Introduction
Human immunodeficiency virus (HIV) infection is
propagated primarily via blood transfusion (before 1985),
intravenous drug use, professional exposure in medical institutes,
sexual transmission and mother-to-child transmission (1,2).
Historically, hepatitis C virus (HCV) infection has
developed in three waves: i) Medical care through needles and
syringes reused without sterilization; ii) blood transfusion before
1991; iii) and through intravenous drug injection or the sharing of
straws for cocaine inhalation (3).
HCV infection is often observed among HIV-infected
persons, with one-third of HIV-infected Americans and 7 million
worldwide being coinfected (4,5). HIV
coinfection deteriorates HCV disease, increasing the likelihood of
cirrhosis and HCV-related mortality (6,7).
There are 33.3 million people globally living with
HIV infection (8). It is estimated
that 20–30% of HIV patients are also infected with hepatitis C
(HCV) (9). In China, small sample
studies suggest that 8.25–56.9% of HIV-infected persons were
simultaneously infected with HCV (10–14).
Delayed testing for HIV and HCV is common among patients at
methadone clinics in Guangdong, with numerous patients experiencing
delays of ≥2 months (13,14). Early diagnosis and enrollment in care
is particularly crucial in a highly populated country such as
China; thus, expediting HIV and HCV testing is important (15).
Guangxi province is a high endemic area of HIV
infection in China [the cumulated report number of HIV-infected
persons is ranked second in the country (16)], which also has a high incidence of
viral liver disease and hepatocellular carcinoma (17). Based on frequent HIV/HCV coinfection
in other areas, there is a crucial requirement to investigate the
HCV infection status in HIV-infected people. Furthermore, there are
likely to be numerous HCV-infected persons among the HIV-infected
population for which the diagnosis of HCV has been missed due to
the diagnosis being made only on the basis of HCV antibody
detection.
Materials and methods
Ethics statement
This study was approved by the Health Bureau of
Guangxi Province and the Ethics Review Committee at Guangxi Medical
University (Nanning, China). Written informed ethical consent was
provided by clinical directors who answered the questionnaire. All
patients gave their written consent for their information to be
used for research.
Study population
This study was performed between August 2008 and
January 2011. The participants were recruited through acquired
immune deficiency syndrome (AIDS) voluntary counselling and testing
clinics, prisons, addiction treatment centers, hospitals and
communities from Liuzhou and Qinzhou cities in Guangxi province. A
total of 300 individuals with HIV-1 infection were recruited. All
participants or their guardians voluntarily signed a consent form.
Written informed ethical consent was provided by professionals who
answered the questionnaire. A total of 41 patients who were only
infected with hepatitis C were recruited from the First Affiliated
Hospital of Guangxi Medical University to serve as the control
group.
All participants were analyzed to characterize their
serological status toward both HIV [screening and confirmation
tests (18)] and HCV [screening,
serological confirmation, search for HCV-RNA by polymerase chain
reaction (PCR) and genotyping] as described below. Samples of serum
and plasma (10 ml) were prepared from venous whole blood.
Reagents
Hepatitis C virus antibody diagnostic kits were
purchased from Shanghai Kehua Bio-Engineering Co., Ltd., (Shanghai,
China). QIAamp Viral RNA Mini kit was obtained from Qiagen (Hilden,
Germany). A Revert Aid First Strand cDNA Synthesis kit was
purchased from Fermentas, Inc., (Burlington, ON, Canada).
Restriction endonucleases BsrBI, HaeII, HinfI,
BstUI, HaeШ and Apol were obtained from New
England Biolabs (Ipswich, MA, USA).
Enzyme-linked immunosorbent assay
(ELISA)
Flavicheck-HCV, a commercial third generation,
rapid, qualitative, two-site sandwich immunoassay test device was
employed according to the manufacturer's instructions (Tulip
Diagnostics (P), Ltd., Goa, India). The kit was used to detect
total antibodies specific to HCV in serum or plasma by using a
multiepitope recombinant peptide antigen that is broadly
cross-reactive to all major HCV genotypes.
Nested-PCR and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from 140 µl plasma using a
QIAamp Viral RNA Mini kit according to the manufacturer's
instructions (Qiagen). RNA samples were DNase treated using an RQ1
RNase-Free DNase kit (Promega, Madison, WI, USA), according to the
manufacturer's instructions. Nucleic acids were then aliquoted, and
one aliquot was reverse-transcribed according to the SuperScript
III First-Strand cDNA Synthesis SuperMix kit protocol using random
hexamers (Invitrogen; ThermoFisher Scientific, Inc., Carlsbad, CA,
USA). cDNA was synthesized from total RNA using reverse
transcriptase using a First Strand cDNA synthesis kit (Fermentas,
Inc.). The cDNA was amplified by nested PCR using a Tiangen 2X PCR
mix kit (Tiangen Biotech Co., Ltd., Bejing, China) or by RT-qPCR
using SYBR Green SuperMix (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) according to the manufacturers' instructions. The PCR
cycling was conducted as follows: In the first round, the
amplification mixture comprised 5.0 µl cDNA, 12.5 µl pre-mixed
solution of Tiangen 2X PCR mix and 0.5 µl of each primer (P1 and
P2). The enzyme was diluted with 6.5 µl water, and the total
reaction volume was 25.0 µl. The reaction conditions were as
follows: 94°C for 5 min; 94°C for 45 sec, 62°C for 30 sec, and 72°C
for 20 sec for 35 cycles; followed by 72°C for 5 min. In the second
round, the amplification mixture comprised 3.0 µl product from the
first round, 12.5 µl pre-mixed solution of Tiangen 2X PCR mix, 0.4
µl each primer (P3 and P4). The enzyme was diluted with 8.7 µl
water, and the total reaction volume was 25.0 µl. The extension
time in the second round was 15 sec, and all other times were as in
the first round. For the RT-qPCR analysis, all experiments were
performed in triplicate. The sample input was normalized against
its own critical threshold (Cq) value of the housekeeping gene
GAPDH. Primer sequences and PCR conditions used in this study are
listed in Table I. The
2−ΔΔCq method (19) was
used to calculate the expression levels.
 | Table I.HCV primer sequences, annealing
temperatures and product sizes for the nested PCR analyses. |
Table I.
HCV primer sequences, annealing
temperatures and product sizes for the nested PCR analyses.
| Primer | Primer sequence | Annealing temperature
(°C) | Product size
(bp) |
|---|
| Nested outer
primer | Forward:
5′-TGAGGAACTACTGTCTTCACG-3′ | 62 | 258 |
|
| Reverse:
5′-AGCACCCTATCAGGCAGTACC-3′ |
|
|
| Nested inner
primer | Forward:
5′-GGGAGAGCCATAGTGGTCTG-3′ | 62 | 186 |
|
| Reverse:
5′-CACTCGCAAGCACCCTATC-3′ |
|
|
PCR product sequencing
The nested PCR products and sense primer were used
for sequencing reaction. PCR products were recovered according to
the manufacturer's instructions (Fermentas, Inc.). The sequencing
was performed using the Sanger dideoxy method (20). This was followed by detection of the
HCV sequence using GeneBank (https://www.ncbi.nlm.nih.gov/genbank/), BBLAST
(http://blast.ncbi.nlm.nih.gov/Blast.cgi), Clustalx
(http://www.clustal.org/clustal2/) and
BioEdit software (http://www.mbio.ncsu.edu/BioEdit/page2.html).
HCV genotype test
Restriction fragment length polymorphism (RFLP)
analysis was conducted using the method described by Chinchai et
al (21,22). In RFLP, the nested PCR products of
RNA positive samples (20–30 µl) were digested using the restriction
enzymes Acc1, Mbol and BstN1 and incubated at
37°C for overnight in a specific endonuclease buffer (Shanghai
Biological Engineering Co., Ltd., Shanghai, China). The digested
product was loaded onto 3% nusieve agarose gel and the restriction
pattern was analyzed using a Gel-Doc 2000 System (Bio-Rad
Laboratories, Inc.).
Data analysis
Data were analyzed using SPSS software version 17.0
(SPSS, Inc., Chicago, IL, USA). Chi-square (χ2) test was
utilized to compare variables. P<0.05 was considered to indicate
a statistically significant difference. Odds ratio (OR) and 95%
confidence interval (CI) were used to measure the strength of
association.
Results
Demographics and epidemiological
characteristics
Demographic data were collected regarding the
participant's gender, age, ethnicity, education, employment and
marital status (Table II). Among
the 300 HIV positive participants, 195 were male and 105 were
female; the oldest patient was 88 years old and the youngest was 2
years old; the mean age was 37.39±13.73 years. Among all
participants, 201 were of Han ethnicity and 99 were of minority
ethnicity. Among the viral hepatitis C and HIV-infected
participants, 103 were Han, while 43 patients were of the Zhuang
ethnicity. The majority of the participants had no fixed occupation
(54.7%); the majority of the participants were unmarried or single
(53.33%); among the HIV positive patients in Liuzhou and Qinzhou,
there were no significant differences in HCV infection rate
(Table II).
 | Table II.Sociodemographic characteristics of
HIV/HCV coinfected persons. |
Table II.
Sociodemographic characteristics of
HIV/HCV coinfected persons.
|
|
|
| HCV |
|---|
|
|
|
|
|
|---|
| Parameter | Participants
(n) | Participants
(%) | Positive (n) | Positive rate | χ2 | P-value |
|---|
| Gender |
|
|
|
| 13.37 | <0.01 |
|
Male | 195 | 65.0 | 110 |
59.46 |
|
|
|
Female | 105 | 35.0 | 36 |
34.29 |
|
|
| Age (years) |
|
|
|
| 13.69 | 0.02 |
|
<20 | 6 |
2.0 | 2 |
33.33 |
|
|
|
20–29 | 89 | 29.7 | 42 |
47.19 |
|
|
|
30–39 | 105 | 35.0 | 63 | 60.0 |
|
|
|
40–49 | 50 | 16.7 | 25 | 50 |
|
|
|
50–59 | 22 |
7.3 | 6 |
27.27 |
|
|
|
≥60 | 28 |
9.3 | 8 |
28.57 |
|
|
| Ethnicity |
|
|
|
| 1.62 | 0.20 |
|
Han | 201 | 67.0 | 103 |
51.24 |
|
|
|
Minority | 99 | 33.0 | 43 |
43.43 |
|
|
| Occupation |
|
|
|
| 21.94 | <0.01 |
|
Unemployed | 164 | 54.7 | 100 |
60.98 |
|
|
|
Employed | 136 | 45.3 | 46 |
33.82 |
|
|
| Education |
|
|
|
| 9.00 | 0.03 |
|
≤Primary school | 51 |
17.0 | 30 |
58.82 |
|
|
| Junior
high school | 201 |
67.0 | 94 |
46.77 |
|
|
| High
school/polytechnic | 30 |
10.0 | 18 | 60.0 |
|
|
|
≥College | 18 |
6.0 | 4 |
22.22 |
|
|
| Marital status |
|
|
|
| 18.85 | <0.01 |
|
Married | 95 |
31.67 | 33 |
34.74 |
|
|
|
Unmarried | 165 |
53.33 | 99 | 60.0 |
|
|
|
Divorced/widowed | 40 |
15.0 | 14 | 35.0 |
|
|
| Region |
|
|
|
| 0.21 | 0.91 |
|
Liuzhou | 141 | 47.0 | 68 |
48.23 |
|
|
|
Qinzhou | 159 | 53.0 | 78 |
49.06 |
|
|
Predominant route of transmission in
HIV and HCV coinfected participants
The prevalence of HCV infection was 48.67% (146/300)
among the HIV infected study population. Among the 101 individuals
who were drug users, 93 (92.08%) exhibited HIV/HCV coinfection. One
of the six subjects that had received a blood transfusion was
infected with HCV (16.67%). One of the five subjects suffered HIV
from vertical transmission was infected with HCV (20%). Among the
188 subjects that suffered from a history of sexually transmitted
diseases, 51 persons were infected with HCV (27.13%). With respect
to sexual behavior, ~69.18% of the subjects had more than one
sexual partner. The rate of coinfection differed between
transmission route according to the results of the χ2
test (Table III).
 | Table III.Distribution of HIV/HCV coinfected
patients in different transmission routes. |
Table III.
Distribution of HIV/HCV coinfected
patients in different transmission routes.
| Transmission
route | Participants
(n) | HIV/HCV
coinfection | Coinfection rate
(%) | χ2 | P-value |
|---|
| Drug-related | 101 | 93 | 92.08 |
|
|
| Sex-related | 188 | 51 | 27.13 |
|
|
| Vertical
transmission |
5 |
1 | 20.0 | 127.4 | <0.001 |
| Blood
transfusion |
6 |
1 | 16.67 |
|
|
| Total | 300 | 146 | 48.67 |
|
|
Gene sequencing of HCV positive serum
samples
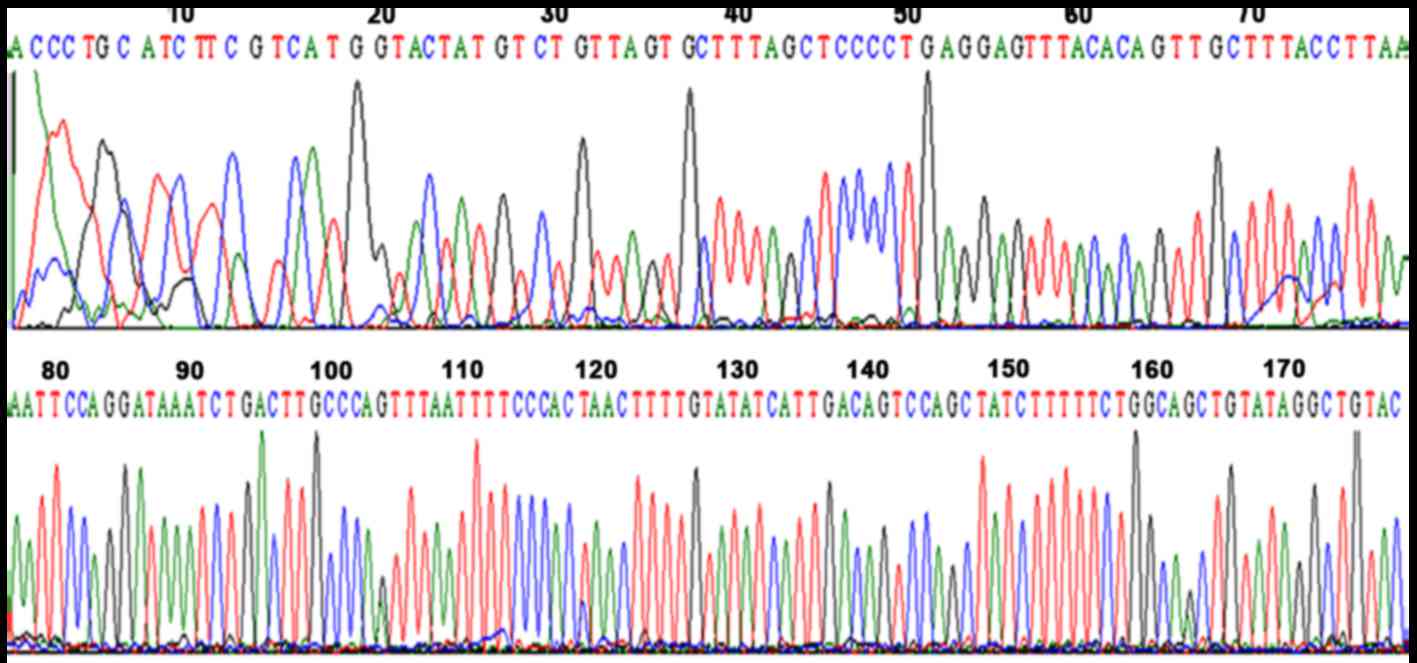
Five cases of HCV RNA-positive serum samples were
randomly selected, and nested PCR products were sequenced. The
results showed that the sequences of the samples were the same as
HCV 5′ untranslated region in GeneBank by blasting HCV database
(Fig. 1).
HCV prevalence at the RNA level
HCV RNA quantification was conducted using nested
PCR. The results showed that among the 146 HCV antibody-positive
serum samples, there were 114 HCV RNA positive samples. However,
there were 41 HCV RNA positive samples among the HCV
antibody-negative serum samples (Fig.
2).
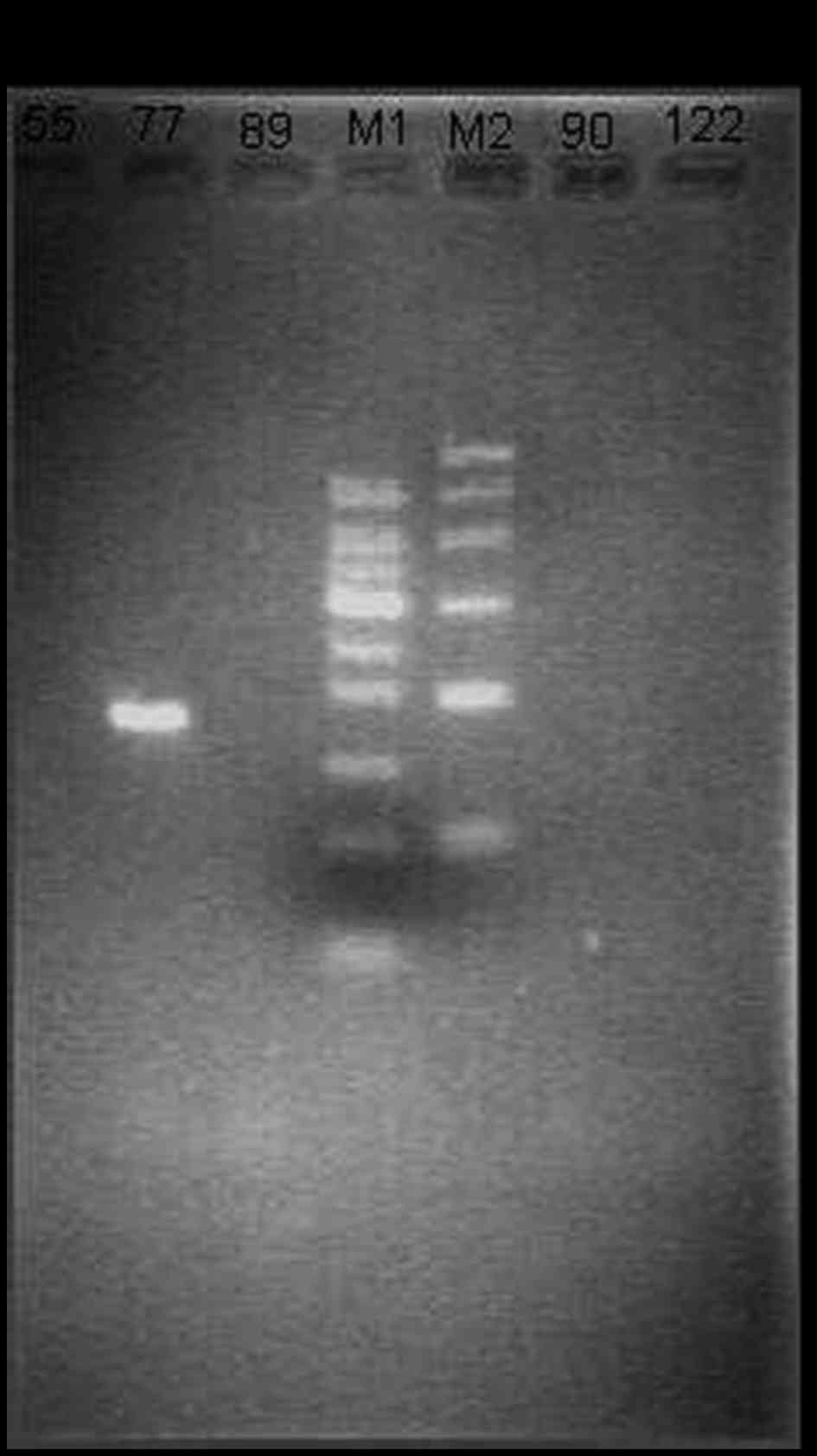 | Figure 2.Electrophoretogram of hepatitis C
virus RNA. M1 is Marker1 (bp): 50, 100, 150, 200, 250, 300, 350,
400 and 500. M2 is Marker2 (bp): 100, 200, 300, 500, 700 and 1,000.
The length of electrophoretic bands of the nested PCR amplified
product is 186 bp. The three lanes to the left and right of the
markers are patient samples (nos. 55, 77, 89, 90 and 122),
respectively. |
HCV genotypes in the Guangxi province
population
The digested results showed that among the 41 study
participants positive for HCV RNA but negative for HCV antibody,
the prevalence rates of the 1b, 1a, 3a and 4a genotypes were 22
(53.7%), 3 (7.3%), 6 (14.6%) and 5 (12.2%), respectively. Among the
146 study participants that were positive for HCV antibody, the
prevalence of the 1b, mixed, 6a, 3b, 1a, 3a, 2a and 2b genotypes
were 31 (27.2%), 27 (23.7%), 18 (15.8%), 14 (12.3%), 10 (8.8%), 5
(4.4%), 5 (4.4%) and 4 (3.5%), respectively (Table IV).
 | Table IV.Comparison of HCV genotype
distribution between two groups (n=155). |
Table IV.
Comparison of HCV genotype
distribution between two groups (n=155).
|
| Genotype |
|
|---|
|
|
|
|
|---|
| HCV antibody | 1a | 1b | 2a | 2b | 3a | 3b | 4a | 6a | Mixed | Total |
|---|
| Negative | 3 | 22 | 0 | 2 | 6 | 1 | 5 | 2 | 0 | 41 |
| Positive | 10 | 31 | 5 | 4 | 5 | 14 | 0 | 18 | 27 | 114 |
| Total | 13 | 53 | 5 | 6 | 11 | 15 | 5 | 20 | 27 | 155 |
Differences between positive HCV
antibody and HCV RNA test results
Among the 300 study participants tested for anti-HCV
antibodies and HCV RNA, 114 were positive for both, accounting for
38%. A total of 41 participants were anti-HCV negative and HCV RNA
positive, accounting for 13.7%. A total of 32 participants were
anti-HCV positive and HCV RNA negative, accounting for 10.67%
(32/300) of all patients. Among the 300 individuals analyzed, 113
were negative for anti-HCV and HCV RNA, accounting for 37.67%.
Among the anti-HCV negative participants, 41 were HCV RNA positive,
accounting for 13.7% (41/300) of all patients (Table V).
 | Table V.Comparison of positive HCV antibody
and HCV RNA (n=300). |
Table V.
Comparison of positive HCV antibody
and HCV RNA (n=300).
|
| HCV RNA |
|
|---|
|
|
|
|
|---|
| HCV antibody | Positive | Negative | Total |
|---|
| Positive | 114 | 32 | 146 |
| Negative | 41 | 113 | 154 |
| Total | 155 | 145 | 300 |
Characteristics of the HCV antibody
negative but HCV RNA positive participants
Among the 41 study participants positive for HCV RNA
but negative for HCV antibody, there were 23 male and 18 female
subjects, with an average age of 39.51±16.32 years, and the viral
load differed, although not significantly, among age groups
(Table VI). The prevalence of HCV
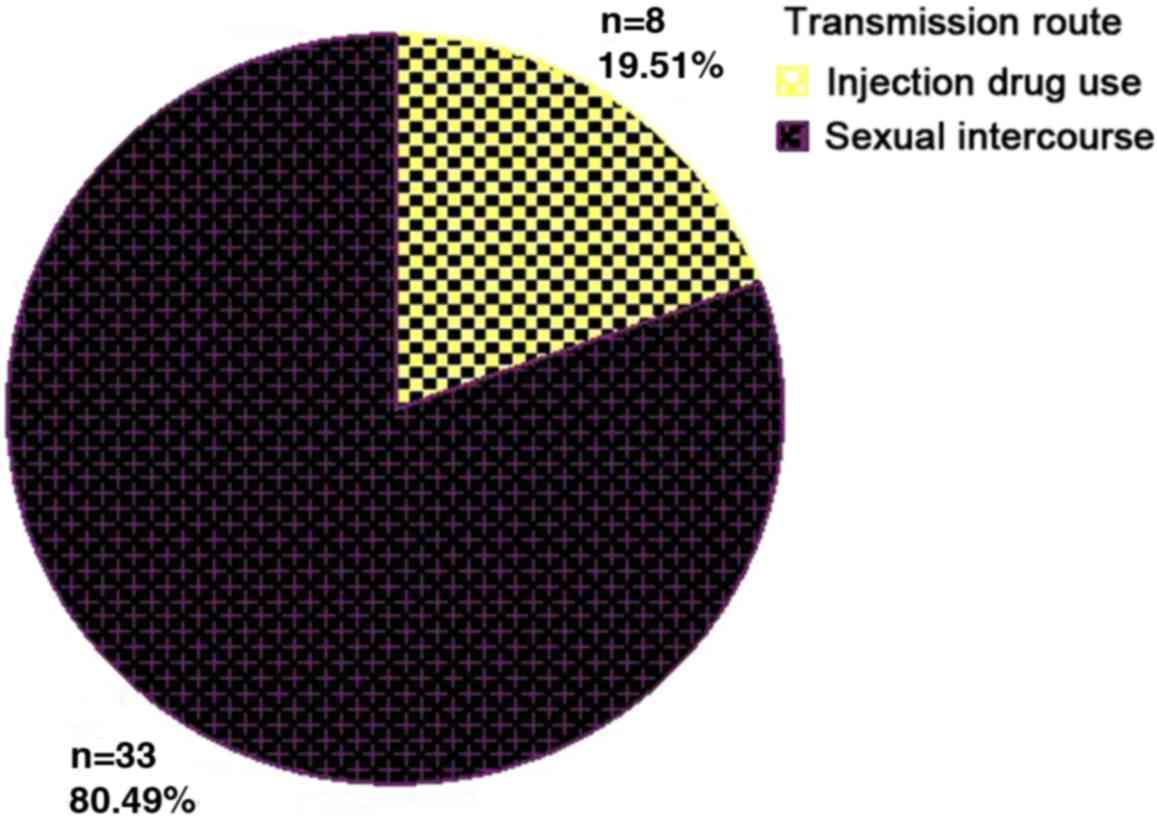
RNA differed according to transmission route; most patients
contracted HCV sexually and the second most prevalent route was
drug-related transmission (Fig. 3);
however, no significant differences in HCV RNA load were observed
between the different transmission routes (Table VII).
 | Table VI.Comparison of HCV RNA load between
three age groups (n=41). |
Table VI.
Comparison of HCV RNA load between
three age groups (n=41).
| Age (years) | Cases | HCV RNA load
(log) | F | P-value |
|---|
| <30 | 15 | 5.84±1.29 |
|
|
| 30–45 | 12 | 6.40±0.76 | 0.83 | 0.44 |
| >45 | 14 | 5.84±1.52 |
|
|
 | Table VII.Comparison of HCV RNA load in
different transmission routes (n=41). |
Table VII.
Comparison of HCV RNA load in
different transmission routes (n=41).
| Transmission | Cases | HCV RNA load
(log) | t | P-value |
|---|
| Sex | 33 | 5.94±1.24 | 0.31 | 0.74 |
| Drugs | 8 | 6.39±0.94 |
|
|
Discussion
This study was aimed at determining the HCV
infection prevalence and HCV genotype in HIV infected patients as
well as obtaining demographic and epidemiological characteristics
of HIV/HCV coinfection patients in the Guangxi province of
China.
With respect to demographic characteristics, the
majority of the HIV/HCV coinfection participants were males, and
aged 20–40 years. The majority of the HIV/HCV coinfection
participants had no fixed occupation, were unmarried or single, and
61.6% were drug users. The results showed that sexually active,
young, single individuals with no fixed occupation were at the
highest risk of HIV/HCV coinfection.
Hepatitis C virus (HCV) is an RNA virus which has
been known to cause acute and chronic necroinflammatory disease of
the liver (23,24). HCV is the leading cause of end-stage
liver disease and hepatocellular carcinoma (25). HIV is known to have a negative impact
on the natural disease outcome and immune response of HCV
infection, whereas the reverse remains unclear (26). HIV infection is propagated primarily
via blood transfusion (before 1985), intravenous drug use,
professional exposure in medical personnel, sexual transmission and
mother-to-child transmission (1,2).
Furthermore, HCV contamination is caused by intravenous drug use
and blood transfusion (before 1991), long term hemodialysis, organ
transplantation and receipt of tattoos from unsanitary facilities
(27). According to above
statements, HIV and HCV share numerous transmission routes, and
thus coinfection with HCV and HIV is quite common. In the present
study, drug addiction, particularly intravenous drug use, was the
primary transmission route in HIV/HCV coinfection subjects, which
indicated that it is necessary to screen for HCV in HIV positive
intravenous drug users. A number of studies have suggested that the
presence of HIV infection accelerates the course of HCV-related
liver disease in HCV/HIV coinfected patients (26,28,29). The
frequency of HCV transmission to sexual partners is significantly
higher when HIV virus is also transmitted (30). HIV is known to impair the T-helper
type 1 immune response, which in turn changes the response of
immune cells to HCV (31). This
facilitates increased HCV replication and consequently easier
infection. Epidemiologic studies have identified HIV as an
independent factor in HCV transmission and acquisition since the
earliest days of the HIV epidemic (32). HIV-infected persons are less likely
to spontaneously clear HCV, and their HCV RNA set point tends to be
higher, making them more infectious to their partners compared with
HCV-monoinfected individuals (33).
A previous study showed that coinfected men were more likely than
HIV-uninfected men to produce HCV RNA in their semen (34). HIV-infected individuals may have
compromised gastrointestinal mucosal barriers and be more likely to
have chronic inflammation, facilitating HCV transmission (35). The present results indicate that
intravenous drug use, sexual transmission and blood transfusion are
the primary transmission routes of HIV/HCV coinfection in patients
from Guangxi.
ELISA is the conventional method used to detect HCV
antibody; however, the shortcoming of ELISA is the long
window-period prior to antibodies becoming detectable and screening
hepatitis C only by ELISA in HIV-infected persons is likely to miss
the identification of some cases. Therefore, HCV antibody can not
be used as an early diagnostic marker of HCV infection. However,
serum HCV RNA detection could be a reliable indicator due to its
shorter window-period and higher sensitivity. It has been reported
that ~11% of HIV/HCV coinfection patients are HCV antibody negative
(36). Liu et al showed that
in HIV/HCV coinfection patients in Beijing, the nucleic acid
positive rate was 19.5% in HCV antibody-negative group (37). In the present study, it was found
that in HIV/HCV coinfection persons in Guangxi, the nucleic acid
positive rate was 26.62% in HCV antibody-negative group, which was
higher than reported previously.
Although the coinfection of HIV with HCV has been
recognized worldwide, few studies have been conducted to
investigate HIV/HCV coinfection prevalence in Guangxi and the
association between prevalence and transmission route, particularly
in patients infected via sexual transmission. The present study was
conducted on different populations coinfected with HIV/HCV,
providing important reference data for prevention and treatment.
The data show that HIV/HCV coinfection prevalence was ~48.67%,
which is higher reports from outside of China (30,38–41), and
higher than the prevalence of HCV among domestic general
populations (42), which indicates
that HIV/HCV coinfection is a serious health concern. In addition,
the results indicate that besides HCV, AIDS infected persons are
also liable to coinfect with other hepatitis virus, tuberculosis
and syphilis, which all indicate that it is important to improve
approaches for the management of HIV coinfection.
In conclusion, the present data indicate that
HIV/HCV coinfection is a serious health concern in the Guangxi
province of China. Therefore, it may be necessary to manage and
control HIV/HCV progression to prevent their potential outbreak or
spread among the general population, on the basis of infection
status, transmission routes and risk behaviors.
Acknowledgements
This study was financially supported by the fourth
round of the Global Fund/Sino-British AIDS Project (grant no.
2008OR52).
References
|
1
|
Semba RD, Kumwenda N, Hoover DR, Taha TE,
Quinn TC, Mtimavalye L, Biggar RJ, Broadhead R, Miotti PG, Sokoll
LJ, et al: Human immunodeficiency virus load in breast milk,
mastitis, and mother-to-child transmission of human
immunodeficiency virus type 1. J Infect Dis. 180:93–98. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Attia S, Egger M, Müller M, Zwahlen M and
Low N: Sexual transmission of HIV according to viral load and
antiretroviral therapy: Systematic review and meta-analysis. AIDS.
23:1397–1404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Macías J, Palacios RB, Claro E, Vargas J,
Vergara S, Mira JA, Merchante N, Corzo JE and Pineda JA: High
prevalence of hepatitis C virus infection among noninjecting drug
users: Association with sharing the inhalation implements of crack.
Liver Int. 28:781–786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sulkowski MS, Mast EE, Seeff LB and Thomas
DL: Hepatitis C virus infection as an opportunistic disease in
persons infected with human immunodeficiency virus. Clin Infect
Dis. 30:(Suppl 1). S77–S84. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Soriano V, Vispo E, Labarga P, Medrano J
and Barreiro P: Viral hepatitis and HIV co-infection. Antiviral
Res. 85:303–315. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
de Lédinghen V, Barreiro P, Foucher J,
Labarga P, Castéra L, Vispo ME, Bernard PH, Martin-Carbonero L,
Neau D, García-Gascó P, et al: Liver fibrosis on account of chronic
hepatitis C is more severe in HIV-positive than HIV-negative
patients despite antiretroviral therapy. J Viral Hepat. 15:427–433.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cockerham L, Scherzer R, Zolopa A, Rimland
D, Lewis CE, Bacchetti P, Grunfeld C, Shlipak M and Tien PC:
Association of HIV infection, demographic and cardiovascular risk
factors with all-cause mortality in the recent HAART era. J Acquir
Immune Defic Syndr. 53:102–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hernandez MD and Sherman KE: HIV/hepatitis
C coinfection natural history and disease progression. Curr Opin
HIV AIDS. 6:478–482. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Bao ZY, Sui HS, Liu SY, Zhuang DM,
Liu YJ, Miao WQ, Li Z and Li JY: Investigation on HCV co-infection
in HIV-infected people in some areas of China. Zhongguo Ai Zi Bing
Xing Bing. 14:9–11. 2008.(In Chinese).
|
|
11
|
Yin N, Mei S and Zhang LQ: Outbreak of HIV
and HCV co-infection among intravenous drug users and illegal blood
donors in China. Zhongguo Kang Gan Ran Hua Liao Za Zhi. 2:67–69.
2002.(In Chinese).
|
|
12
|
Ma JX, Wang JR, Shen YZ, Zhang RF, Liu XN,
Jiang XY, Sun HQ and Lu HZ: Clinical epidemiology studies on
HIV-1/AIDS subjects co-infected with HBV and/or HCV in Shanghai.
Wei Sheng Wu Yu Gan Ran. 1:207–210. 2006.(In Chinese).
|
|
13
|
Xia YH, McLaughlin MM, Chen W, Ling L and
Tucker JD: HIV and hepatitis C virus testing delays at methadone
clinics in Guangdong Province, China. PLoS One. 8:e667872013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia YH, Chen W, Tucker JD, Wang C and Ling
L: HIV and hepatitis C virus test uptake at methadone clinics in
Southern China: Opportunities for expanding detection of bloodborne
infections. BMC Public Health. 13:8992013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Caliendo AM, Gilbert DN, Ginocchio CC,
Hanson KE, May L, Quinn TC, Tenover FC, Alland D, Blaschke AJ,
Bonomo RA, et al: Infectious Diseases Society of America (IDSA):
Better tests, better care: Improved diagnostics for infectious
diseases. Clin Infect Dis. 57:(Suppl 3). S139–S170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Su Q, Liang H, Cen P, Bi Z and Zhou P: HIV
type 1 subtypes based on the pol gene and drug resistance mutations
among antiretroviral-naive patients from Guangxi, Southern China.
AIDS Res Hum Retroviruses. 28:725–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yeh FS, Yu MC, Mo CC, Luo S, Tong MJ and
Henderson BE: Hepatitis B virus, aflatoxins, and hepatocellular
carcinoma in southern Guangxi, China. Cancer Res. 49:2506–2509.
1989.PubMed/NCBI
|
|
18
|
Tian D, Li L, Liu Y, Li H, Xu X and Li JD:
Different HCV genotype distributions of HIV-infected individuals in
Henan and Guangxi, China. PLoS One. 7:e503432012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sanger F, Nicklen S and Coulson AR: DNA
sequencing with chain-terminating inhibitors. Proc Natl Acad Sci
USA. 74:5463–5467. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chinchai T, Labout J, Noppornpanth S,
Theamboonlers A, Haagmans BL, Osterhaus AD and Poovorawan Y:
Comparative study of different methods to genotype hepatitis C
virus type 6 variants. J Virol Methods. 109:195–201. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moreira S, Garcia RF, Gutberlet A, Bertol
BC, Ferreira LE, Mde S Pinho and de França PH: A straightforward
genotyping of the relevant IL28B SNPs for the prediction of
hepatitis C treatment outcome. J Virol Methods. 184:93–97. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fattovich G, Stroffolini T, Zagni I and
Donato F: Hepatocellular carcinoma in cirrhosis: incidence and risk
factors. Gastroenterology. 27:(Suppl 1). S35–S50. 2004. View Article : Google Scholar
|
|
24
|
Moradpour D and Blum HE: Pathogenesis of
hepatocellular carcinoma. Eur J Gastroenterol Hepatol. 17:477–483.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Williams R: Global challenges in liver
disease. Hepatology. 44:521–526. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taye S and Lakew M: Impact of hepatitis C
virus co-infection on HIV patients before and after highly active
antiretroviral therapy: an immunological and clinical chemistry
observation, Addis Ababa, Ethiopia. BMC Immunol. 17:14–23.
2013.
|
|
27
|
Bouare N, Gothot A, Delwaide J, Bontems S,
Vaira D, Seidel L, Gerard P and Gerard C: Epidemiological profiles
of human immunodeficiency virus and hepatitis C virus infections in
Malian women: Risk factors and relevance of disparities. World J
Hepatol. 5:196–205. 2013.PubMed/NCBI
|
|
28
|
Merchante N, Girón-González JA,
González-Serrano M, Torre-Cisneros J, García-García JA, Arizcorreta
A, Ruiz-Morales J, Cano-Lliteras P, Lozano F, Martínez-Sierra C, et
al: Grupo Andaluz para el Estudio de las Enfermedades Infecciosas:
Survival and prognostic factors of HIV-infected patients with
HCV-related end-stage liver disease. AIDS. 20:49–57. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
García-Samaniego J, Rodríguez M, Berenguer
J, Rodríguez-Rosado R, Carbó J, Asensi V and Soriano V:
Hepatocellular carcinoma in HIV-infected patients with chronic
hepatitis C. Am J Gastroenterol. 96:179–183. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hadush H, Gebre-Selassie S and Mihret A:
Hepatitis C virus and human immunodeficiency virus coinfection
among attendants of voluntary counseling and testing centre and HIV
follow up clinics in Mekelle Hospital. Pan Afr Med J. 14:1072013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chang KM, Thimme R, Melpolder JJ, Oldach
D, Pemberton J, Moorhead-Loudis J, McHutchison JG, Alter HJ and
Chisari FV: Differential CD4(+) and CD8(+) T-cell responsiveness in
hepatitis C virus infection. Hepatology. 33:267–276. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Riestra S, Fernández E, Rodríguez M and
Rodrigo L: Hepatitis C virus infection in heterosexual partners of
HCV carriers. J Hepatol. 22:509–510. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sherman KE, Shire NJ, Rouster SD, Peters
MG, Koziel M James, Chung RT and Horn PS: Viral kinetics in
hepatitis C or hepatitis C/human immunodeficiency virus-infected
patients. Gastroenterology. 128:313–327. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Briat A, Dulioust E, Galimand J, Fontaine
H, Chaix ML, Letur-Könirsch H, Pol S, Jouannet P, Rouzioux C and
Leruez-Ville M: Hepatitis C virus in the semen of men coinfected
with HIV-1: Prevalence and origin. AIDS. 19:1827–1835. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Taylor LE, Swan T and Mayer KH: HIV
coinfection with hepatitis C virus: Evolving epidemiology and
treatment paradigms. Clin Infect Dis. 55:(Suppl 1). S33–S42. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Podlekareva D, Bannister W, Mocroft A,
Abrosimova L, Karpov I, Lundgren JD and Kirk O: EuroSIDA Study
Group: The EuroSIDA study: Regional differences in the HIV-1
epidemic and treatment response to antiretroviral therapy among
HIV-infected patients across Europe-a review of published results.
Cent Eur J Public Health. 16:99–105. 2008.PubMed/NCBI
|
|
37
|
Liu Z, Jiang Y, Wu Hao, Xiao Y, Zhang YH,
Pan PL and Xiang Z: Mutual effect of HIV/HCV Co-infection on
laboratory diagnosis. Chin J Lab Med. 28:691–693. 2005.
|
|
38
|
Mboto CI, Fielder M, Davies-Russell A and
Jewell AP: Prevalence of HIV-1, HIV-2, hepatitis C and co-infection
in the gambia. West Afr J Med. 28:16–19. 2009.PubMed/NCBI
|
|
39
|
Onakewhor JU and Okonofua FE: The
prevalence of dual human immunodeficiency virus/hepatitis C virus
(HIV/HCV) infection in asymptomatic pregnant women in Benin City,
Nigeria. Afr J Reprod Health. 13:97–108. 2009.PubMed/NCBI
|
|
40
|
Amin J, Kaye M, Skidmore S, Pillay D,
Cooper DA and Dore GJ: HIV and hepatitis C coinfection within the
CAESAR study. HIV Med. 5:174–179. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Walusansa V and Kagimu M: Screening for
hepatitis C among HIV positive patients at Mulago hospital in
Uganda. Afr Health Sci. 9:143–146. 2009.PubMed/NCBI
|
|
42
|
Chinese Society of Hepatology, . Chinese
Society of Infectious Diseases and Parasitic Diseases. Principles
for prevention and treatment of hepatitis C. Chin J Int Med (Chin).
43:551–555. 2004.
|