Introduction
Nasopharyngeal carcinoma (NPC) is a rare malignancy
in most parts of the world, but occurs with a high prevalence in
areas of Southern China, Southeast Asia and North Africa where it
comprises a substantial health burden (1–3). NPC is
a highly malignant tumor because of its frequent metastasis and
poor prognosis (2,4). Therefore, it is important to gain a
better understanding of the molecular mechanisms of NPC invasion
and migration in order to improve the prognosis of patients with
NPC.
MicroRNAs (miRNAs) are an evolutionarily conserved
family of small non-coding RNA molecules, comprising ~22
nucleotides, that are found in plants, animals and some viruses and
function to silence RNA and suppress gene expression at a
post-transcriptional level. They are increasingly considered as
important gene expression regulators in multiple cellular
progresses, including tumori-genesis and metastasis in variety of
tumors (5). In NPC, a number of
miRNAs have been reported to serve as oncogenes or suppressor genes
in the development and progression of NPC, including miR-10b, which
was shown to promote the metastasis of NPC cells in previous
studies (6–8). However, miR-200a has been reported to
be downregulated in NPC, and to act as an inhibitor of migration
and invasion (9).
Several reports have suggested that miR-143 is
downregulated in many types of cancers, including colorectal,
gastric, osteosarcoma, bladder and epithelial cancers (10–14).
Upregulated miR-143 transcribed by nuclear factor-κB has been
reported to increase the metastasis of hepatocellular carcinoma by
targeting fibronectin expression (15). In addition, miR-143 has been found to
be significantly downregulated in clinical samples from NPC
patients, and to inhibit NPC proliferation in vivo and in
vitro (16). However, the
downregulation of miR-143 in NPC tissues and cell lines requires
further investigation and the correlations of miR-143 with invasion
and migration are not yet known.
In the present study, the aim was to investigate the
following: i) Whether miR-143 expression is changed in NPC tissues
and cell lines; ii) the role of miR-143 in tumor proliferation,
invasion and migration; and iii) the functional target(s) of
miR-143 involved in tumor growth, invasion and migration.
Materials and methods
Clinical specimens and cell
culture
Paired human NPC tissues and matched normal tissues
were collected from 40 patients (15 women and 25 men; age range,
44–85 years; median age, 71 years), who had undergone standard
surgical procedures in the Renmin Hospital of Wuhan University
(Wuchang, China), with the informed consent of the patients. The
tissue samples were collected from February 2010 to October 2013.
Parts of tissue samples were immediately snap-frozen in liquid
nitrogen, and sections were fixed in formalin for histological
examination. The disease stage and lymph node metastasis were
determined via the pathological examination of histology slides in
the patient cohort. The experimental protocols were approved by the
Institutional Review Committees of Wuhan University.
Human NPC cell lines CNE-1, CNE-2Z and NP69,
obtained from the Cell Bank of Academia Sinica (Shanghai, China),
were cultured in RPMI-1640 medium supplemented with 10% fetal
bovine serum (FBS; both Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 1% penicillin-streptomycin solution in a 37°C
incubator containing 5% CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was obtained from NPCs, normal tissue
samples and cell lines using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. Complementary DNA was synthesized from the RNA using
a cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The RT-qPCR
reactions were run on a 7500 Real-Time PCR machine (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Maxima SYBR Green/ROX
qPCR Master mix (K0223; Finnzymes; Thermo Fisher Scientific, Inc.)
was used, according to the manufacturer's protocol. The qPCR
cycling conditions were as follows: 95°C for 10 min, followed by 40
cycles at 95°C for 15 sec and 60°C for 45 sec, and a final
extension step of 95°C for 15 sec, 60°C for 1 min, 95°C for 15 sec
and 60°C for 15 sec. The miRNA expression level was normalized to
the expression level of U6 small nuclear RNA (RNU6B). Primers used
for hsa-miR-143 were purchased from Applied Biosystems. Their
sequences were as follows: hsa-miR-143, forward:
5′-ACACTCCAGCTGGGGGTGCAGTGCTGCATC-3′ and reverse:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACCAGA-3′; RNU6B, forward:
5′-CTTCGGCAGCACATATAC-3′ and reverse: 5′-GGCCATGCTAATCTTCTC-3′. All
reactions were performed in triplicate and included a negative
control lacking cDNA. The relative expression values were
calculated using the ΔΔCq method (17).
Transfection
CNE-1, CNE-2Z and NP69 cells were transfected with
double stranded synthetic syn-hsa-miR-143 mimics and scrambled
controls (Thermo Fisher Scientific, Inc.) using Lipofectamine 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol for overexpression. Briefly, ~1.5×105 cells
were seeded and cultured in 6-well plates the day prior to
transfection. miRNA mimic with a concentration of 1, 2.5 or 5 nM
and scrambled controls (NC) were each transfected into CNE-1 and
CNE-2Z cells. After transfection for 72 h, the functions of the
cells were examined. All groups were performed in triplicate.
Cell viability and cell apoptosis
assay
CNE-1 and CNE-2Z cells transfected with miRNA mimics
or scrambled controls were harvested at 24, 48 and 72 h after
transfection and seeded in 96-well plates (5×103 cells/well). Then,
10 µl Cell Counting kit-8 (CCK-8) assay solution (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added to each well and the
plate was incubated for 1 h at 37°C. The absorbance was then
measured at 450 nm using a microplate reader.
For the cell apoptosis assay, the transfected CNE-1
and CNE-2Z cells were seeded in 6-well plates (5×105 cells). At 72
h after transfection the cells were harvested and stained with
Annexin V-fluorescein isothiocyanate (BD Biosciences, Franklin
Lakes, NJ, USA) and propidium iodide (BD Biosciences) for 15 min in
the dark at room temperature followed by flow cytometric analysis
using a BD Accuri C6 Flow Cytometer equipped with software version
1.0.264.21 (BD Biosciences, San Diego, CA, USA).
In vitro invasion and migration
assays
The CNE-2Z cell line, which has high invasiveness,
was selected for analysis using in vitro migration and
invasion assays. In Transwell migration and invasion assays, cells
were serum-starved for 24 h, following which 1×104
transfected cells in serum-free RPMI-1640 were seeded into the
upper well of the Transwell chamber, onto non-coated or
Matrigel-coated membrane (BD Biosciences), respectively. RPMI-1640
medium supplemented with 10% FBS (750 µl) was added to the lower
well of the chamber. The chamber was maintained at 37°C in a 5% CO2
incubator for 48 h. The uninvaded/unmigrated cells were removed
with a cotton swab. The cells on the lower surface of the membrane
were then fixed with 4% paraformaldehyde (25°C for 10 min), stained
with 0.5% crystal violet (25°C for 30 min) and then counted.
Protein extraction and western
blotting
Transfected CNE-2Z cells were harvested and lysed on
ice for 30 min in radioimmunoprecipitation assay buffer (Beyotime
Institute of Biotechnology, Haimen, China) supplemented with 1 mM
phenylmethylsulfonyl fluoride. Total protein extracts were
separated by electrophoresis on 8% SDS-PAGE gels and transferred to
polyvinylidene fluoride membranes. Primary antibodies against
extracellular-signal-regulated kinase 5 (ERK5; 1:1,000; cat. no.
12950; Cell Signaling Technology, Inc., Danvers, MA, USA), B-cell
lymphoma 2 (Bcl-2; 1:1,000; cat. no. 3498; Cell Signaling
Technology, Inc.), Kirsten rat sarcoma viral oncogene homolog
(KRAS; 1:1,000; cat. no. 3339; Cell Signaling Technology, Inc.),
caspase 3 (1:1,000; cat. no. 9662; Cell Signaling Technology, Inc.)
and β-actin (1:500; cat. no. sc-47778; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) were incubated with the membranes at 4°C
overnight. After washing with PBS three times, the membranes were
incubated with the secondary antibodies horseradish
peroxidase-conjugated goat anti-rabbit IgG (1,000; cat. no. A0208;
Beyotime Institute of Biotechnology) and goat anti-mouse IgG
(1:1,000; cat. no. A0216; Beyotime Institute of Biotechnology) for
1 h at 37°C. The membranes were washed again, and the
antigen-antibody reaction was visualized using an Amersham ECL
detection system (GE Healthcare Life Sciences, Chalfont, UK).
Protein levels were determined by assesing the signal intensity of
the bands using ImageJ 1.46 software (National Institutes of
Health, Bethesda, MD, USA).
Statistical analysis
Data are presented as the mean ± standard deviation.
The paired, two-tailed Student's t-test was used to analyze the
significance of differences between groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-143 is downregulated in NPC
tissues
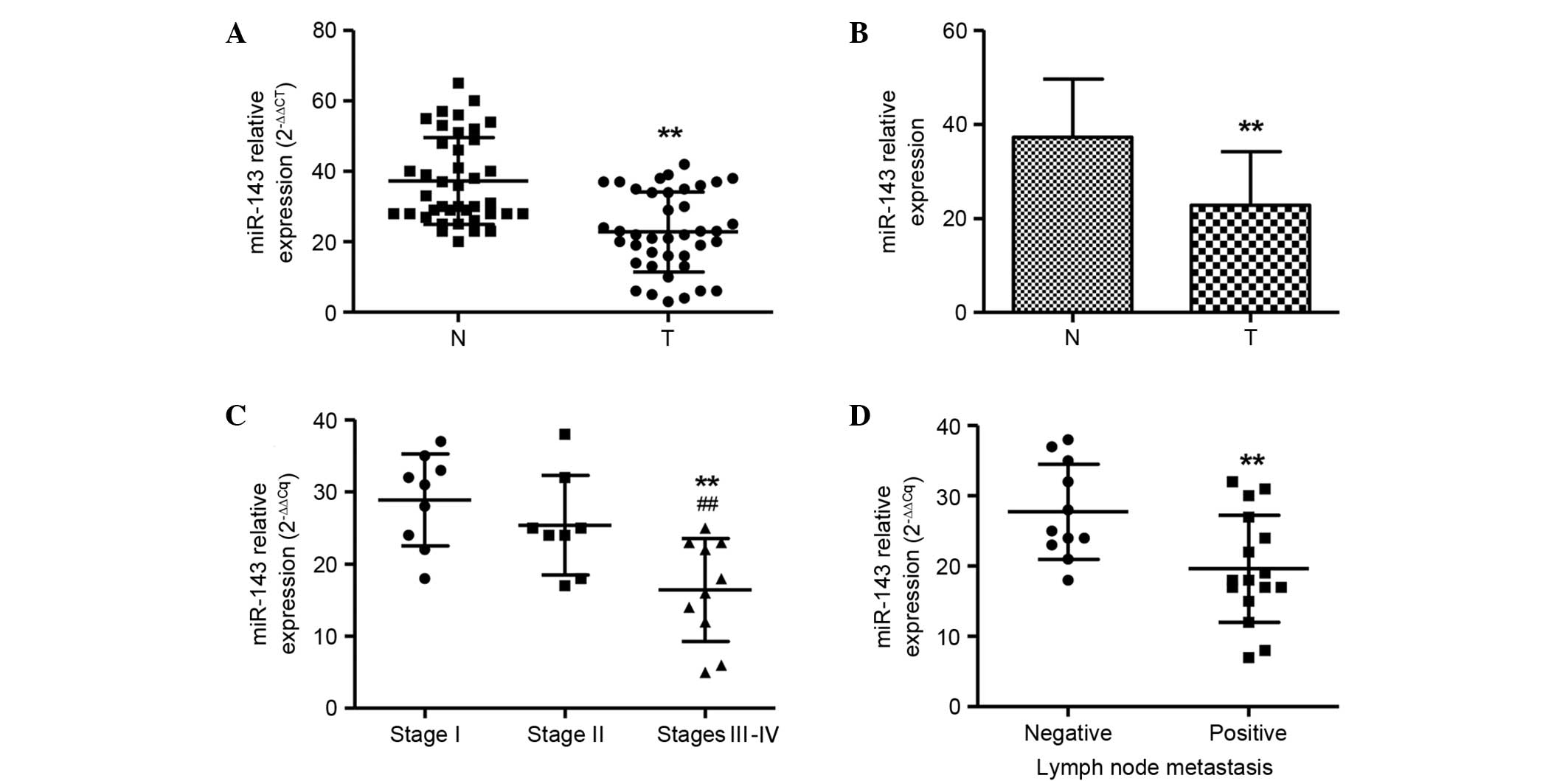
To investigate the role of miR-143 in NPC
tumorigenosis, the expression levels of miR-143 in 40 clinical
samples of NPC and matched normal tissue samples were first
evaluated by TaqMan qPCR (Fig. 1A).
RNU6B was used as an internal standard. A significant
downregulation of miR-143 expression was found in NPCs in
comparison with normal tissue samples (P<0.01). The mean level
of miR-143 in NPCs was decreased to ~51.6% of that in the matched
normal tissue samples (Fig. 1B). The
miR-143 expression levels were also compared among NPC samples of
different stages. The number of samples of each stage was as
follows: Stage I (n=12), stage II (n=12) and stages III–IV (n=16)
The miR-143 expression levels in cancer tissues were negatively
correlated with the stage of the NPC patients. The early stages I
and II showed significantly higher miR-143 expression levels than
those in the late stages III and IV (P<0.01; Fig. 1C). Furthermore, miR-143 expression
levels were also compared among NPC samples with (n=24) and without
(n=16) lymph node metastasis. miR-143 levels were markedly lower in
the patients with lymph node metastasis than in the patients
without lymph node metastasis (Fig.
1D), which is consistent with the aforementioned result, since
lymph node metastasis commonly occurs in stages III and IV, but not
I and II.
miR-143 is downregulated in NPC cells
and involved in cell proliferation and apoptosis
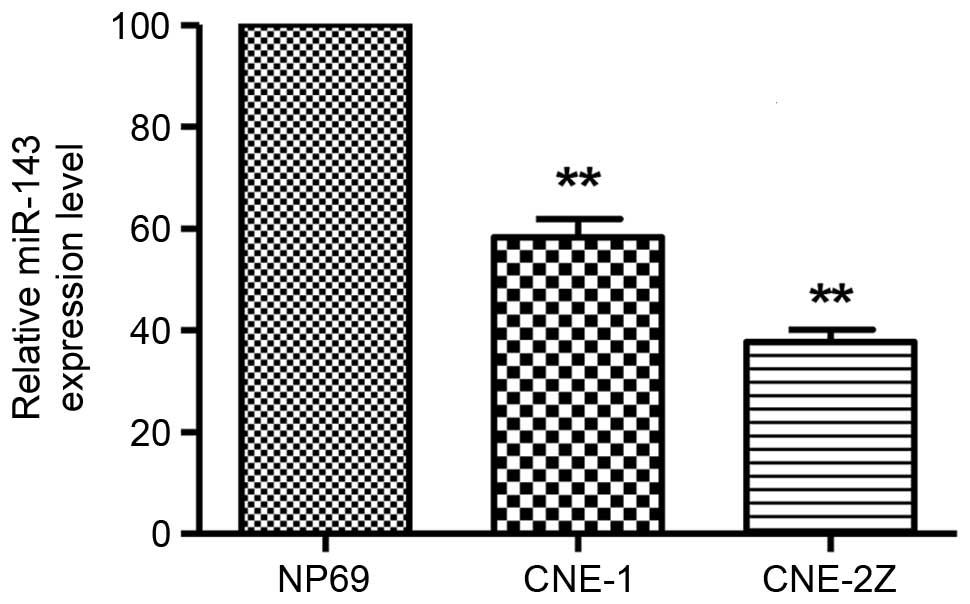
In the two NPC cell lines, miR-143 was substantially
downregulated compared with that in the non-malignant nasopharynx
cell line NP69 (P<0.01; Fig. 2).
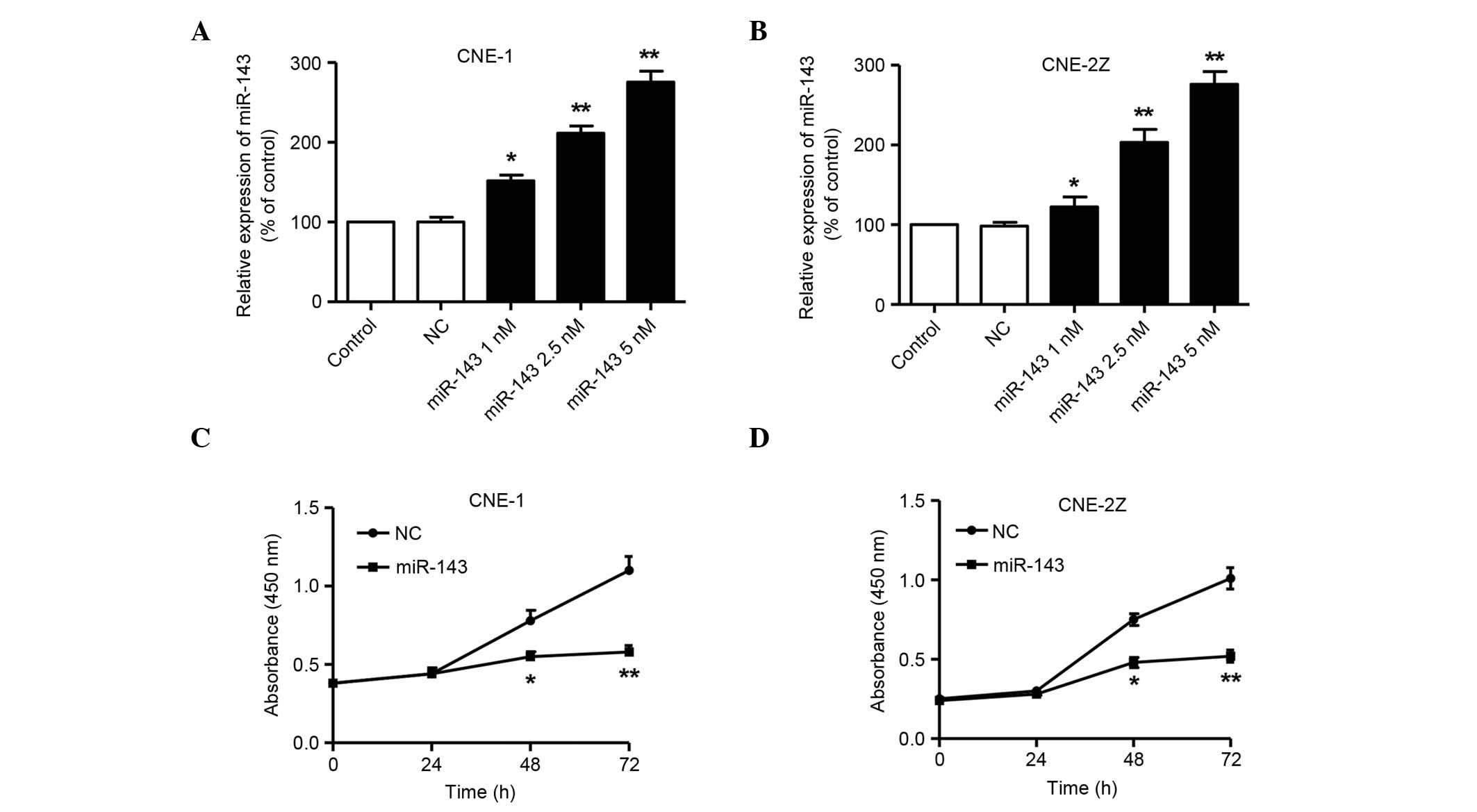
The effects of miR-143 on the NPC cell lines were then examined
in vitro by transfection. As shown in Fig. 3A and B, the expression levels of
mature miR-143 in CNE-1 and CNE-2Z cells transfected with miR-143
mimics were increased in a dose-dependent manner, compared with
those in the cells transfected with NC miRNA 72 h after
transfection. The CCK-8 assay showed a significant reduction of
cell proliferation following transfection with 5 nM miR-143 in
comparison with NC miRNA at 48 and 72 h in the two cell lines
(P<0.05 at 48 h and P<0.01 at 72 h; Fig. 3C and D). These results suggest that
miR-143 significantly inhibits the proliferation of NPC cells.
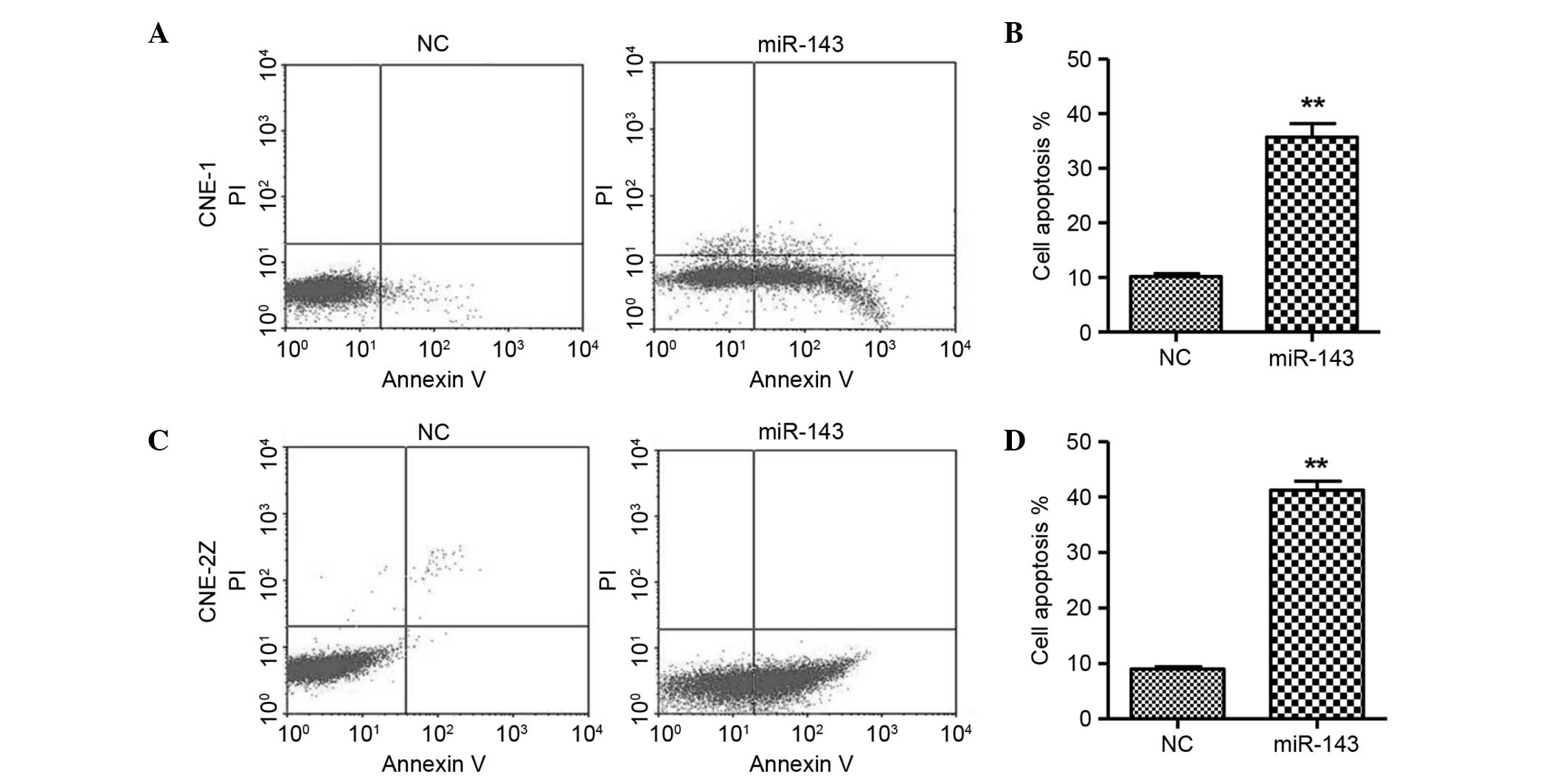
Moreover, flow cytometric analysis revealed that the overexpression
of miR-143 resulted in a significant increase in the apoptosis of
CNE-1 and CNE-2Z cells (Fig. 4),
indicating that miR-143 may lead to apoptosis.
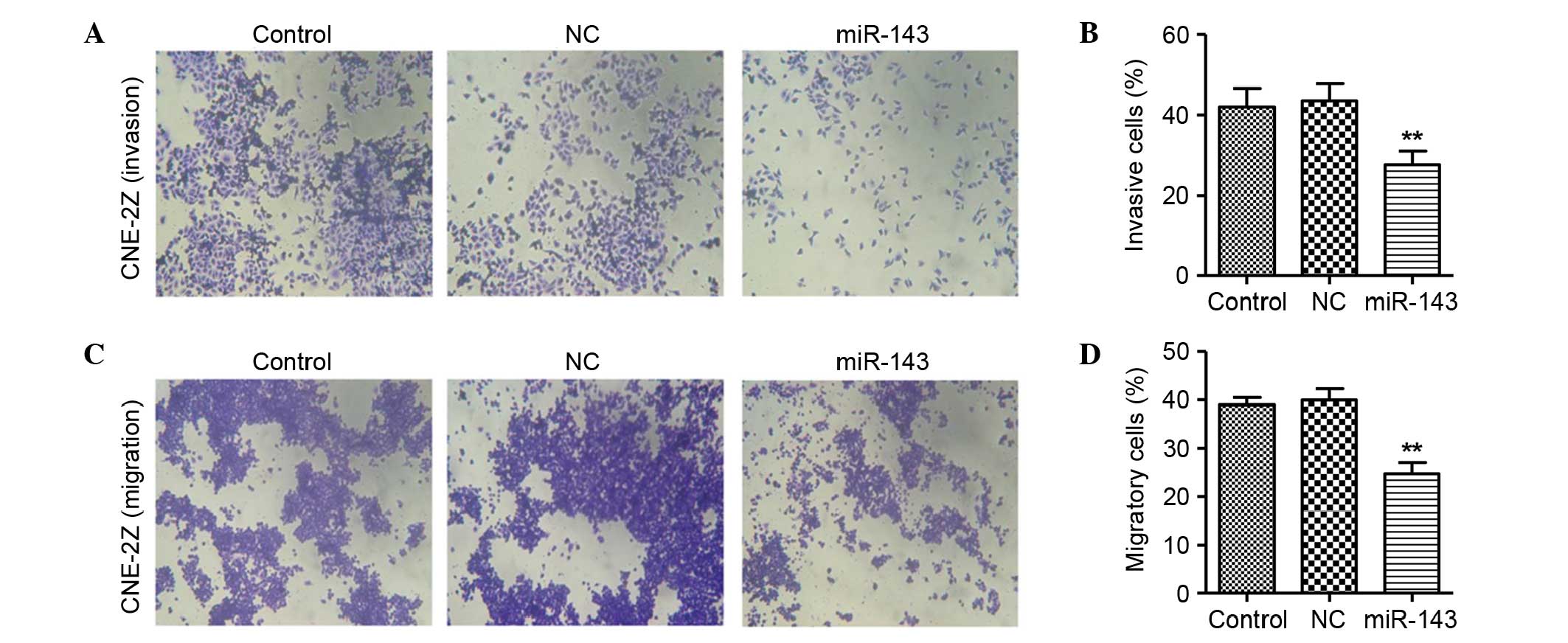
miR-143 inhibits NPC cell invasion and
migration
In the invasion assay, the invasive rate of the NC
miRNA group of CNE-2Z cells was 43.53% at 48 h after transfection.
However, the invasive rate of CNE-2Z cells was significantly
decreased by transfection with miR-143 (27.56% at 48 h after
transfection; Fig. 5A and B),
compared with the CNE-2Z cells without miR-143 transfection. No
significant difference between the NC miRNA and untreated groups
was observed.
In the migration assay, the migratory rate of CNE-2Z
cells in the NC miRNA group was 39.93% at 48 h after transfection.
However, the migratory rate of CNE-2Z cells was significantly
decreased by transfection with miR-143 (24.7% at 48 h after
transfection; Fig. 5C and D)
compared with the NC miRNA group. There was no evident difference
between the NC miRNA and the untreated groups.
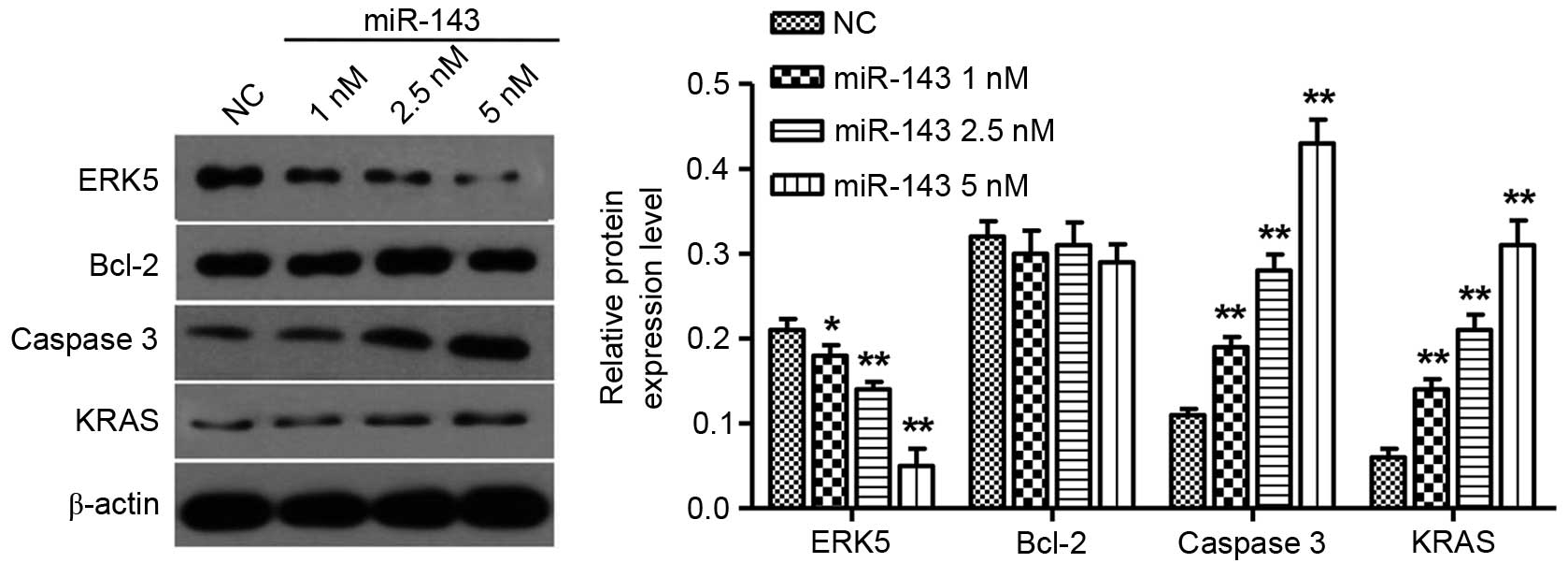
miR-143 inhibits ERK5 and promotes
caspase 3 and KRAS expression in NPC cells
It has been reported that miR-143 targets the
expression of ERK5 and Bcl-2 in prostate and cervical cancer
(18,19). To further investigate the targets of
miR-143 in NPC, the expression levels of ERK5, KRAS, caspase 3 and
Bcl-2 in CNE-2Z cells following miR-143 transfection were explored.
The ERK5 protein levels were significantly reduced in a
dose-dependent manner 72 h after miR-143 transfection, but caspase
3 as well as KRAS expression levels were increased in a
dose-dependent manner 72 h after miR-143 transfection. Notably,
overexpression of miR-143 did not have an impact on Bcl-2 protein
expression (Fig. 6).
Discussion
miRNAs are regarded as important regulators involved
in the regulation of protein post-transcription and can function as
either tumor suppressors or promoters in cancer depending on the
specific genes they target, and the abnormal expression of miRNAs
may contribute to human carcinogenesis (20,21).
miR-143 is downregulated in various human malignancies, such as
colon (10), gastric (11) and prostate cancer (22). In the present study, the expression
levels of miR-143 in NPC tissues and matched normal tissues were
examined, and it was found that miR-143 expression levels were
significantly lower in NPC tissues than in normal tissues. In
addition, the results also demonstrated that the downregulation of
miR-143 expression was associated with later clinical cancer stages
and lymph node metastasis in patients with NPC.
To the best of our knowledge, this is the first
study to report that the overexpression of miR-143 contributes to
the inhibition of cell proliferation, invasion and migration, and
to apoptosis in NPC. In agreement with these findings, previous
reports showed that miR-143 is associated with bone metastasis of
prostate cancer (22) and invasion
in esophageal squamous cell carcinoma (23). Clinical data in the present study
indicate that miR-143 may function as a metastatic suppressor by
inhibiting cell invasion and migration and inducing apoptosis,
which affects multiple cellular processes, including
carcinogenesis, invasion and lymph node metastasis, vital for the
development and progression of cancer.
miRNAs target various genes involved in multiple
cellular signaling pathways. miR-143 has been shown to downregulate
Bcl-2 and KRAS in cervical cancer, osteosarcoma and colon cancer
(24–26). In addition, miR-143 has been reported
to regulate the 3′ untranslated region of fascin actin-bundling
protein 1 in esophageal cancer cells (27,28).
However, little is known about the effects of miR-143 on these
targets in NPC. The results of the present study demonstrated that
different doses of miR-143 reduced ERK5 protein levels and
increased caspase 3 and KRAS expression levels in a dose-dependent
manner, but the Bcl-2 level was not affected. The ERK5 protein is a
protein kinase of the mitogen-activated protein kinase family,
involved in the signaling processes downstream of various
receptors. In addition, it has been indicated to be involved in
sustaining cell proliferation, resisting cell apoptosis and
promoting metastasis in a variety of malignancies (29,30). The
overexpression of miR-143 has been demonstrated to inhibit cell
growth through the reduction of ERK5 expression in colon cancer,
esophageal cancer cells and adipocytes (24–32).
Caspase 3 is a member of the cysteine-aspartic acid protease
(caspase) family. Sequential activation of caspases plays a central
role in the execution-phase of cell apoptosis. A previous study
showed that the overexpression of miR-143 was associated with
decreased expression levels of Bcl-2 and increased caspase 3
activation in human colon cancer (24). By contrast, the expression of Bcl-2
was not significantly affected by miR-143 overexpression,
suggesting that the targets of miR-143 differed between different
cancers. The findings of the present study provide evidence that
miR-143 may function as a tumor suppressor in NPC by inhibiting
ERK5 protein expression and promoting caspase 3 and KRAS
expression.
In conclusion, this study has shown that miR-143 is
frequently decreased in NPC tissues and cells and acts as a
potential tumor suppressor in NPC. miR-143 represses ERK5
expression and promotes caspase 3 and KRAS expression, which may
play a role in NPC progression. Therefore miR-143 may serve as a
biomarker and therapeutic target for NPC.
References
|
1
|
Hildesheim A and Levine PH: Etiology of
nasopharyngeal carcinoma: A review. Epidemiol Rev. 15:466–485.
1993.PubMed/NCBI
|
|
2
|
Feng BJ, Huang W, Shugart YY, Lee MK,
Zhang F, Xia JC, Wang HY, Huang TB, Jian SW, Huang P, et al:
Genome-wide scan for familial nasopharyngeal carcinoma reveals
evidence of linkage to chromosome 4. Nat Genet. 31:395–399.
2002.PubMed/NCBI
|
|
3
|
Sobin LH and Fleming ID: Union
Internationale Contre le Cancer and the American Joint Committee on
Cancer: TNM classification of malignant tumors, fifth edition
(1997). Cancer. 80:1803–1804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang J, Guo LP, Chen LZ, Zeng YX and Lu
SH: Identification of cancer stem cell-like side population cells
in human nasopharyngeal carcinoma cell line. Cancer Res.
67:3716–3724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li G, Wu Z, Peng Y, Liu X, Lu J, Wang L,
Pan Q, He ML and Li XP: MicroRNA-10b induced by Epstein-Barr
virus-encoded latent membrane protein-1 promotes the metastasis of
human nasopharyngeal carcinoma cells. Cancer Lett. 299:29–36. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong TS, Man OY, Tsang CM, Tsao SW, Tsang
RK, Chan JY, Ho WK, Wei WI and To VS: MicroRNA let-7 suppresses
nasopharyngeal carcinoma cells proliferation through downregulating
c-Myc expression. J Cancer Res Clin Oncol. 137:415–422. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li
JJ, Röcken C, Ebert MP, Kwok TT and Sung JJ: MicroRNA-143 targets
DNA methyltransferases 3A in colorectal cancer. Br J Cancer.
101:699–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
−145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Noguchi S, Yasui Y, Iwasaki J, Kumazaki M,
Yamada N, Naito S and Akao Y: Replacement treatment with
microRNA-143 and −145 induces synergistic inhibition of the growth
of human bladder cancer cells by regulating PI3K/Akt and MAPK
signaling pathways. Cancer Lett. 328:353–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J, Sun Q, Zhang Z, Ge S, Han ZG and
Chen WT: Loss of microRNA-143/145 disturbs cellular growth and
apoptosis of human epithelial cancers by impairing the MDM2-p53
feedback loop. Oncogene. 32:61–69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu YF, Li YQ, Guo R, He QM, Ren XY, Tang
XR, Jia WH, Kang TB, Zeng MS, Sun Y, et al: Identification of
miR-143 as a tumour suppressor in nasopharyngeal carcinoma based on
microRNA expression profiling. Int J Biochem Cell Biol. 61:120–128.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clapé C, Fritz V, Henriquet C, Apparailly
F, Fernandez PL, Iborra F, Avancès C, Villalba M, Culine S and
Fajas L: miR-143 interferes with ERK5 signaling, and abrogates
prostate cancer progression in mice. PloS One. 4:e75422009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu L, Yu X, Guo X, Tian Z, Su M, Long Y,
Huang C, Zhou F, Liu M, Wu X and Wang X: miR-143 is downregulated
in cervical cancer and promotes apoptosis and inhibits tumor
formation by targeting Bcl-2. Mol Med Rep. 5:753–760.
2012.PubMed/NCBI
|
|
20
|
Zhang Y, Wang Z, Chen M, Peng L, Wang X,
Ma Q, Ma F and Jiang B: MicroRNA-143 targets MACC1 to inhibit cell
invasion and migration in colorectal cancer. Mol Cancer. 11:232012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu S, Wu H, Wu F, Nie D, Sheng S and Mo
YY: MicroRNA-21 targets tumor suppressor genes in invasion and
metastasis. Cell Res. 18:350–359. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng X, Guo W, Liu T, Wang X, Tu X, Xiong
D, Chen S, Lai Y, Du H, Chen G, et al: Identification of miRs-143
and −145 that is associated with bone metastasis of prostate cancer
and involved in the regulation of EMT. PloS One. 6:e203412011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M
and Baba H: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Borralho PM, Simões AE, Gomes SE, Lima RT,
Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE and Rodrigues
CM: miR-143 overexpression impairs growth of human colon carcinoma
xenografts in mice with induction of apoptosis and inhibition of
proliferation. PLoS One. 6:e237872011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wang K, Wang G, Ba Y, et al: Role of miR-143
targeting KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
27
|
Liu R, Liao J, Yang M, Sheng J, Yang H,
Wang Y, Pan E, Guo W, Pu Y, Kim SJ and Yin L: The cluster of
miR-143 and miR-145 affects the risk for esophageal squamous cell
carcinoma through co-regulating fascin homolog 1. PloS One.
7:e339872012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu BL, Xu LY, Du ZP, Liao LD, Zhang HF,
Huang Q, Fang GQ and Li EM: MiRNA profile in esophageal squamous
cell carcinoma: Downregulation of miR-143 and miR-145. World J
Gastroenterol. 17:79–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Drew BA, Burow ME and Beckman BS:
MEK5/ERK5 pathway: The first fifteen years. Biochim Biophys Acta.
1825:37–48. 2012.PubMed/NCBI
|
|
30
|
Lochhead PA, Gilley R and Cook SJ: ERK5
and its role in tumour development. Biochem Soc Trans. 40:251–256.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang
G, Liu Q and Du J: MicroRNA-143 functions as a tumor suppressor in
human esophageal squamous cell carcinoma. Gene. 517:197–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Esau C, Kang X, Peralta E, Hanson E,
Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et
al: MicroRNA-143 regulates adipocyte differentiation. J Biol Chem.
279:52361–52365. 2004. View Article : Google Scholar : PubMed/NCBI
|