Introduction
The prevalence of renal cell carcinoma (RCC), a
renal neoplasm accounting for ~3% of adult malignancies, has been
increasing in the recent years (1,2). Less
than 10% of patients with RCC are reported to have ≥5-year survival
rates due to the aggressive nature of the neoplasm, lack of early
detection and poor responses to clinical treatments (2). Among the histological subtypes of RCC,
clear-cell RCC (ccRCC) is the most common type, accounting for
75–80% of all RCC cases (3). In the
past decades, studies on ccRCC have mainly focused on the genome
mutations, expression of protein-coding genes as well as epigenetic
changes. However, increasing evidence has indicated that
dysregulation of certain microRNAs (miRNAs) is also closely
associated with the pathogenesis of ccRCC (4,5).
miRNAs are a class of non-coding RNAs that exert
post-transcriptional control of gene expression by specifically
binding to the 3′-untranslation region (3′-UTR) of their target
genes (6). miRNAs have been
acknowledged to have important roles in a wide range of biological
functions, including cellular proliferation, differentiation,
development, apoptosis and cellular metabolism (7). Furthermore, miRNAs have been reported
to have crucial roles in the pathogenesis and development of
cancer. Accumulating evidence revealed aberrant expression of
numerous miRNAs in a number of human malignancies (8,9). For
instance, miR-155, miR-210, miR-21, miR-17-5p, miR-122 and miR-20
were found to be upregulated in ccRCC, while miR-9, miR-200bc,
miR-141, miR-455-5p, miR-363 and miR-429 were reported to be
downregulated (10). In addition,
apoptosis was induced in cancer cells subjected to overexpression
of tumor suppressor miRNAs or silencing of oncogenic miRNAs
(11). Taken together, it is
reasonable to speculate that miRNAs are novel targets for cancer
therapy.
miR-155, localized within a genomic region known as
B cell integration cluster, has important roles in immune responses
and cancer as well as aberrant proliferation (12–15).
Extensive studies revealed that miR-155 has crucial roles in the
formation of hematopoietic cells, inflammation and immune reactions
as well as in the pathogenesis and development of cancer (16–18).
miR-155 was found to be upregulated in ccRCC and may play have an
oncogenic effect in RCC (16);
however, the exact molecular mechanisms underlying its function in
the pathogenesis of ccRCC has remained to be fully elucidated.
Forkhead box O3a (FOXO3a), a target gene of miR-155,
is a family member of forkhead transcriptional factor. It was
reported to be distributed in the nucleus, and to have crucial
roles in cellular apoptosis through upregulating B-cell lymphoma
2-interacting mediator of cell death and Fas (19). Furthermore, miR-155 was shown to
enhance the expression of the gene growth arrest and
DNA-damage-inducible alpha (GADD45A) (20). As FOXO3a is known to regulate GADD45A
expression (21), it is reasonable
to speculate that miR-155 may be involved in the pathogenesis and
progression of ccRCC through targeting FOXO3a. The present study
demonstrated that miR-155 is a determinant of cell proliferation
and invasion by targeting FOXO3a in ccRCC.
Materials and methods
Specimens
A total of 20 ccRCC tissue specimens and matched
normal kidney tissues were obtained from patients admitted to
Binzhou Medical University Hospital (Binzhou, China) from January
2013 to January 2014 immediately after radical nephrectomy. None of
these patients received anti-tumor treatment prior to surgery and
the diagnosis as ccRCC was histologically confirmed. Tissue samples
were immediately frozen in liquid nitrogen after resection and
stored at −80°C prior to RNA extraction. Written informed consent
was obtained from each patient. The study protocols were approved
by the Institutional Review Board of Binzhou Medical University
Hospital (Binzhou, China).
Cell culture
The ccRCC cell lines ACHN, 786-0 and CAKI-1 were
purchased from the Institute of Biochemistry and Cell Biology
(Shanghai, China). The HK-2 human kidney tubular epithelial cell
line was purchased from the American Type Culture Collection
(Manassas, VA, USA). ACHN and CAKI-1 cells were cultured in Eagle's
Minimum Essential Medium supplemented with 10% fetal bovine serum
(FBS) and 786-0 cells were cultured in RPMI-1640 medium
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The HK-2 cell line was cultured in KSF medium
(Gibco; Thermo Fisher Scientific, Inc.) containing epidermal growth
factor (PeproTech, Inc., Rocky Hill, NJ, USA). All cells were
cultured at 37°C in a humidified incubator with 5%
CO2.
Cell transfection with miR-155
inhibitor
miR-155 inhibitor, single-stranded chemically
modified oligonucleotides, was purchased from GenePharma Biological
Technology (Shanghai, China). The sequence of the oligonucleotides
was 5′-CCCCTATCACGATTAGCATTAA-3′. Cells were transfected with
miR-155 inhibitor using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Following 24, 48 or 72 h of transfection, cells were
harvested and used for further study. Scrambled
sequence-transfected cells served as a negative control. The
scrambled sequence (GenePharma Biological Technology) was
5′-CATTAATGTCGGACAAC-3′.
RNA extraction and
reverse-transcription quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Complementary DNA
synthesis was performed using 2 µg RNA. The RT reaction mixture,
which was provided in the PrimeScript 1st Strand cDNA Synthesis kit
(cat. no. D6110A; Takara Biotechnology Co., Ltd., Dalian, China)
consisted of 1 ml oligo dT primer, 1 ml dNTP mixture, 2 µg total
RNA, RNase-free dH2O. The mixture was incubated at 65°C
for 5 min, and 5X PrimeScript Buffer, RNase inhibitor (0.5 µl),
PrimeScript RTase (1 µl) and RNase free dH2O (4.5 µl)
was subsequently added to the upper reaction mixture. The mixture
was further incubated at 42°C for 30 min, followed by 95°C for 5
min. The real-time PCR reaction mixture (SYBR Premix Ex Taq; cat.
no. RR041A; Takara Biotechnology Co., Ltd.) was prepared with
Takara Ex Taq HS DNA polymerase, dNTP mixture, Mg+, RNase H and
SYBR Green I. PCR reactions were performed on an ABI 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.)
under the following conditions: 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 1 min. All values were
normalized to an endogenous U6 control. The sequences of primers
used (Sangon Biotech Co., Ltd., Shanghai, China) were as follows:
miR-155; forward, 5′-GCGGTTAATGCTAATCGTGAT-3′, and reverse,
5′-GTGCAGGGTCCGAGGT-3′; and U6, forward 5′-CTCGCTTCGGCAGCAC-3′ and
reverse 5′-AACGCTTCACGAATTTGCGT-3′. The quantification of the PCR
results was performed using the 2−ΔΔCq method as
previously described (22).
Cell proliferation assay
ACHN cells (3×104 cells/well) were
cultured in 96-well plates overnight and then transfected with
miR-155 inhibitor. At 24, 48 and 72 h after transfection, cell
growth was examined using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT)
assay according to the manufacturer's instructions (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany). The absorbance of samples was
recorded at 490 nm using a microplate spectrophotometer (Model 680;
Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Colony formation assay
ACHN cells were transfected with miR-155 inhibitor
or control for 48 h and seeded into 6-well plates at a density of
1,000 cells/well. After incubation at 37°C for 10 days, the cells
were fixed and stained with crystal violet. The number of colonies
containing >50 cells was counted and images were captured using
a light microscope (Nikon Corp., Tokyo, Japan) for three
independent replicates.
Flow cytometric cell cycle
analysis
ACHN cells were cultured in 6-well plates overnight
and then transfected with miR-155 inhibitor or control as described
above. After 48 h of incubation, the cell cycle distribution was
determined using flow cytometry. In brief, the cells were
collected, washed with ice-cold PBS twice and fixed with 70% cold
ethanol at 4°C overnight. After incubation in 100 µg/ml RNase A
(Sigma-Aldrich; Merck Millipore) at 37°C for 30 min, the cells were
stained with 50 µg/ml propidium iodide (PI; Sigma-Aldrich; Merck
Millipore). Flow cytometric analysis was performed using a
FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ,
USA). All experiments were performed at least in triplicate.
Apoptosis assay
ACHN cells were seeded in 6-well plates overnight
and then transfected with miR-155 inhibitor or control. After 48 h
of incubation, the cells were harvested and washed twice with cold
PBS. A total of 1.0×105 cells were re-suspended in 100
µl binding buffer and mixed with 5 µl fluorescein
isothiocyanate-labeled Annexin V and 5 µl PI at room temperature
for 15 min in the dark. After addition of 400 µl binding buffer,
apoptosis was analyzed by flow cytometry.
Wound healing assay
ACHN cells were seeded in 6-well plates overnight
and then transfected with miR-155 inhibitor or negative control.
After 48 h of incubation, the cells were scratched with a 200-µl
pipette tip and then washed three times with PBS to clear cell
debris. Fresh medium supplemented with 10% FBS was added and the
cells were allowed to close the wound for 48 h under normal
incubation conditions. Images were captured at the position of the
generated wound using a computer-assisted microscope (Nikon
Corp.).
Cell invasion assay
Cellular migration assays were performed using a
Boyden chamber containing 24-well Transwell plates (Corning Inc.,
Corning, NY, USA) with 8-mm pore membranes. ACHN cells were
transfected with miR-155 inhibitor or negative control. After 48 h
of incubation, ~5×104 cells in 200 µl culture medium
supplemented with 5% FBS were seeded into the upper chamber. The
lower chamber was filled with complete medium (with 10% FBS) as a
chemoattractant. After 12 h of incubation, the cells on the lower
side of the membranes were fixed and stained with crystal violet.
Images of the lower surfaces of the membranes were captured at ×100
magnification. Five fields of view were randomly selected for the
determination of cell migration using NIS-Elements 2.1 software
(Nikon Corp.).
Immunohistochemistry
Tissue samples were fixed with formalin and embedded
using paraffin. Then 4-µm sections were cut and stained using the
avidin biotin complex method. The slides were pre-treated by
microwaving in 10 mmol/l citrate buffer (pH 6.0) for antigen
retrieval. Endogenous peroxidase activity was blocked by incubating
with 3% hydrogen peroxide for 10 min. After blocking non-specific
protein binding, tissue sections were incubated with primary
antibody to FOXO3a (1:1,000; cat. no. 12829; Cell Signaling
Technology, Inc., Danvers, MA, USA) at 4°C overnight. After rinsing
for 5 min with PBS three times, sections were treated with
horseradish peroxidase (HRP)-conjugated rabbit anti-mouse
immunoglobulin G (1:5,000; cat. no. 7074; Cell Signaling
Technology, Inc.) for 30 min at room temperature, followed by
incubation with streptavidin biotin complex for 15 min. Subsequent
to incubation of the sections with diaminobenzidine for 5 min, they
were lightly counterstained with hematoxylin. A four-grade scoring
system was used to evaluate the degree of immunostaining under a
light microscope (Nikon Corp.): 0, <5%; 1, 5–25%; 2, 25–50%; and
3, >50% of cells with immunostaining.
Western blot analysis
Cells were lysed with radioimmunoprecipitation
buffer and the quantity of the protein was determined using the
bicinchoninic acid method (Bicinchoninic Acid Kit for Protein
Determination; cat. no. BCA1-1KT; Sigma-Aldrich; Merck Millipore).
Protein (80 µg per lane) was subjected to 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis. Subsequently, the
samples were transferred to a Hybond™ polyvinylidene difluoride
membrane (Roche Diagnostics, Indianopolis, IN, USA), which was
blocked with 5% non-fat milk and incubated with mouse anti-human
FOXO3a monoclonal antibody (1:1,000; cat. no. 12829; Cell Signaling
Technology, Inc.) followed by HRP-conjugated secondary antibody
(1:5,000; cat. no. ab191866;, Cambridge, MA, USA). Protein
expression was detected using an enhanced chemiluminescence kit
(Pierce™ ECL Western Blotting Substrate; Thermo Fisher Scientific,
Inc.). GAPDH (1:5,000; cat. no. 5174; Cell Signalling Technology,
Inc.) served as a loading control. The images were captured on
X-ray film and quantified using Image J 1.41 software (National
Institutes of Health, Bethesda, MD, USA).
Bioinformatics analysis
Potential targets of miR-155 in ccRCC were evaluated
using Targetscan software (www.targetscan.org). Via predicted pairing of target
region and miR-155, this revealed that FOXO3a was one of the most
likely targets.
Statistical analysis
SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA)
was used to perform statistical analysis. Each experiment was
performed at least in triplicate. All values are expressed as the
mean ± standard deviation. Student's t test or analysis of
variance was used for inter-group comparison. Pearson correlation
analysis was used to evaluate the correlation between miR-155
expression and FOXO3a expression. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-155 is upregulated in ccRCC
tissues and cell lines
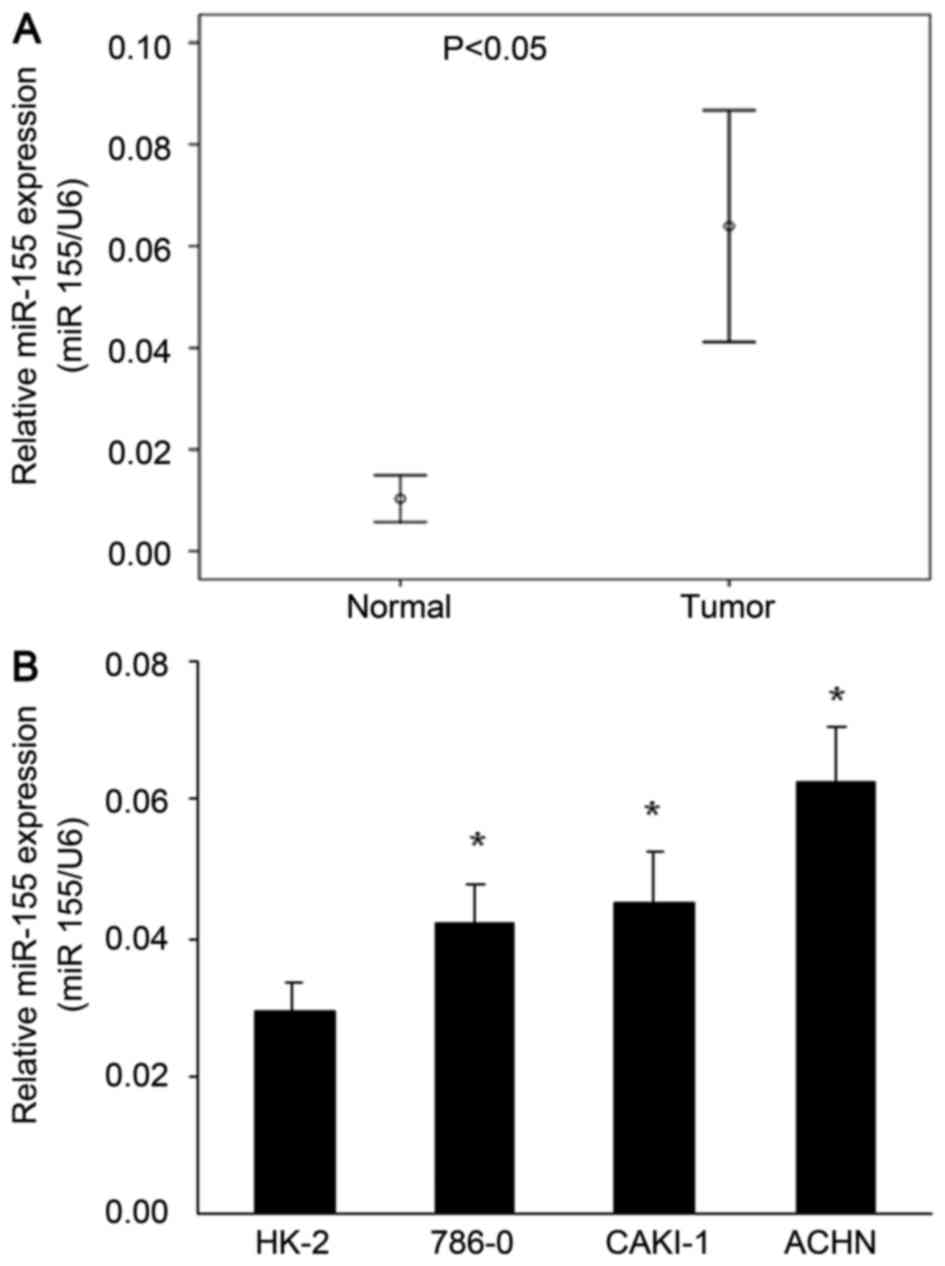
To investigate the potential roles of miR-155 in the
pathogenesis of ccRCC, the miR-155 expression in 20 ccRCC samples
and paired adjacent normal kidney tissues was determined by
RT-qPCR. The results showed that miR-155 expression was
significantly upregulated (5.6-fold) in ccRCC compared with
adjacent normal kidney tissues (P<0.05; Fig. 1A). Moreover, this pattern was also
observed in cell lines in vitro, as miR-155 was
significantly upregulated in the ACHA, CAKI-1 and 786-0 human ccRCC
cell lines compared with the HK-2 human kidney tubular epithelial
cell (P<0.05; Fig. 1B).
Inhibition of miR-155 reduces the
proliferation of ACHN cells
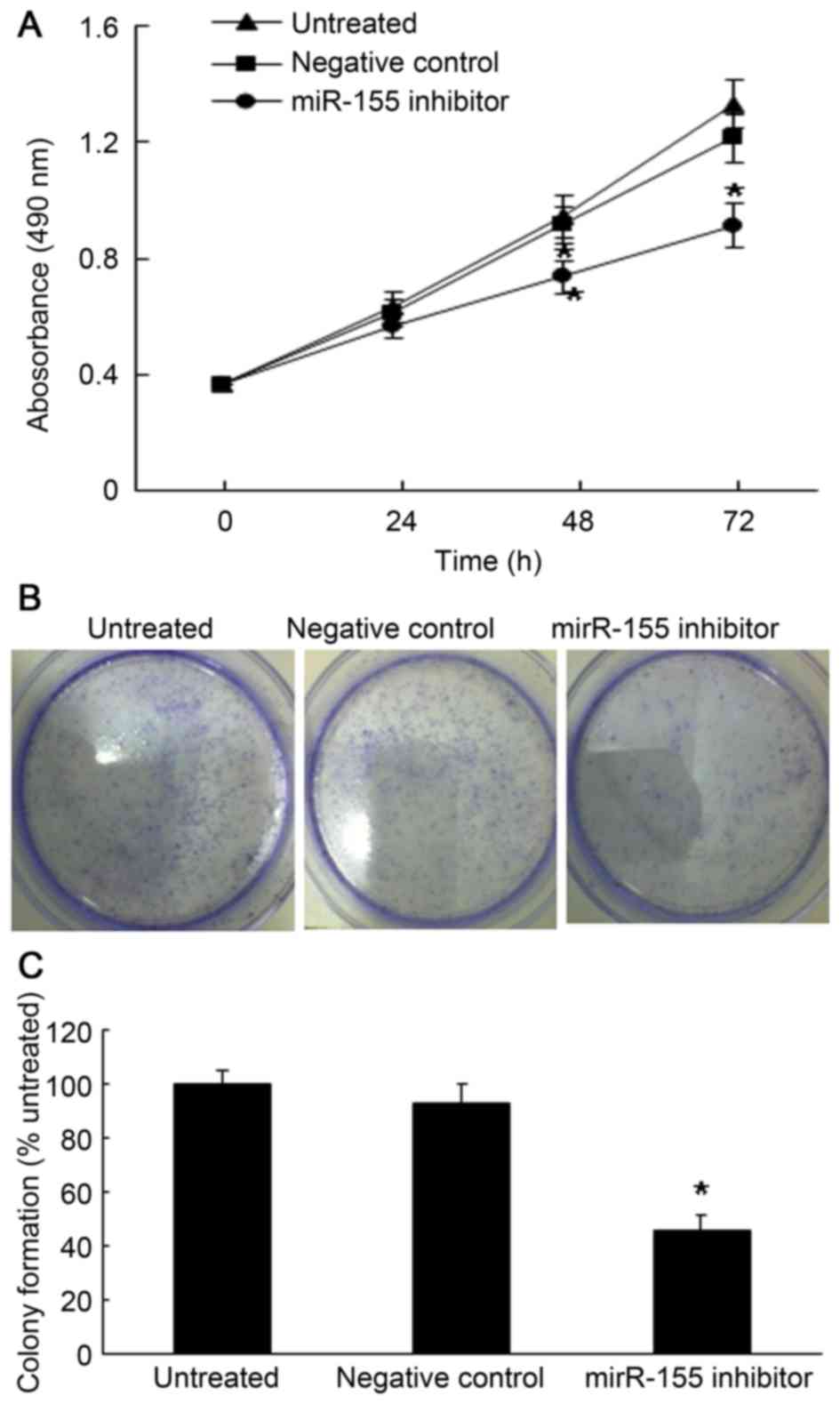
To investigate the role of miR-155 in ccRCC, the
influence of miR-155 inhibition on the proliferation of the ACHN
ccRCC cell line was examined. First, the cellular proliferation
rate was determined by an MTT assay after the cells were
transfected with miR-155 inhibitor or negative control for 24, 48
or 72 h. As shown in Fig. 2A,
miR-155 inhibition induced a significant decrease on the growth
rate of ACHN cells (P<0.05). In line with this, the colony
formation assay revealed that miR-155 inhibition significantly
decreased the colony sphere formation after 10 days of culture
(Fig. 2B).
Inhibition of miR-155 expression
induces apoptosis and cell cycle arrest in ccRCC cells
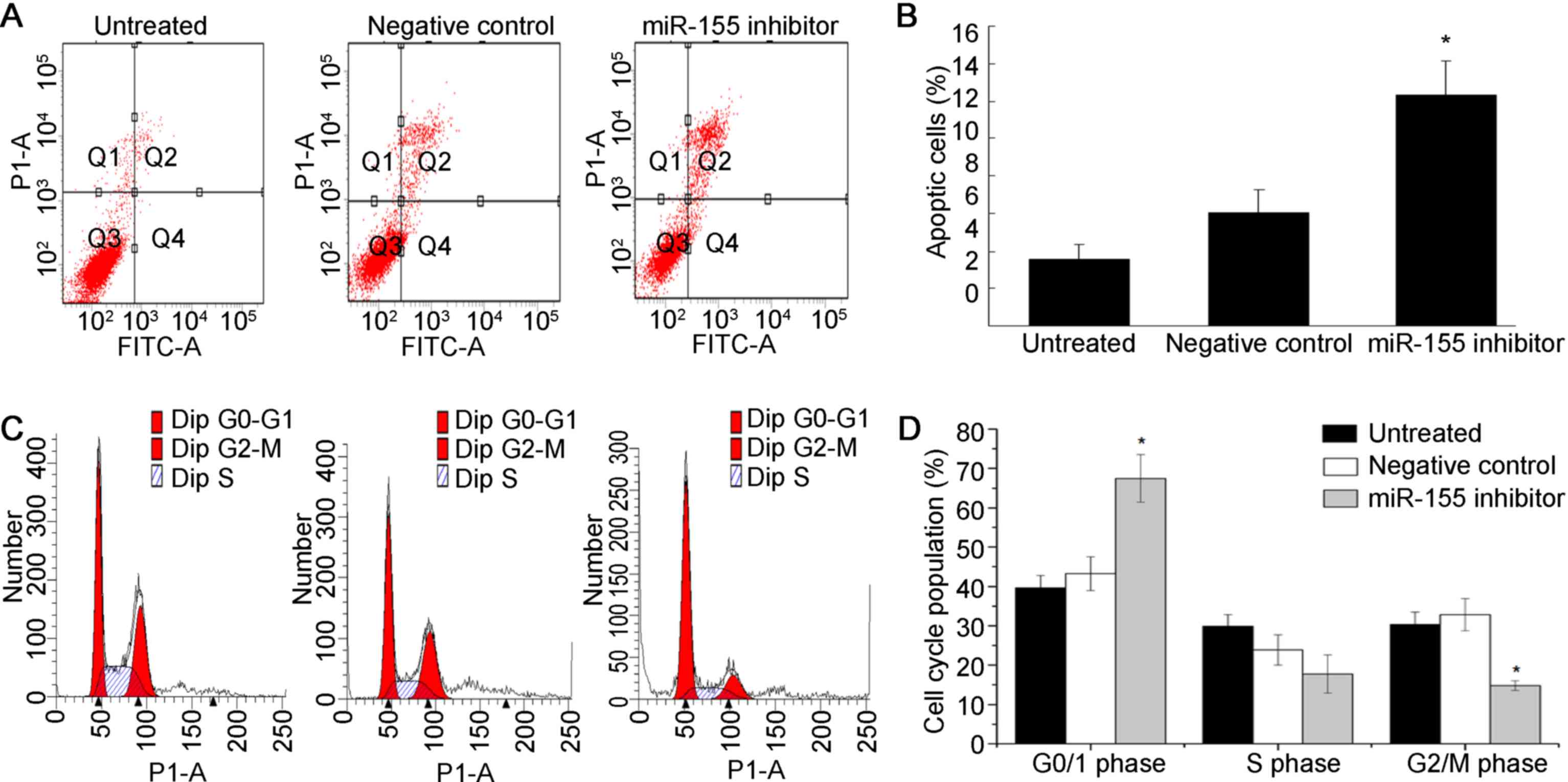
In the present study, flow cytometric analysis was
performed to evaluate the roles of miR-155 in apoptosis of ccRCC
cells. As shown in Fig. 3A and B,
flow cytometric analysis revealed that the rate of apoptosis was
significantly increased in ACHN cells transfected with miR-155
inhibitor (P<0.05). Taken together, inhibition of miR-155 caused
apoptosis in ccRCC cells.
To further analyze the mechanisms by which miR-155
expression affects cell growth, flow cytometric analysis was
performed to examine the cell cycle distribution of ccRCC cells
after transfection with miR-155 inhibitor. As shown in Fig 3C and D, miR-155 inhibition markedly
decrease the percentage of cells in S phase, while the percentage
of cells arrested in G1/G0 phase was obviously increased.
Collectively, these results indicated that inhibition of miR-155
resulted in G1/G0 arrest and suppressed ccRCC cell proliferation
in vitro.
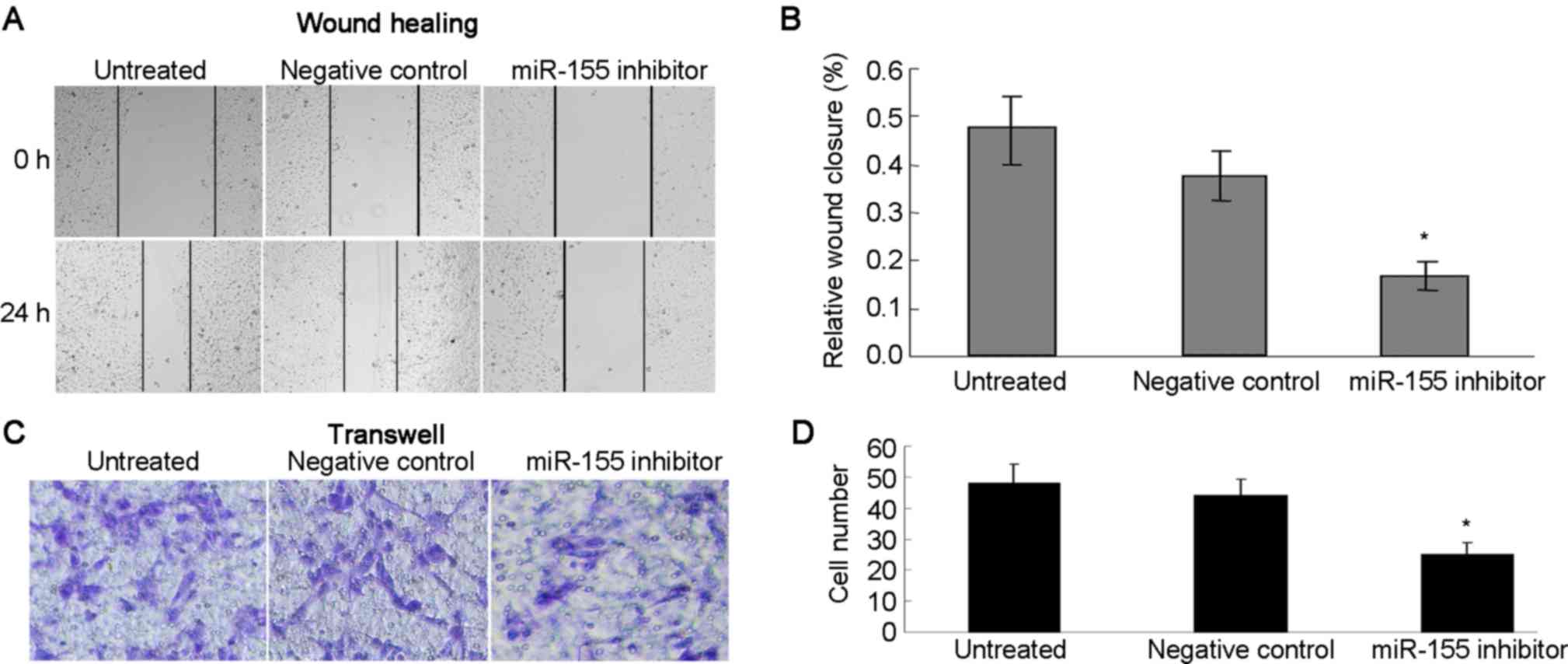
Inhibition of miR-155 reduces migration and invasion
of ccRCC cells. The potential effects of miR-155 on cell migration
and invasion were assessed using wound healing and Transwell
assays. The wound healing assay demonstrated that inhibition of
miR-155 reduced the migratory capacity of the ANCH cells (Fig. 4A and B). The Transwell assay showed
that miR-155 inhibition resulted in a reduction of ANCH cell
invasion compared with that in the negative control and untreated
groups (Fig. 4C and D). Taken
together, it is reasonable to conclude that miR-155 may have an
important role in the migration and invasion of RCC cells.
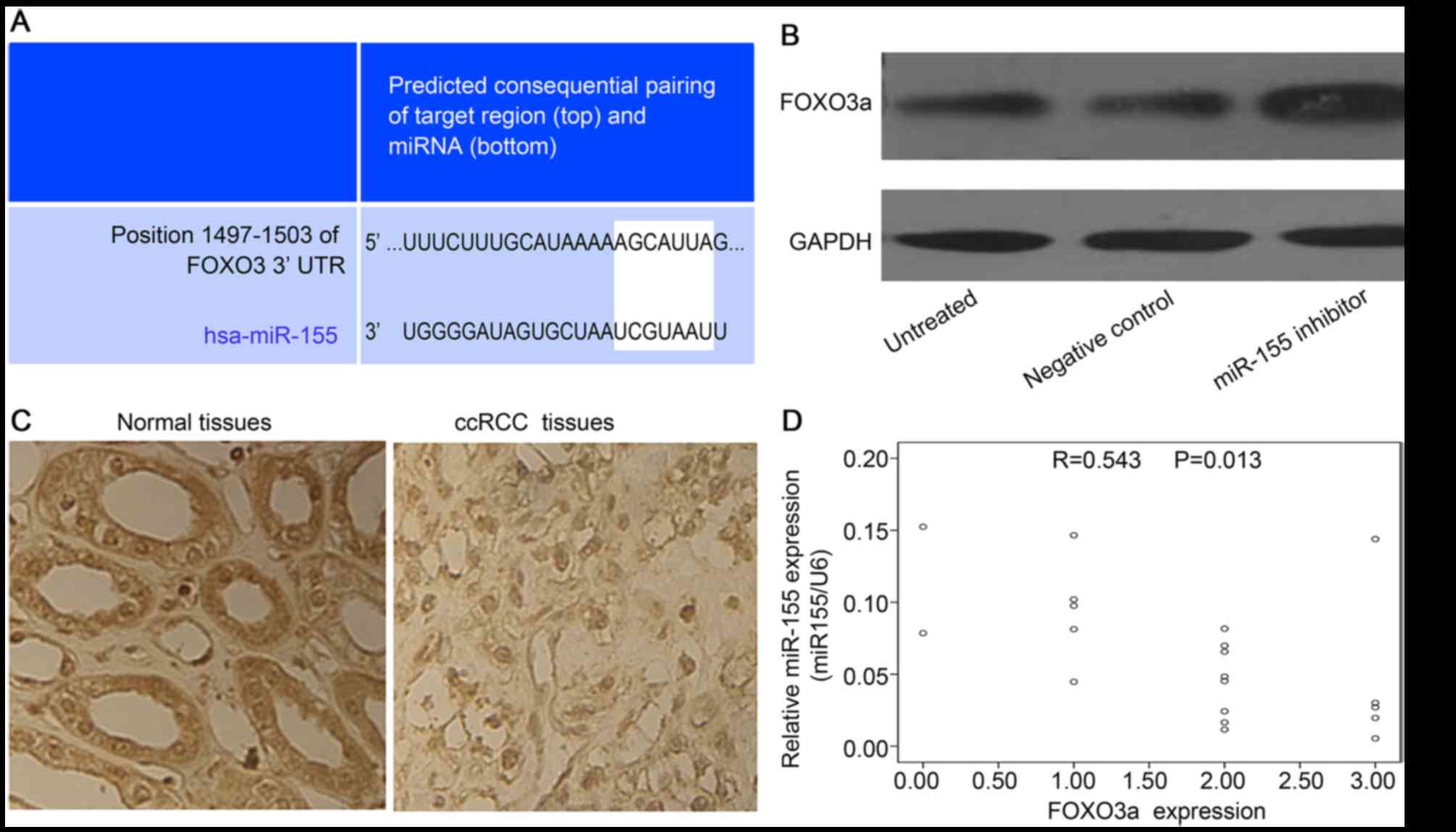
FOXO3a is a target gene of miR-155 in
ccRCC cells
In order to investigate the underlying mechanism by
which miR-155 may influence the progression of ccRCC, the potential
targets of miR-155 were analyzed using Targetscan software. It was
demonstrated via the predicted consequential pairing of target
region and miRNA that FOXO3a was the target of miR-155 (Fig. 5A). In order to further confirm that
miR-155 targeted FOXO3a in ccRCC, the influence of altered miR-155
levels on FOXO3a expression was determined using RT-qPCR and
immunoblot analysis. The results indicated FOXO3a protein showed a
2.0-fold increase after miR-155 inhibitor treatment (Fig. 5B).
To further validate the negative regulation of
miR-155 on FOXO3a in vivo, the expression of FOXO3a protein
was determined using immunohistochemistry in the same ccRCC tissues
in which miR-155 expression was detected. As shown in Fig 5C, FOXO3a was mainly localized in the
cellular nucleus. Strong staining signals were observed in the
adjacent normal kidney cells, while it was extremely low in the
ccRCC cells. Furthermore, the expression of miR-155 was negatively
correlated with the expression of FOXO3a (R=0.534; P=0.013;
Fig. 5D).
Discussion
miRNAs are closely linked with the pathogenesis of
tumors and malignant processes (17,18).
miR-155 has been shown to be overexpressed in a wide range of
malignancies, including carcinomas of breast, lung, pancreas, head
and neck (18). However, the
molecular mechanisms by which miR-155 exerts its oncogenic role in
ccRCC has remained poorly understood. The present study showed that
miR-155 expression was significantly upregulated in ccRCC tissues
compared with that in corresponding non-tumor tissues. In addition,
the levels of miR-155 expression in ccRCC cell lines were
significantly higher than that in a normal renal cell line. All of
these results are consistent with the notion that miR-155 functions
as an oncogenic miRNA in human cancer.
Given that miR-155 is overexpressed in ccRCC and
that it acts as an oncomiR, the present study further investigated
the functions of miR-155 in ccRCC cells in vitro. The
results demonstrated that inhibition of miR-155 significantly
decreased ccRCC cell proliferation, colony formation, and induced
G1 arrest and apoptosis in vitro. Previous studies have
shown that miR-155 enhances malignant tumor phenotypes by promoting
cell proliferation. For instance, Cai et al (23) showed that overexpression of miR-155
promoted cell proliferation, while inhibition of miR-155 expression
induced cell cycle arrest and promoted apoptosis in prostate cancer
cells. Lao et al (24)
demonstrated that inhibition of miR-155 promoted apoptosis of the
cervical cancer cell lines Hela and SiHa and increased the
percentage of cells in G1 phase. The present study expanded the
current knowledge by highlighting the role of miR-155 in
proliferation of ccRCC cells and confirmed the oncogenic role of
miR-155 in ccRCC via targeting FOXO3a.
Metastasis is an important step in the progression
of ccRCC. Localized and metastatic ccRCC considerably differ in
terms of prognosis and therapeutic approach. Indeed, the 5-year
survival rate is <27.1% for metastatic ccRCC, but >70% for
non-metastatic ccRCC (25). Early
detection of metastatic ccRCC is difficult due to a lack of
reliable molecular markers. The present study further evaluated the
role of miR-155 in the metastasis of ccRCC cells. It was revealed
that when miR-155 was downregulated, ccRCC cell invasion and
migration were inhibited as indicated by wound healing and
Transwell assays. These results indicated that miR-155 exerts a
promoting effect in the metastasis of ccRCC and may serve as a
metastatic marker.
An increasing number of studies have confirmed that
miR-155 has crucial roles in the regulation of cancer pathogenesis.
For instance, miR-155 was shown to drive telomere fragility in
human breast cancer by targeting telomeric repeat factor 1
(26). In addition, it contributed
to the proliferation of prostate cancer cells via targeting annexin
7 (23). Furthermore, miR-155 was
shown to regulate the proliferation and cell cycle distribution of
colorectal cancer cells by targeting E2F transcription factor 2
(27). Each miRNA can have multiple
targets, which vary depending on the cell type in which a given
miRNA is expressed. To explore the molecular mechanisms underlying
the oncogenic effect of miR-155 in ccRCC, FOXO3a was identified as
a potential target of miR-155 through a bioinformatics analysis
(28,29). FOXO3a is a well studied
transcriptional factor that contains a forehead DNA binding domain
and has a crucial role in cell growth and apoptosis by
transcriptional regulation of a number of genes associated with
these processes (30–32). Activation of FOXO3a has a tumor
suppressor effect, promoting cell-cycle arrest and apoptosis in RCC
cell lines. Recently, a study revealed that downregulation of
FOXO3a promotes tumor metastasis and is negatively associated with
metastasis-free survival in patients with ccRCC (33). FOXO3a is considered to be a major
tumor suppressor in ccRCC. The present study showed that FOXO3a is
expressed in adjacent normal kidney tissues and is significantly
downregulated in the majority of primary ccRCC tissues.
Furthermore, expression of miR-155 was negatively correlated with
that of FOXO3a in ccRCC tissues. In addition, downregulation of
miR-155 increased FOXO3a expression at the protein level in ACHN
cells. These results indicated that miR-155 promotes the
progression of ccRCC at least in part by targeting FOXO3a. In order
to further comfirm that FOXO3a was directly regulated by miR-155 in
ccRCC, a luciferase reporter assay should be performed in future
studies (34).
In conclusion, miR-155 was shown to be upregulated
in ccRCC and to function as an oncogene in ccRCC by directly
targeting FOXO3a. Targeting miR-155 may provide an effective
therapeutic approach to treat ccRCC.
Acknowledgements
This work was supported by Binzhou Medical
University (Binzhou, China; grant nos. BY2010KYQD01, BY2011KJ018
and BY2011KJ001), Yantai Municipal Scientific Program (grant no.
2014ZH093), and Beinzhou Municipal Scientific Program (grant no.
2014ZC0106).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Curti BD: Renal cell carcinoma. JAMA.
292:97–100. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ge YZ, Wu R, Xin H, Zhu M, Lu TZ, Liu H,
Xu Z, Yu P, Zhao YC, Li MH, et al: A tumor-specific microRNA
signature predicts survival in clear cell renal cell carcinoma. J
Cancer Res Clin Oncol. 141:1291–1299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishihara T, Seki N, Inoguchi S, Yoshino H,
Tatarano S, Yamada Y, Itesako T, Goto Y, Nishikawa R, Nakagawa M
and Enokida H: Expression of the tumor suppressive miRNA-23b/27b
cluster is a good prognostic marker in clear cell renal cell
carcinoma. J Urol. 192:1822–1830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cummins JM and Velculescu VE: Implications
of micro-RNA profiling for cancer diagnosis. Oncogene.
25:6220–6227. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alvarez-Garcia I and Miska E: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Luo M, Shen D, Wang W and Xian J: Aberrant
expression of microRNA-26b and its prognostic potential in human
cervical cancer. Int J Clin Exp Pathol. 8:5542–5548.
2015.PubMed/NCBI
|
|
9
|
Ha TY: MicroRNAs in human diseases: From
cancer to cardiovascular disease. Immune Netw. 11:135–154. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li M, Wang Y, Song Y, Bu R, Yin B, Fei X,
Guo Q and Wu B: MicroRNAs in renal cell carcinoma: A systematic
review of clinical implications (Review). Oncol Rep. 33:1571–1578.
2015.PubMed/NCBI
|
|
11
|
Calvo E, Schmidinger M, Heng DY, Grünwald
V and Escudier B: Improvement in survival end points of patients
with metastatic renal cell carcinoma through sequential targeted
therapy. Cancer Treat Rev. 50:109–117. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
O'Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rodriguez A, Vigorito E, Clare S, Warren
MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska
EA, et al: Requirement of bic/microRNA-155 for normal immune
function. Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thai TH, Calado DP, Casola S, Ansel KM,
Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et
al: Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tili E, Croce CM and Michaille JJ:
miR-155: On the crosstalk between inflammation and cancer. Int Rev
Immunol. 28:264–284. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hale JS, Nelson LT, Simmons KB and Fink
PJ: Bcl-2-interacting mediator of cell death influences
autoantigen-driven deletion and TCR revision. J Immunol.
186:799–806. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bouamar H, Jiang D, Wang L, Lin AP, Ortega
M and Aguiar RC: MicroRNA 155 control of p53 activity is context
dependent and mediated by Aicda and Socs1. Mol Cell Biol.
35:1329–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amente S, Zhang J, Lavadera ML, Lania L,
Avvedimento EV and Majello B: Myc and PI3K/AKT signaling
cooperatively repress FOXO3a-dependent PUMA and GADD45a gene
expression. Nucleic Acids Res. 39:9498–9507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai ZK, Chen Q, Chen YB, Gu M, Zheng DC,
Zhou J and Wang Z: microRNA-155 promotes the proliferation of
prostate cancer cells by targeting annexin 7. Mol Med Rep.
11:533–538. 2015.PubMed/NCBI
|
|
24
|
Lao G, Liu P, Wu Q, Zhang W, Liu Y, Yang L
and Ma C: Mir-155 promotes cervical cancer cell proliferation
through suppression of its target gene LKB1. Tumour Biol.
35:11933–11938. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Novara G, Ficarra V, Antonelli A, Artibani
W, Bertini R, Carini M, Cosciani Cunico S, Imbimbo C, Longo N,
Martignoni G, et al: Validation of the 2009 TNM version in a large
multi-institutional cohort of patients treated for renal cell
carcinoma: Are further improvements needed? Eur Urol. 58:588–595.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dinami R, Ercolani C, Petti E, Piazza S,
Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, et
al: miR-155 drives telomere fragility in human breast cancer by
targeting TRF1. Cancer Res. 74:4145–4156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Yang J, Lv X, Liu K, Gao C, Xing Y
and Xi T: miR-155 regulates the proliferation and cell cycle of
colorectal carcinoma cells by targeting E2F2. Biotechnol Lett.
36:1743–1752. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong W, He L, Coppola M, Guo J, Esposito
NN, Coppola D and Cheng JQ: MicroRNA-155 regulates cell survival,
growth and chemosensitivity by targeting FOXO3a in breast cancer. J
Biol Chem. 285:17869–12879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Spinetti G, Cordella D, Fortunato O,
Sangalli E, Losa S, Gotti A, Carnelli F, Rosa F, Riboldi S, Sessa
F, et al: Global remodeling of the vascular stem cell niche in bone
marrow of diabetic patients: Implication of the microRNA-155/FOXO3a
signaling pathway. Circ Res. 112:510–522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anderson MJ, Viars CS, Czekay S, Cavenee
WK and Arden KC: Cloning and characterization of three human
forkhead genes that comprise an FKHR-like gene subfamily. Genomics.
47:187–199. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tran H, Brunet A, Grenier JM, Datta SR,
Fornace AJ Jr, DiStefano PS, Chiang LW and Greenberg ME: DNA repair
pathway stimulated by the forkhead transcription factor FOXO3a
through the Gadd45 protein. Science. 296:530–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sunters A, de Fernández Mattos S, Stahl M,
Brosens JJ, Zoumpoulidou G, Saunders CA, Coffer PJ, Medema RH,
Coombes RC and Lam EW: FoxO3a transcriptional regulation of Bim
controls apoptosis in paclitaxel-treated breast cancer cell lines.
J Biol Chem. 278:49795–49805. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ni D, Ma X, Li HZ, Gao Y, Li XT, Zhang Y,
Ai Q, Zhang P, Song EL, Huang QB, et al: Downregulation of FOXO3a
promotes tumor metastasis and is associated with metastasis-free
survival of patients with clear cell renal cell carcinoma. Clin
Cancer Res. 20:1779–1790. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ling N, Gu J, Lei Z, Li M, Zhao J, Zhang
HT and Li X: microRNA-155 regulates cell proliferation and invasion
by targeting FOXO3a in glioma. Oncol Rep. 30:2111–2118.
2013.PubMed/NCBI
|