTGTTTAGGTTTGGATTG | 105/57 |
|
| UR |
ACCAAAAATCCCAACCCAAAATAACCTCAAAACA |
|
| CYP1B1 | MF |
CGCGTTTTTAAGTCGAGC | 125/60 |
|
| MR |
ACCCACGTTTCCATTATACG |
|
|
| UF |
GGGTGTGTTTTTAAGTTGAGT | 125/55 |
|
| UR |
ACCCACATTTCCATTATACAATA |
|
Statistical analysis
Comparisons between APC2 and CYP1B1 promoter
methylation were performed using the correction formula of a χ
2 test. Statistical analyses were performed using
the SPSS 18.0 Evaluation version software for Windows (SPSS Inc.,
Chicago, IL, USA). MSP results were compared in samples pre- and
post-chemotherapy.
Results
Chemotherapy
As shown in Table I,
the chemotherapy agents used included Ara-C, ATRA,
As2O3, HHT, G-CSF, IDA, ACLA and DNR. A total
of 22 patients in remission and eight patients with a poorer
prognosis were treated with these 13 agents, either in single or
combined regimens.
MSP was performed on pre- and post-chemotherapy bone
marrow samples from 30 AML patients in order to determine whether
the chemotherapy treatment was able to alter APC2 and
CYP1B1 promoter methylation levels. APC2 promoter
methylation status remained unchanged by chemotherapy. In contrast,
seven patients demonstrated CYP1B1 promoter changes during
chemotherapy.
Regimen-based subgroup analyses of
CYP1B1 promoter methylation changes in AML patients
Seven patients treated with six regimens exhibited
CYP1B1 promoter methylation changes, including five samples
with hypermethylation and two samples demonstrating
hypomethylation. Of the five patients with induced
hypermethylation, three patients in remission were treated with
Ara-C, HHT + Ara-C and IDA + Ara-C + ACLA + G-CSF + HHT regimens,
and two patients treated with As2O3 + DNR +
ATRA. Ara-C + ACLA + G-CSF regimens resulted in a poor patient
prognosis. Two patients with induced hypomethylation were treated
with Ara-C + ACLA + G-CSF, and IDA + Ara-C regimens and had a
poorer prognosis.
Analyses of CYP1B1 promoter
methylation changes in patients based on AML subtypes
The results of the present study were obtained from
three patients diagnosed with the M1 AML subtype, eight with M2,
seven with M3, six with M4, three with M5 and three with M6.
Outcomes associated with chemotherapy-induced CYP1B1
promoter methylation changes varied according to the subtypes.
Chemotherapy-induced methylation changes were more often observed
in M3 patients (57.1%, 4/7) compared with patients diagnosed with
other subtypes (M1: 33.3%, 1/3; M2: 12.5%, 1/8; M4: 16.7%, 1/6; M5:
0%, 0/3; and M6: 0%, 0/3).
Age-based subgroup analyses of CYP1B1
promoter methylation changes in AML patients
As shown in Table I,
of the 24 patients aged ≤60 years, 20 patients exhibited induced
hypermethylation and four patients showed induced hypomethylation.
Among the six patients aged >60 years old, five exhibited
induced hypermethylation and one demonstrated induced
hypomethylation. By further categorizing patients according to age,
chemotherapy-induced methylation changes were observed to be more
often present among AML patients >60 years of age (33.3%, 2/6)
compared with patients ≤60 years of age (20.8%, 5/24). Among the
patients aged ≤60 years, one patient of the M1 subtype (Ara-C +
ACLA + G-CSF; aged 59 years) exhibited an induced hypermethylation
and worse consequence, one patient of M3 subtype
(As2O3 + DNR + ATRA; aged 40 years) showed an
induced hypermethylation along with a poor prognosis, two M3
patients (HHT + Ara-C; aged 59 years and Ara-C; aged 30 years)
demonstrated induced hypermethylation along with remission and one
patient of the M3 subtype (IDA + Ara-C; aged 42 years) exhibited
induced hypomethylation along with a poor prognosis. As for
patients >60 years of age, one M2 subtype patient (Ara-C + ACLA
+ G-CSF, aged 76 years) exhibited an induced hypomethylation along
with a poor prognosis and one M4 subtype patient (IDA + Ara-C +
ACLA + G-CSF + HHT, aged 67 years) demonstrated hypermethylation
along with an improved prognosis.
Gender-based subgroup analyses of
CYP1B1 promoter methylation changes in AML patients
As shown in Table I,
4/13 male and 3/17 female patients demonstrated CYP1B1
promoter methylation status changes. Among them, one M3 subtype
male patient (HHT + Ara-C) and one M4 subtype male patient (IDA +
Ara-C + ACLA + G-CSF + HHT) displayed induced hypermethylation
along with remission. One M3 subtype male patient
(As2O3 + DNR + ATRA) exhibited induced
hypermethylation along with a poor prognosis, and one M2 subtype
male patient (Ara-C + ACLA + G-CSF) demonstrated induced
hypomethylation along with a poor prognosis. The remaining nine
male patients did not exhibit any methylation changes induced by
chemotherapy.
In the female subgroup, one M1 subtype patient
(Ara-C + ACLA + G-CSF) exhibited induced hypermethylation with a
poor prognosis, one M3 subtype patient (IDA + Ara-C) exhibited
induced hypomethylation with a poor prognosis, and one M3 subtype
patient (Ara-C) demonstrated induced hypermethylation along with
remission. However, the remaining 14 female patients did not show
any methylation changes following chemotherapy.
Discussion
The present study aimed to explore the
chemotherapy-induced methylation changes of the APC2 and
CYP1B1 promoter in bone marrow samples from AML patients and
their association with the treatment outcome.
Consistent with its role in the Wnt signaling
pathway, APC2 promoter methylation was associated with the
progression of various cancers, particularly colorectal cancer
(12). However, APC2
methylation is rarely observed in epithelial tumors (6). The results of the present study
demonstrated that APC2 promoter methylation remained
unchanged by various chemotherapy treatments.
CYP1B1 contributes to the development and
progression of various diseases, including tumorigenesis and
multidrug resistance (13).
CYP1B1 promoter methylation is downregulated in colorectal
cancer (14) and stratifies
prognosis in patients with tamoxifen- and non tamoxifen-treated
breast cancer (15). The results
demonstrated that CYP1B1 promoter methylation changed
following treatment with various chemotherapy regimens, indicating
complex regulation of CYP1B1 methylation in the bone marrow.
In addition, different AML subtypes displayed distinct responses to
multiple chemotherapy regimens, which may influence the changes of
CYP1B1 methylation and the outcome of chemotherapy. Among
the AML subtypes, M3 patients often revealed chemotherapy-induced
changes in CYP1B1 methylation. The present study
demonstrated that CYP1B1 hypermethylation in M3 patients may
be associated with an improved prognosis if treated with Ara-C, HHT
+ Ara-C or IDA + Ara-C chemotherapy regimens. This observation
provides a response tendency that is potentially useful for
individualized AML therapy.
The incidence rates of acute leukemia are
significantly higher in males compared with females (16). The present study demonstrated that
male patients were more susceptible to induced methylation changes
compared with female patients following chemotherapy. Furthermore,
the CYP1B1 promoter methylation changes during chemotherapy
may serve as a potential biomarker to predict the outcome of
therapy in male patients.
However, there are some limitations in the present
study. Firstly, a relatively small sample size was used. The sample
size of 30 patients used may prevent powerful statistical
significance. Therefore, a larger sample size is required to
confirm our observations in the future. Secondly, a conclusion was
drawn through patients treated with 13 types of chemotherapy
regimens, however, future investigation with more samples and a
patient cohort treated with the same chemotherapy are required.
Thirdly, the main population of the present study were people from
Ningbo and studies in other regions are required in order to
elucidate an integrated conclusion on the association between
changeable gene methylation levels and chemotherapeutic
outcomes.
In conclusion, the results of the present study
demonstrated that chemotherapy-induced changes in CYP1B1
promoter methylation were associated with the AML subtype, gender
and age of the patients. However, future analyses on the mechanisms
by which CYP1B1 promoter methylation is altered by
chemotherapeutic regimens are required.
Acknowledgements
The present study was supported by the grants from
the National Natural Science Foundation of China (grant nos.
31100919 and 81371469), the Zhejiang Provincial Natural Science
Foundation (grant no. LR13H020003), the Ningbo City Medical Science
and Technology Projects (grant no. 2014A20) and the K. C. Wong
Magna Fund in Ningbo University.
References
|
1
|
Boultwood J and Wainscoat JS: Gene
silencing by DNA methylation in haematological malignancies. Br J
Haematol. 138:3–11. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Powers HR, Bachar M, Savage N, Toscano M
and Dainer PM: Azacitidine as salvage therapy for acute myeloid
leukemia in a severely ill patient. Hematol Rep. 6:55162014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galm O, Herman JG and Baylin SB: The
fundamental role of epigenetics in hematopoietic malignancies.
Blood Rev. 20:1–13. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rowe JM: The increasing genomic complexity
of acute myeloid leukemia. Best Pract Res Clin Haematol.
27:209–213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sierra J, Yoshida T, Joazeiro CA and Jones
KA: The APC tumor suppressor counteracts beta-catenin activation
and H3K4 methylation at Wnt target genes. Genes Dev. 20:586–600.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Griffiths EA, Gore SD, Hooker C, McDevitt
MA, Karp JE, Smith BD, Mohammad HP, Ye Y, Herman JG and Carraway
HE: Acute myeloid leukemia is characterized by Wnt pathway
inhibitor promoter hypermethylation. Leuk Lymphoma. 51:1711–1719.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Faiq MA, Ali M, Dada T, Dada R and Saluja
D: A novel methodology for enhanced and consistent heterologous
expression of unmodified human cytochrome P450 1B1 (CYP1B1). PLoS
One. 9:e1104732014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nagai F, Hiyoshi Y, Sugimachi K and Tamura
HO: Cytochrome P450 (CYP) expression in human myeloblastic and
lymphoid cell lines. Biol Pharm Bull. 25:383–385. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
DiNardo CD, Gharibyan V, Yang H, Wei Y,
Pierce S, Kantarjian HM, Garcia-Manero G and Rytting M: Impact of
aberrant DNA methylation patterns including CYP1B1 methylation in
adolescents and young adults with acute lymphocytic leukemia. Am J
Hematol. 88:784–789. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fasan A, Alpermann T, Haferlach C,
Grossmann V, Roller A, Kohlmann A, Eder C, Kern W, Haferlach T and
Schnittger S: Frequency and prognostic impact of CEBPA proximal,
distal and core promoter methylation in normal karyotype AML: A
study on 623 cases. PLoS One. 8:e543652013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen C, Wang L, Liao Q, Huang Y, Ye H,
Chen F, Xu L, Ye M and Duan S: Hypermethylation of EDNRB promoter
contributes to the risk of colorectal cancer. Diagn Pathol.
8:1992013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mokarram P, Zamani M, Kavousipour S,
Naghibalhossaini F, Irajie C, Sarabi M Moradi and Hosseini SV:
Different patterns of DNA methylation of the two distinct
O6-methylguanine-DNA methyltransferase (O6-MGMT) promoter regions
in colorectal cancer. Mol Biol Rep. 40:3851–3857. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Shi J, Xu L, Shi B, Hou P and Ji
M: Aberrant DNA methylation of drug metabolism and transport genes
in nodular goiter. Thyroid Res. 4:152011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Habano W, Gamo T, Sugai T, Otsuka K,
Wakabayashi G and Ozawa S: CYP1B1, but not CYP1A1, is downregulated
by promoter methylation in colorectal cancers. Int J Oncol.
34:1085–1091. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Widschwendter M, Siegmund KD, Müller HM,
Fiegl H, Marth C, Müller-Holzner E, Jones PA and Laird PW:
Association of breast cancer DNA methylation profiles with hormone
receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shahab F and Raziq F: Clinical
presentations of acute leukemia. J Coll Physicians Surg Pak.
24:472–476. 2014.PubMed/NCBI
|