Introduction
Medulloblastoma (MB) is the most common pediatric
malignant brain tumor, and has a poor clinical outcome (1,2). With
currently available multimodality therapies, including surgery,
radiotherapy and chemotherapy, numerous children have a favorable
prognosis; however, the majority of patients suffer from
considerable long-term disabilities and morbidity following
aggressive multimodal therapy (3–5).
Attempts to further improve the outcomes have been restricted by
the cytotoxicity of conventional medication and the nature of the
disease. Therefore, an increased understanding of the mechanisms
underlying MB is crucial in the development of novel therapeutic
approaches.
Aberrant activation of the sonic hedgehog (SHH)
signaling pathway has been implicated in the development of MB
(6–8). The Gli family zinc finger 1 (Gli1)
transcription factor is considered to be a mediator of the SHH
signaling pathway in MB, although its tumorigenic nature and its
relative contribution to tumorigenesis remain poorly understood
(9).
CyclinD1 is a key protein in the cyclin family that
regulates the G1/S transition and is highly expressed in multiple
types of tumors (10,11). This protein is regulated by a complex
system of signal transduction pathways (12,13).
CyclinD1 expression is known to be regulated by Gli1 in MB.
Furthermore, GANT61 is a specific Gli1 inhibitor, which has been
shown to inhibit the DNA binding activity of Gli1 by binding to the
zinc-finger domain (14–16).
In order to examine the role of Gli1 in MB, our
previous studies screened for genes preferentially regulated by
Gli1 in MB cells (17,18). CyclinD1 plays important role in tumor
proliferation, and thus the expression of CyclinD1 was investigated
in MB cells.
Materials and methods
Reagents and antibodies
GANT61 (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) was dissolved in dimethyl sulfoxide (DMSO) and stored at
−20°C until required for use. The final DMSO concentration in all
cultures, including the vehicle control groups, was 0.1% in RPMI
1640 medium (Gibco; Thermo Fisher Scientific, Inc., Grand Island,
NY, USA). Fetal bovine serum (FBS) and 0.25% trypsin/EDTA were
purchased from Gibco (Thermo Fisher Scientific, Inc.). The
hematoxylin and eosin (HE) staining kit (G1060) was purchased from
SuoLaibao Technology Co., Ltd. (Beijing, China), and the
FITC-Annexin V kit from Abcam (ab14150; Cambridge, MA, USA). The
cell counting kit-8 (CCK-8) assay for cell proliferation analysis
was purchased from Dojindo Chemical Research Institute (Tokyo,
Japan), while the PrimeScript RT Master Mix and reverse
transcription (RT) kit (RR014A) was obtained from Takara Bio, Inc.
(Shiga, Japan; PrimeScript RT Master Mix). In addition, SYBR Green
I was purchased from Beijing Noble Ryder Technology Co., Ltd.
(Beijing, China). Antibodies against Gli1 (ab49314) and CyclinD1
(ab187364) were acquired from Abcam, while β-actin antibody
(AP0060) was purchased from Bioworld Technology, Inc. (Louis Park,
MN, USA). The secondary antibody of Gli1 (BL003A) and CyclinD1
(BL001A) were acquired from Biosharp (Wuhan, China) (19).
Cell culture
Daoy, an MB cell line, was purchased from ATCC
(Manassas, VA, USA). The Daoy cells were maintained in RPMI 1640
medium supplemented with 10% fetal bovine serum (500 ml; Gibco),
100 µg/ml penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc., Carlsbad, CA, USA) at 37°C with 5%
CO2. Prior to each experiment, trypan blue staining
(Sigma-Aldrich) was used to define the cell vitality. The cell
activity was determined to be >98%.
Cell proliferation analysis
CCK-8 assay was performed to investigate the cell
proliferation, according to the manufacturer's instructions of the
kit. Briefly, Daoy cells in exponential growth phase were pipetted
into single cells following trypsin digestion. Cells were seeded in
a 96-well plate at a density of 8×103 cells/well. RPMI
1640 medium containing 10% FBS was used to culture the cells for 24
h prior to replacing with serum-free medium. Next, the cells were
starved for 6 h and then incubated in RPMI 1640 medium supplemented
with 1% FBS. The cell culture groups included three groups treated
with different concentrations of GANT61 (10, 20 and 40 µM) and a
negative untreated control group with normal growing cells, while
wells with no cells acted as the blank control. A total of six
replicates per group were investigated. The cells were continually
cultured in the incubator for a further 24 h before the culture
medium was discarded. Subsequently, 100 µl fresh RPMI 1640 medium
and 10 µl CCK-8 solution were added into each well. The cells were
placed in the incubator to avoid light exposure, and the absorbance
at 450 nm (A450) was measured at 0.5, 1, 2 and 4 h, with
a Bio-Rad 680 microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).. The proliferation inhibition rate was
calculated as follows: Proliferation inhibition (%) =
[A450 (negative control group) - A450
(GANT61-treated group)] / A450 (negative control group)
×100%.
HE staining
Daoy cells in the exponential growth phase were
digested into a concentration of 1×106 cells/ml, added
to glass coverslips and cultured for 24 h in an incubator. The
medium was replaced, followed by addition of different
concentrations of GANT61 (10, 20 and 40 µM), while the group
without GANT61 treatment served as the control. Subsequently, the
cells were extracted after culturing for 24 h, washed with
phosphate-buffered saline (PBS) for three times and fixed in 4%
paraformaldehyde for 60 min, followed by washing three times with
PBS. The cells were then stained with hematoxylin for 5 min and
washed by tap water. Following incubation in differentiation buffer
for a few seconds and washing with water, eosin was added for 10
min. After washing with tap water, the stained sample was
dehydrated, sealed and prepared for microscopic observation.
Flow cytometry
In order to investigate the cell cycle progression,
flow cytometry analysis was performed using the FITC-Annexin V kit,
according to the manufacturer's instructions. Briefly,
2×104 cells were transferred into 10-ml centrifuge
tubes, and centrifuged for 5 min at 250–500 × g at 4°C.
After the culture medium was discarded, cells were washed once with
the binding buffer and centrifuged for 5 min at at 250–500 ×
g at 4°C. The final concentration of 1 µg/ml propidium
iodide (PI) with FITC-Annexin V (included in the kit) was dissolved
in incubation buffer. Resuspended cells were labeled in the dark
for 10–15 min with 100 µl solution buffer at room temperature.
Cells were then precipitated by centrifugation at at 250–500 × g at
4°C for 5 min and washed with incubation buffer. The sample was
then incubated at the 4°C for 20 min in the dark without vibration.
Detection and quantification of apoptotic cells was obtained by
flow cytometry. This test was performed according to the
manufacturer's instructions
RT-polymerase chain reaction (PCR)
array analysis
Daoy cells were seeded in RPMI 1640 medium
supplemented with 10% FBS, followed by exposure to different
concentrations of GANT61 for 24 h, while the control was not
treated with any GANT61. Total RNA was extracted from the cells
with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
after 24 h, according to the manufacturer's instructions. The total
RNA extracted was then treated with the PrimeScript RT Master Mix
for removal of contaminating DNA and for reverse transcription into
cDNA. Briefly, Primers specific for each of the signaling molecules
were designed using NCBI/Primer-BLAST and used to generate the PCR
products. The following primers were used: GLI1-Forward:
5′-GGGAGGAAAGCAGACTGACT-3′; GLI1-Reverse:
5′-TGGAGAGGTCTTCAGTGCTG-3′; CyclinD1-Forward:
5′-GCATGTTCGTGGCCTCTAAG-3′; CyclinD1-Reverse:
5′-CGTGTTTGCGGATGATCTGT-3′; GAPDH-Forward:
5′-CTCTCTGCTCCTCCCTGTTC-3′; GAPDH-Reverse:
5′-CAATCTCCACTTTGCCACTGC-3′. Target sequences were amplified at
95°C for 1 min, followed by 40 cycles of 95°C for 5 sec and 60°C
for 30 sec. GAPDH was used as endogenous normalization control.
Subsequently, the samples were investigated by PCR array. Data were
analyzed by the ΔΔCq method to determine the mRNA expression
levels, as previously described (20,21). The
experiment was performed in triplicate and repeated three
times.
Western blot analysis
Daoy cells were synchronized in RPMI 1640 medium
with 10% FBS, followed by exposure to different concentrations of
GANT61 for 24 h, while the control was not treated with any GANT61.
The protein profile in the samples was examined by western blot
analysis. Briefly, cells were collected and washed three times with
PBS. Next, the cells were lysed in fresh radioimmunoprecipitation
assay protein lysis buffer containing phenylmethylsulfonyl fluoride
(ratio, 100:1) on ice. The total protein concentration was
determined by the BCA method (ab102536; Abcam). Following
separation by 10% SDS-PAGE, the samples were transferred to
polyvinylidene difluoride films. Protein blots were visualized by
Ponceau S staining. The films were subsequently blocked with 5%
non-fat milk for 2 h at room temperature. Anti-Gli1 (1:500) and
anti-CyclinD1 (1:1,000) protein antibodies were added and incubated
overnight at 4°C. The films were then incubated with the secondary
antibody (1:10,000) at room temperature for 1 h and washed three
times with Tris-buffered saline/Tween 20 buffer. An enhanced
chemiluminescence reagent (WBKLS0500; Merck Millipore, Billerica,
MA, USA) was used to detect the protein levels, which were scanned
using a Bio-Rad exposure system, and Image Lab 3.0 software used
for quantification (Bio-Rad Laboratories, Inc.).
Immunofluorescence analysis
Daoy cells (5×103) were seeded on glass
coverslips and treated with different concentrations of GANT61. At
24 h after incubation, the cells were fixed with 4%
paraformaldehyde for 10 min and permeabilized with 1% Triton X-100
in PBS for 10 min. Next, the cells were incubated with rabbit
anti-Gli1 and mouse anti-CyclinD1 antibodies at 37°C for 1 h and
washed with PBS. Subsequently, incubation for 1 h with
DyLight594-conjugated goat anti-rabbit and FITC conjugated goat
anti-mouse secondary antibodies (111-165-003 and 111-025-003;
1:10,000; Jackson ImmunoResearch Laboratories, Inc., West Grove,
PA, USA) was performed, followed by DAPI staining. The cells were
then mounted and observed under a fluorescence microscope.
Statistical analysis
SPSS version 19.0 (IBM Corp., Armonk, NY, USA)
software was used for statistical analysis. Data were statistically
analyzed by one-way analysis of variance. All experimental data are
expressed as the mean ± standard deviation. P<0.05 indicated a
statistically significant difference.
Results
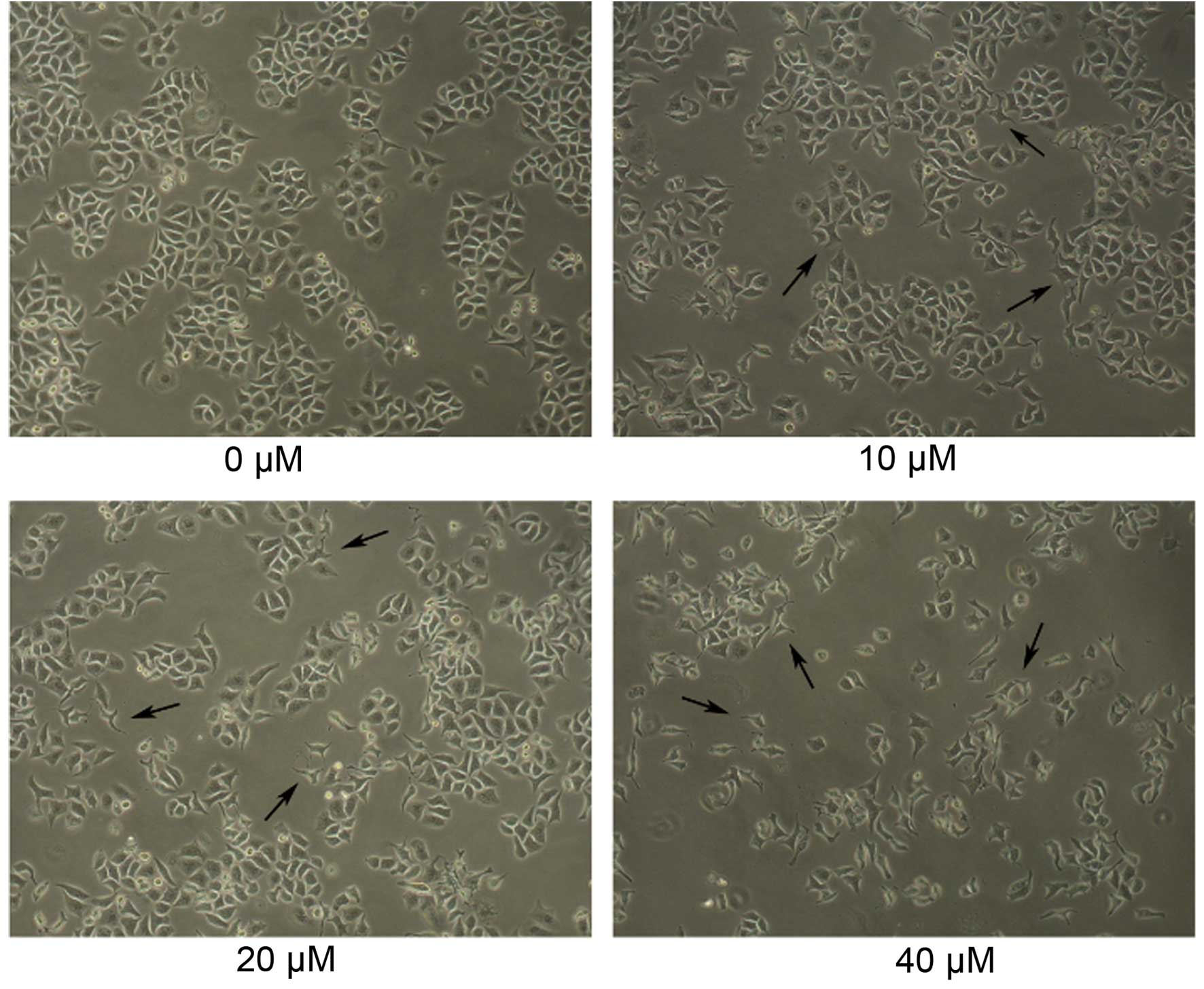
Morphological changes of Daoy cells
following GANT61 treatment
Daoy cells were cultured for 24 h, and then
different concentrations of GANT61 (10, 20 or 40 µM in 0.1% DMSO)
were added to examine the effects of GANT61 on the cell morphology.
The cells were cultured for a further 24 h and then subjected to
inverted microscopic observation. As shown in Fig. 1, the normal, non-adherent Daoy cells
in the untreated control group were spherical in shape. Similarly,
normal adherent cells were intercellular tight, follow flaky
aggregational growth and morphological rules, and their shapes were
rectangular or triangular. Notably, groups treated with increasing
concentrations of GANT61 demonstrated an evident decreased in cell
number, as well as changes in morphology and diversity, which the
cells presented with shrinkage and abnormal form. (Fig. 1).
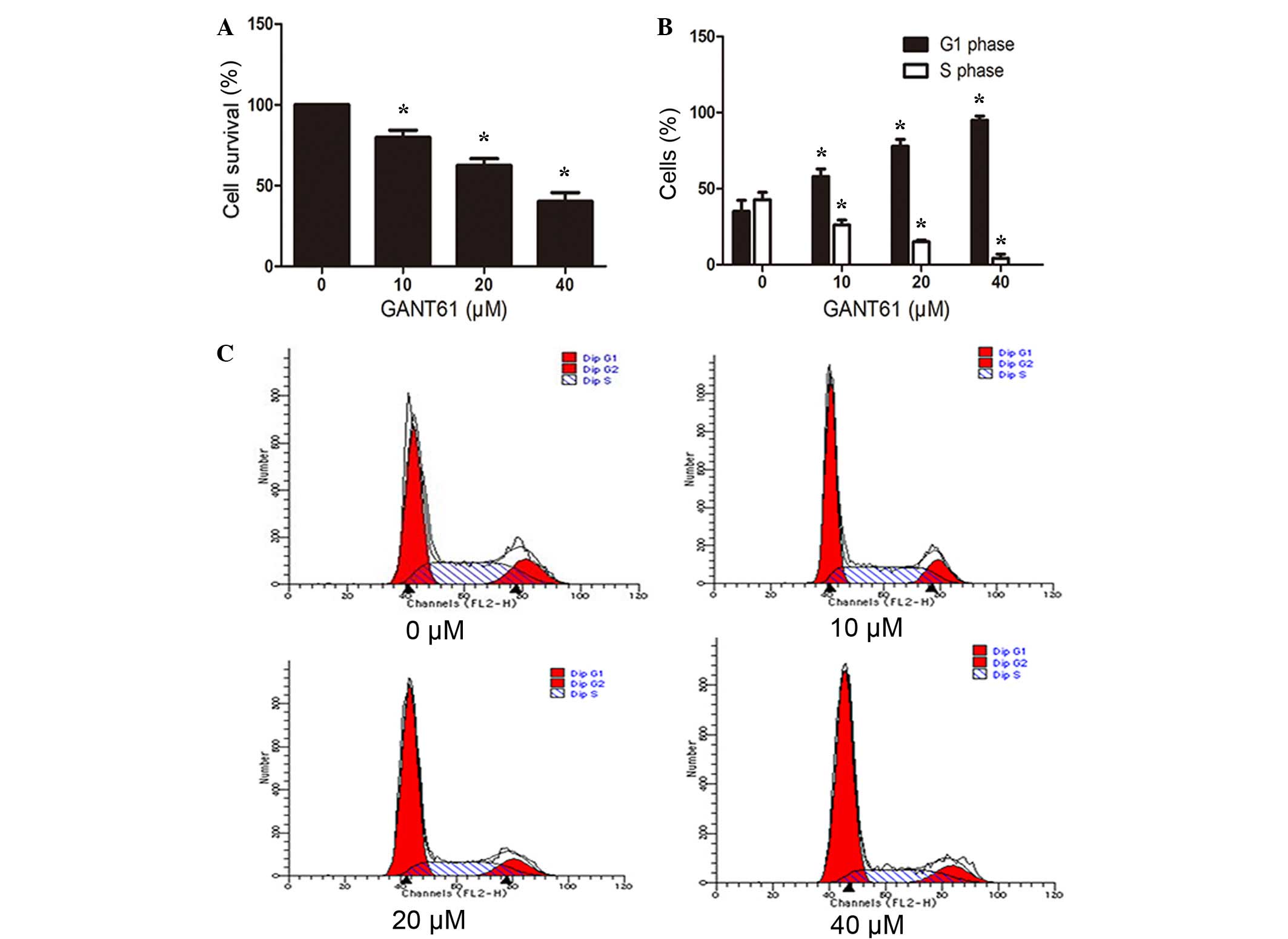
GANT61 inhibits the proliferation and
induces cell cycle arrest of Daoy cells
Marked morphological changes and decreased cell
number was observed following GANT61 treatment (Fig. 1), indicating reduced cell
proliferation or induced cell apoptosis. To elucidate whether cell
proliferation was decreased following treatment with different
concentrations of GANT61 for 24 h, the cell proliferation was
detected by a CCK-8 assay. As shown in Fig. 2A, GANT61 significantly inhibited the
proliferation of Daoy cells. The inhibition of proliferation in
GANT61-treated groups compared with the control group was
dose-dependent (P<0.05; Fig. 2A).
Furthermore, to examine whether the growth inhibition of the cells
was a result of cell cycle arrest, Daoy cells were stained with
FITC-Annexin V and PI, and then subjected to flow cytometry. As
displayed in Fig. 2B and C, the
percentage of cells in G1 phase increased (P<0.05) with
increasing concentration of GANT61 treatment, whereas cells in S
phase decreased in a dose-dependent manner (P<0.05). This
indicated that GANT61 resulted in cell cycle arrest of Daoy cells
at the G1/S transition.
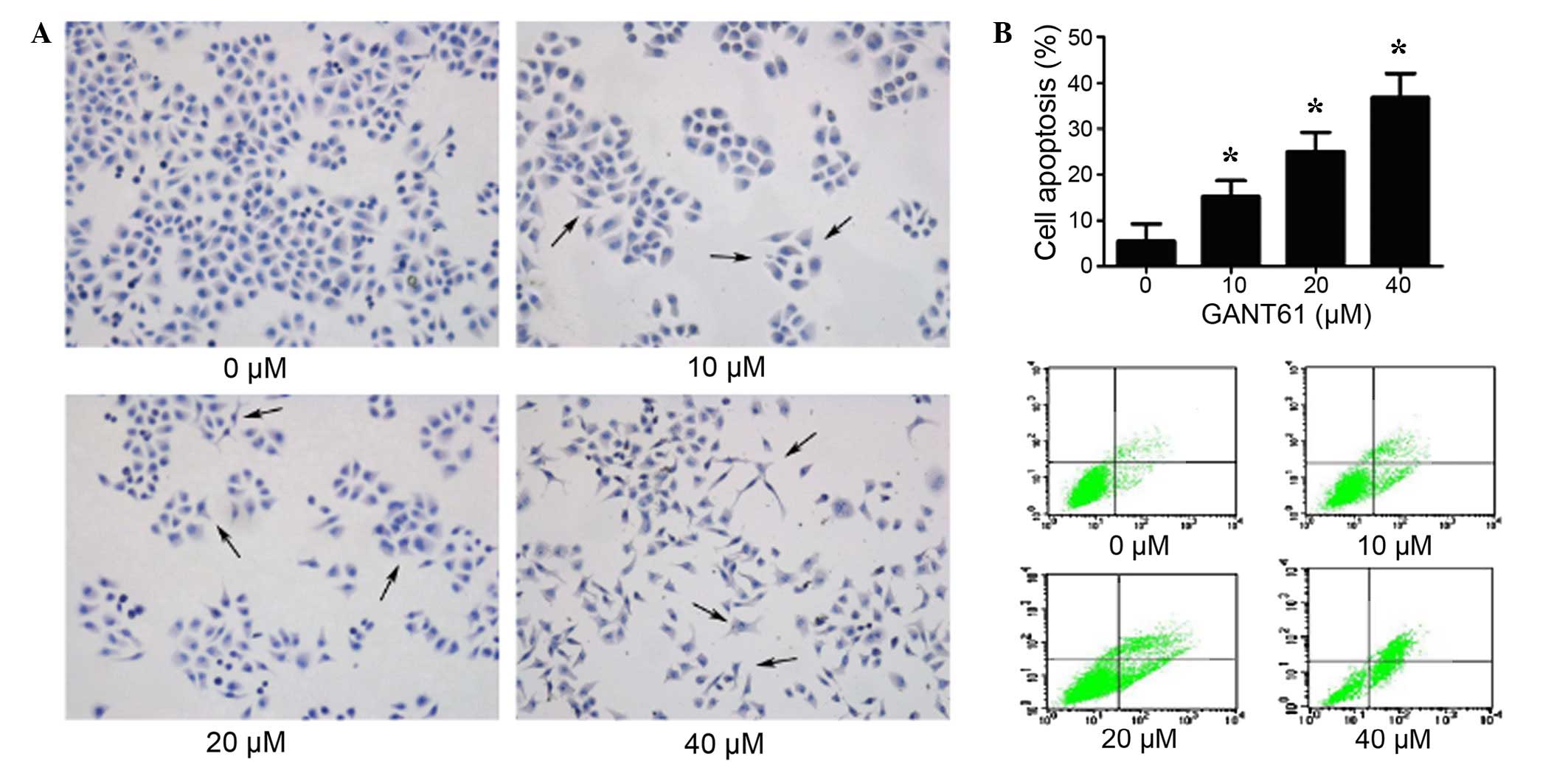
GANT61 promotes cell apoptosis of Daoy
cells
To determine whether GANT61 treatment induced cells
apoptosis, normal growing Daoy cells were treated with various
concentrations of GANT61. After 24 h, the cells were subjected to
HE staining and flow cytometry analysis. As shown in Fig. 3A, the HE staining results
demonstrated that normal cells had a regular morphology. However,
clearly visible abnormal morphologies were observed in Daoy cells
treated with GANT61, with abnormal protuberance observed. The
abnormal protuberance, chromatin condensation and fragmentation
features were more evident at increased concentrations of GANT61,
thus indicating a dose-dependent effect. HE staining also
demonstrated decreased in cell number, increased cell shrinkage and
nuclear fragmentation. As shown in Fig.
3B, the percentage of apoptotic cells increased significantly
in the GANT61-treated cells, compared with the untreated group
(P<0.05). These results verified the prediction that GANT61
induced cell apoptosis in Daoy cells (19).
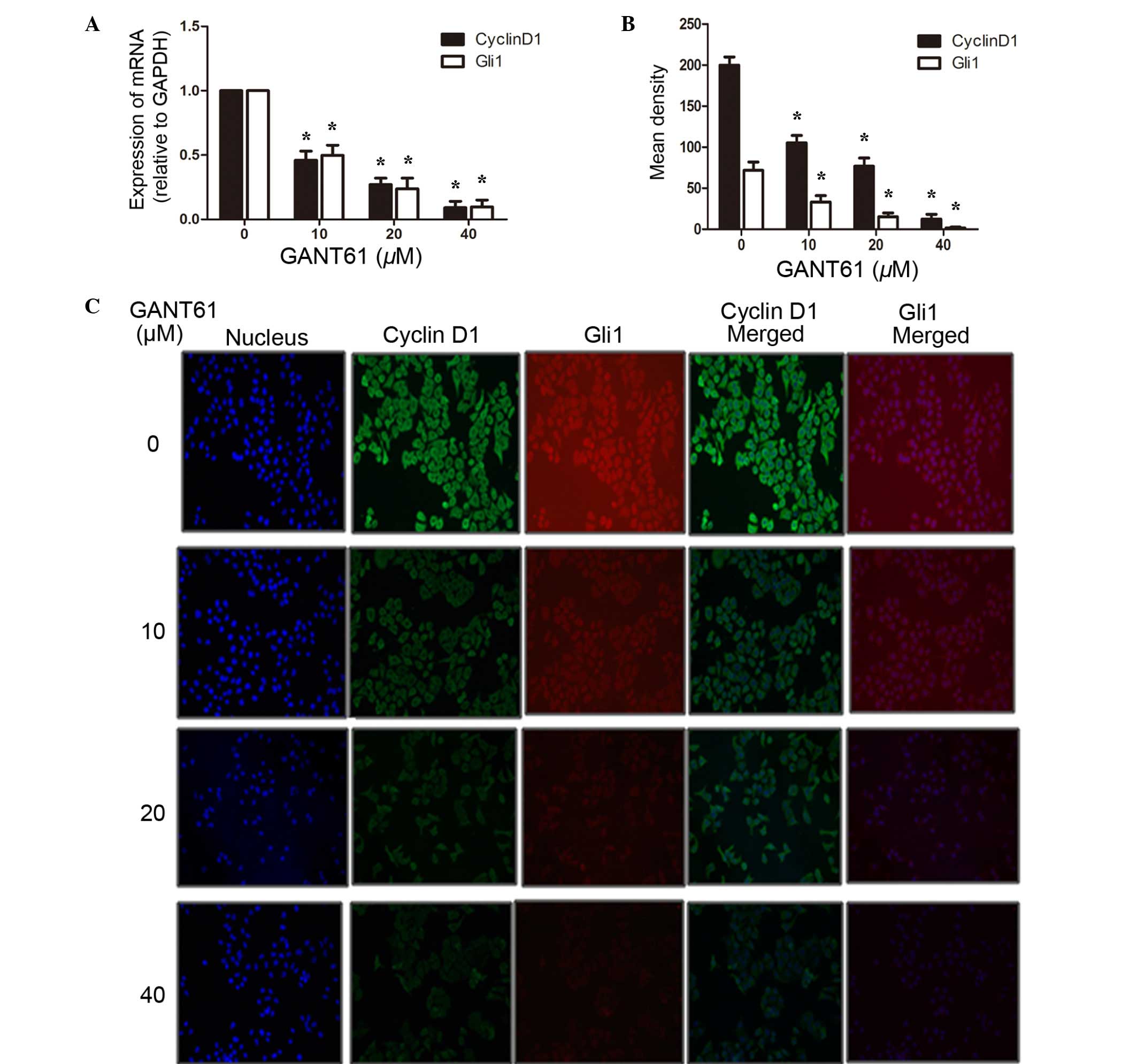
GANT61 inhibits the expression of Gli1
and CyclinD1 in the mRNA and protein level
To examine the underlying mechanism of reduced cell
apoptosis and cell cycle arrest, the total RNA of the cells were
extracted by TRIzol reagent, reverse transcribed into cDNA and then
subjected to PCR. Gli1 is an important transcription factor in the
SHH signaling pathway, regulating the transcription of multiple
downstream target genes, including CyclinD1, the oncogene
controlling cell cycle entry (22,23). As
shown in Fig. 4A, the results
revealed that GANT61 was able to significantly inhibit the gene
expression of Gli1 (P<0.05). Along with the decreased expression
of the Gli1 gene, CyclinD1 mRNA appeared to be downregulated
synchronously (P<0.05). In addition, protein levels were assayed
by immunofluorescence analysis. As indicated in Fig. 4B and C, CyclinD1 was mainly localized
in the cytosol of Daoy cells, whereas Gli1, as a transcription
factor, was located in both the cell cytosol and nucleus. Following
treatment with GANT61 for 24 h, Daoy cells showed decreased levels
of Gli1 protein compared with that in untreated cells (P<0.05).
Subsequently, CyclinD1 was also decreased, as one of the Gli1
transcriptional targets (P<0.05). The inhibition by GANT61 on
Gli1 and CyclinD1 was dose-dependent. To further elucidate the
inhibitory effects of GANT61 on the expression of Gli1 and
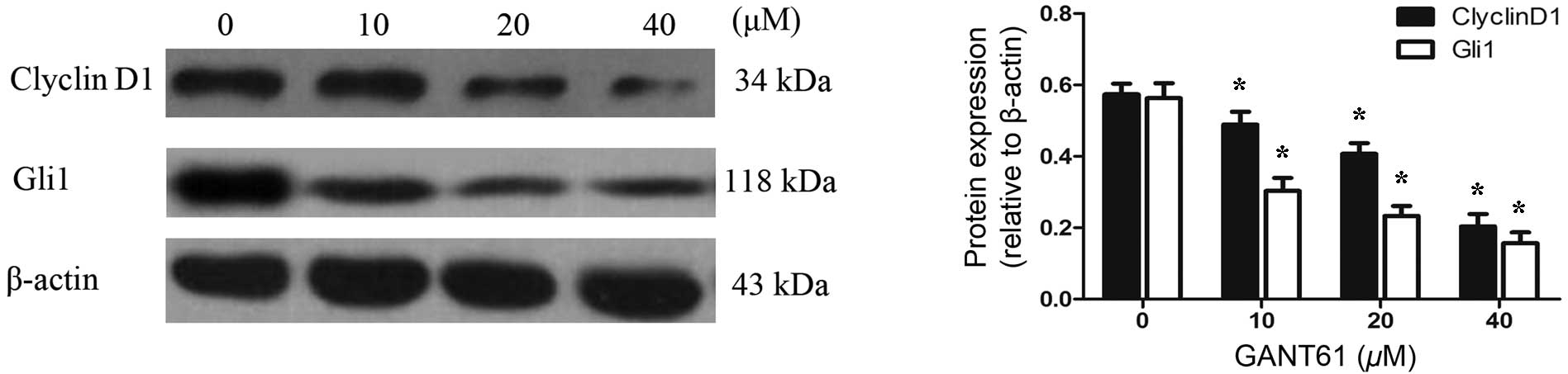
CyclinD1, their protein levels were examined by western blot
analysis. Daoy cells treated with GANT61 for 24 h were lysed and
separated by SDS-PAGE, and the protein expression levels of Gli1
and CyclinD1 were detected using the corresponding antibodies. The
results demonstrated that GANT61 was able to decrease the level of
Gli1 protein (Fig. 5). In line with
the decreased expression of Gli1 protein, CyclinD1 protein also
appeared to be downregulated (P<0.05). The inhibition of Gli1
and CyclinD1 protein levels by GANT61 was in a dose-dependent
manner (P<0.05). These results were consistent with the data
obtained by qPCR and immunofluorescence analyses, indicating that
GANT61 can significantly inhibit Gli1 and CyclinD1 expression at
the mRNA and protein levels.
Discussion
Aberrant activation of the SHH signaling pathway is
implicated in various types of human cancer (24). The SHH signaling pathway is important
in regulating cell proliferation and differentiation in the
embryonic development of the cerebellum (25). MB is characterized by constitutive
activation of the SHH signaling pathway, and is genetically
characterized by mutations in patched homolog 1 (PTCH1), which
blocks the function of smoothened (SMO), or other downstream
pathway mutations (26). Gli1
expression is inhibited by suppressor-of-fused, preventing it from
activating gene transcription. The binding of SHH to PTCH1 or other
mutations releases a basal repression on SMO, which is then
activated (27). Subsequently, Gli1
is released and activates a series of gene transcriptions (28,29).
Inhibitors of the SHH signaling pathway are
currently being developed to mainly target SMO or its upstream
sites (30). Numerous studies using
such inhibitors in MB have demonstrated the efficacy of this
treatment, and these findings have been translated into Phase I and
II clinical trials (31–34). While these therapies have shown
promising results, various significant challenges remain, including
the possible long-term bone marrow suppression and drug toxicity
(35,36). As the majority of targeted therapies
for MB have focused on SMO, it is concerning that only a single
mechanism has been identified and targeted, making resistance a
frequently encountered complication (37). SMO mutation is not the only mechanism
of acquired drug resistance, as the development of other downstream
hedgehog pathway component mutations have since been implicated in
SHH inhibitor resistance. Kool et al (38) indicated that amplifications of Gli
may result in inability to respond to current SMO inhibitors. Such
aberrations include the amplification of Gli and the upregulation
of PI3K/AKT signaling, manifesting in vivo as tumor regrowth
in the same mouse model (39,40).
Gli1 serves a crucial role in the transformation and
proliferation of malignant cells (41). It is also important for preventing
apoptosis and maintaining the malignant proliferation of tumor
cells, and is involved in tumor cell protection against
chemotherapy (42). Berman et
al (43) indicated that the
expression level of Gli1 may reflect the degree of activation of
the SHH signaling pathway. Inhibition of abnormal activation of
this signaling pathway by inhibiting the expression of Gli1 can
inhibit the growth of tumor cells. Gli1, as the main transcription
factor downstream of the SHH signaling pathway, may be able to
inhibit tumor cell proliferation and differentiation through
downregulation of downstream target genes. On the basis of the
pivotal role of Gli1 in malignant cells, it has become increasingly
evident that Gli1 is a promising target for anticancer therapy. A
direct strategy to interfere with Gli1 activity is to induce
selective inhibition of its DNA transcription.
GANT61, an agent that exerts an inhibitory activity
of the SHH signaling pathway, functions by selectively binding to
Gli1 and has been found to suppress proliferation in various tumors
(44,45). In the present study, GANT61 had in
vitro activity against tumor proliferation, and induced cell
cycle arrest and apoptosis. Furthermore, GANT61 was found to
inhibit the Gli1 mRNA and protein expression levels. Dysregulation
of cell cycle progression is considered to serve an important role
in cancer; thus, the current study investigated whether Gli1 is
associated with the typical oncogene CyclinD1 in the cell cycle.
CyclinD1 is a key protein regulating the G1/S transition in the
cell cycle and is highly expressed in multiple types of tumors
(46). CyclinD1 is frequently
deregulated in various cancer types, and is a biomarker of cancer
phenotype and disease progression (46,47).
Overexpressed CyclinD1 accelerates the cell cycle transition,
leading to uncontrolled cell proliferation and the development of
cancer. The present study identified that the mRNA expression of
Gli1 was significantly associated with CyclinD1 expression in MB,
and a similar observation was identified regarding the protein
levels. Suppressing the expression of Gli1 may inhibit the
overexpression of CyclinD1 and the proliferation of tumor cells,
and synchronously promote cell apoptosis. Therefore, blocking the
expression of Gli1 may be an attractive therapeutic strategy for
MB.
In conclusion, SHH signaling pathway can regulate
tumor cell cycle and apoptosis in different molecular levels.
Increased expression of Gli1 induced the upregulation of CyclinD1
expression, thus promoting cell proliferation, which may be one of
the growth patterns of tumor cells. Therefore, Gli1 may be an
important target for MB treatment. Therapies using Gli1-targeted
inhibitors alone or combined with other cytotoxic chemotherapeutics
may become an effective targeted treatment of MB. However, the
association of the SHH signaling pathway and other pathways in MB
cells with the specific mechanism of apoptosis induced by targeted
therapy requires further investigation.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Zhejiang Province (no. LY13H160033),
the Zhejiang Medical and Health Science and Technology Plan Project
(no. 2012RCA043) and the Foundation of Wenzhou Scientific and
Technological Bureau Protect (no. Y20140717).
References
|
1
|
Gerber NU, Mynarek M, von Hoff K,
Friedrich C, Resch A and Rutkowski S: Recent developments and
current concepts in medulloblastoma. Cancer Treat Rev. 40:356–365.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gilbertson RJ: Medulloblastoma: Signalling
a change in treatment. Lancet Oncol. 5:209–218. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Packer RJ and Vezina G: Management of and
prognosis with medulloblastoma: Therapy at a crossroads. Arch
Neurol. 65:1419–1424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rutkowski S, von Hoff K, Emser A, Zwiener
I, Pietsch T, Figarella-Branger D, Giangaspero F, Ellison DW, Garre
ML, Biassoni V, et al: Survival and prognostic factors of early
childhood medulloblastoma: An international meta-analysis. J Clin
Oncol. 28:4961–4968. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moxon-Emre I, Bouffet E, Taylor MD,
Laperriere N, Scantlebury N, Law N, Spiegler BJ, Malkin D, Janzen L
and Mabbott D: Impact of craniospinal dose, boost volume, and
neurologic complications on intellectual outcome in patients with
medulloblastoma. J Clin Oncol. 32:1760–1768. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Archer TC, Weeraratne SD and Pomeroy SL:
Hedgehog-GLI pathway in medulloblastoma. J Clin Oncol.
30:2154–2156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gotschel F, Berg D, Gruber W, Bender C,
Eberl M, Friedel M, Sonntag J, Rüngeler E, Hache H, Wierling C, et
al: Synergism between Hedgehog-GLI and EGFR signaling in
Hedgehog-responsive human medulloblastoma cells induces
downregulation of canonical Hedgehog-target genes and stabilized
expression of GLI1. PLoS One. 8:e654032013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cho YJ, Tsherniak A, Tamayo P, Santagata
S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L,
Eberhart CG, et al: Integrative genomic analysis of medulloblastoma
identifies a molecular subgroup that drives poor clinical outcome.
J Clin Oncol. 29:1424–1430. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mcmillan R and Matsui W: Molecular
pathways: The hedgehog signaling pathway in cancer. Clin Cancer
Res. 18:4883–4888. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shahi MH, Afzal M, Sinha S, Eberhart CG,
Rey JA, Fan X and Castresana JS: Regulation of sonic hedgehog-GLI1
downstream target genes PTCH1, Cyclin D2, Plakoglobin, PAX6 and
NKX2.2 and their epigenetic status in medulloblastoma and
astrocytoma. BMC Cancer. 10:6142010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kenney AM, Cole MD and Rowitch DH: Nmyc
upregulation by sonic hedgehog signaling promotes proliferation in
developing cerebellar granule neuron precursors. Development.
130:15–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Musgrove EA, Caldon CE, Barraclough J,
Stone A and Sutherland RL: Cyclin D as a therapeutic target in
cancer. Nat Rev Cancer. 11:558–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barbash O, Zamfirova P, Lin DI, Chen X,
Yang K, Nakagawa H, Lu F, Rustgi AK and Diehl JA: Mutations in Fbx4
inhibit dimerization of the SCF (Fbx4) ligase and contribute to
cyclin D1 overexpression in human cancer. Cancer Cell. 14:68–78.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fu M, Wang C, Li Z, Sakamaki T and Pestell
RG: Minireview: Cyclin D1: Normal and abnormal functions.
Endocrinology. 145:5439–5447. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Musgrove EA: Cyclins: Roles in mitogenic
signaling and oncogenic transformation. Growth Factors. 24:13–19.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fu J, Rodova M, Roy SK, Sharma J, Singh
KP, Srivastava RK and Shankar S: GANT-61 inhibits pancreatic cancer
stem cell growth in vitro and in NOD/SCID/IL2R gamma null mice
xenograft. Cancer Lett. 330:22–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peukert S and Miller-Moslin K:
Small-molecule inhibitors of the hedgehog signaling pathway as
cancer therapeutics. ChemMedChem. 5:500–512. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mazumdar T, Devecchio J, Shi T, Jones J,
Agyeman A and Houghton JA: Hedgehog signaling drives cellular
survival in human colon carcinoma cells. Cancer Res. 71:1092–1102.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin Z, Li S, Sheng H, Cai M, Ma LY, Hu L,
Xu S, Yu LS and Zhang N: Suppression of GLI sensitizes
medulloblastoma cells to mitochondria-mediated apoptosis. J Cancer
Res Clin Oncol. 142:2469–2478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rafiee M, Keramati MR, Ayatollahi H,
Sadeghian MH, Barzegar M, Asgharzadeh A and Alinejad M:
Down-Regulation of Ribosomal S6 kinase RPS6KA6 in Acute Myeloid
Leukemia Patients. Cell J. 18:159–164. 2016.PubMed/NCBI
|
|
21
|
Floris I, Billard H, Boquien CY,
Joram-Gauvard E, Simon L, Legrand A, Boscher C, Rozé JC,
Bolaños-Jiménez F and Kaeffer B: MiRNA Analysis by Quantitative PCR
in Preterm Human Breast Milk Reveals Daily Fluctuations of
hsa-miR-16-5p. PLoS One. 10:e1404882015. View Article : Google Scholar
|
|
22
|
Huang XB, Shi Y, Wang CS, Wang XD, Cheng J
and Che FF: Synergistic Inhibitory Effect of Arsenic Trioxide
Combined with Itraconazole on Hedgehog Pathway of Multiple Myeloma
NCI-H929 Cells. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 24:1459–1465.
2016.(In Chinese). PubMed/NCBI
|
|
23
|
Du WZ, Feng Y, Wang XF, Piao XY, Cui YQ,
Chen LC, Lei XH, Sun X, Liu X, Wang HB, et al: Curcumin suppresses
malignant glioma cells growth and induces apoptosis by inhibition
of SHH/GLI1 signaling pathway in vitro and vivo. CNS Neurosci Ther.
19:926–936. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakamura M and Katano M: Hedgehog
signaling pathway and its impact on development of cancer therapy.
Fukuoka Igaku Zasshi. 99:102–106. 2008.(In Japanese). PubMed/NCBI
|
|
25
|
Malek R, Matta J, Taylor N, Perry ME and
Mendrysa SM: The p53 inhibitor MDM2 facilitates Sonic
Hedgehog-mediated tumorigenesis and influences cerebellar
foliation. PLoS One. 6:e178842011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sahebjam S, Siu LL and Razak AA: The
utility of hedgehog signaling pathway inhibition for cancer.
Oncologist. 17:1090–1099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X, Venugopal C, Manoranjan B,
McFarlane N, O'Farrell E, Nolte S, Gunnarsson T, Hollenberg R,
Kwiecien J, Northcott P, et al: Sonic hedgehog regulates Bmi1 in
human medulloblastoma brain tumor-initiating cells. Oncogene.
31:187–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang J and Hui CC: Hedgehog signaling in
development and cancer. Dev Cell. 15:801–812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruiz i Altaba A, Palma V and Dahmane N:
Hedgehog-Gli signalling and the growth of the brain. Nat Rev
Neurosci. 3:24–33. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Y, Gholamin S, Schubert S, Willardson
MI, Lee A, Bandopadhayay P, Bergthold G, Masoud S, Nguyen B, Vue N,
et al: Epigenetic targeting of Hedgehog pathway transcriptional
output through BET bromodomain inhibition. Nat Med. 20:732–740.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jimeno A, Weiss GJ, Miller WH Jr,
Gettinger S, Eigl BJ, Chang AL, Dunbar J, Devens S, Faia K, Skliris
G, et al: Phase I study of the Hedgehog pathway inhibitor IPI-926
in adult patients with solid tumors. Clin Cancer Res. 19:2766–2774.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Berlin J, Bendell JC, Hart LL, Firdaus I,
Gore I, Hermann RC, Mulcahy MF, Zalupski MM, Mackey HM, Yauch RL,
et al: A randomized phase II trial of vismodegib versus placebo
with FOLFOX or FOLFIRI and bevacizumab in patients with previously
untreated metastatic colorectal cancer. Clin Cancer Res.
19:258–267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim EJ, Sahai V, Abel EV, Griffith KA,
Greenson JK, Takebe N, Khan GN, Blau JL, Craig R, Balis UG, et al:
Pilot clinical trial of hedgehog pathway inhibitor GDC-0449
(vismodegib) in combination with gemcitabine in patients with
metastatic pancreatic adenocarcinoma. Clin Cancer Res.
20:5937–5945. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
D'Amato C, Rosa R, Marciano R, D'Amato V,
Formisano L, Nappi L, Raimondo L, Di Mauro C, Servetto A, Fulciniti
F, et al: Inhibition of Hedgehog signalling by NVP-LDE225
(Erismodegib) interferes with growth and invasion of human renal
cell carcinoma cells. Br J Cancer. 111:1168–1179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rudin CM, Hann CL, Laterra J, Yauch RL,
Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et
al: Treatment of medulloblastoma with hedgehog pathway inhibitor
GDC-0449. N Engl J Med. 361:1173–1178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kimura H, Ng JM and Curran T: Transient
inhibition of the Hedgehog pathway in young mice causes permanent
defects in bone structure. Cancer Cell. 13:249–260. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Meani RE, Lim SW, Chang AL and Kelly JW:
Emergence of chemoresistance in a metastatic basal cell carcinoma
patient after complete response to hedgehog pathway inhibitor
vismodegib (GDC-0449). Australas J Dermatol. 55:218–221. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kool M, Jones DT, Jäger N, Northcott PA,
Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et
al: Genome sequencing of SHH medulloblastoma predicts
genotype-related response to smoothened inhibition. Cancer Cell.
25:393–405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Samkari A, White J and Packer R: SHH
inhibitors for the treatment of medulloblastoma. Expert Rev
Neurother. 15:763–770. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan S, Wu X, Jiang J, Gao W, Wan Y, Cheng
D, Han D, Liu J, Englund NP, Wang Y, et al: Discovery of
NVP-LDE225, a potent and selective smoothened antagonist. ACS Med
Chem Lett. 1:130–134. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ruiz i Altaba A, Mas C and Stecca B: The
Gli code: An information nexus regulating cell fate, stemness and
cancer. Trends Cell Biol. 17:438–447. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Katoh Y and Katoh M: Hedgehog target
genes: Mechanisms of carcinogenesis induced by aberrant hedgehog
signaling activation. Curr Mol Med. 9:873–886. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Berman DM, Karhadkar SS, Hallahan AR,
Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale
J, Olson JM and Beachy PA: Medulloblastoma growth inhibition by
hedgehog pathway blockade. Science. 297:1559–1561. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Miyazaki Y, Matsubara S, Ding Q, Tsukasa
K, Yoshimitsu M, Kosai K and Takao S: Efficient elimination of
pancreatic cancer stem cells by hedgehog/GLI inhibitor GANT61 in
combination with mTOR inhibition. Mol Cancer. 15:492016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Benvenuto M, Masuelli L, De Smaele E,
Fantini M, Mattera R, Cucchi D, Bonanno E, Di Stefano E, Frajese
GV, Orlandi A, et al: In vitro and in vivo inhibition of breast
cancer cell growth by targeting the Hedgehog/GLI pathway with SMO
(GDC-0449) or GLI (GANT-61) inhibitors. Oncotarget. 7:9250–9270.
2016.PubMed/NCBI
|
|
46
|
Li X, Hao Z, Fan R, Zou X, Jin H, Pan Y,
He L, Du R, Gao L, Liu D and Fan D: CIAPIN1 inhibits gastric cancer
cell proliferation and cell cycle progression by downregulating
CyclinD1 and upregulating P27. Cancer Biol Ther. 6:1539–1545. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Seiler R, Thalmann GN, Rotzer D, Perren A
and Fleischmann A: CCND1/CyclinD1 status in metastasizing bladder
cancer: A prognosticator and predictor of chemotherapeutic
response. Mod Pathol. 27:87–95. 2014. View Article : Google Scholar : PubMed/NCBI
|