Introduction
Cardiovascular diseases (CVDs) account for >17
million cases of mortality globally and annually (30% of all
mortality), 80% of which occur in low- and middle-income countries,
and this figure is expected to grow to 23.6 million by 2030
(1). Coronary artery disease (CAD)
is the largest contributor to CVDs (2). Off-pump coronary artery bypass graft
(CABG) surgery emerged in recent years as a means to avoid the
sequelae of extracorporeal circulation such as whole-body
inflammatory response, coagulation disorders, and multiple organ
dysfunction (3). At present, gas
anesthesia-sevoflurane and intravenous anesthesia-propofol have
been widely used during the CABG (4).
Propofol and sevoflurane both possess certain,
although different, cardioprotective properties. Sevoflurane
appeared to be superior to propofol in patients with little or no
ischemic heart disease, including CABG surgery without severe
preoperative ischemia (5). However,
propofol appeared superior in patients with cardiovascular
instability, severe ischemia or acute/urgent surgery (6). Over the past decades, numerous
experimental strategies (association studies, genome-wide linkage
scan, proteomics and global microarray gene expression analysis
amongst others and large efforts have been applied onto the studies
(7,8). Lucchinetti et al (9) performed direct comparisons between
anesthetic gases and intravenous anesthetics in human hearts at the
gene expression level. These results indicated that
anesthetic-induced and constitutive gene regulatory control of
myocardial substrate metabolism predicts postoperative cardiac
function in patients undergoing off-pump CABG surgery. However, the
underlying mechanisms of these anesthetics on the gene level remain
unclear.
Previous studies have established a constructed gene
co-expression network, which contained genes that exhibited similar
expression patterns across different organisms (10,11). It
has also been demonstrated that functionally related genes were
frequently co-expressed across organisms constituting conserved
transcription modules (12). By
constructing a co-expression network, the underlying regulatory
relationships under different conditions may be estimated (13). In order to define the adjacency
matrix, one makes use of an adjacency function, which transforms
the co-expression similarities into connection strengths (14). The node dissimilarity measure is used
as input of a clustering method to define network modules (clusters
of nodes) (15). Furthermore,
modules are groups of genes whose expression profiles are highly
correlated across the samples (16).
Network modules implement the hypothesis that a network can be
divided into functional modules (17). In this case, significant
interactions, such as key genes in significant pathways can be
tested. Therefore, in the present study modules from the
co-expression network based on genes enriched in significant
pathways were identified, and these modules were defined as
pathway-related modules.
The present study aimed to identify changed
pathway-related modules in CAD patients undergoing CABG under
sevoflurane or propofol anesthesia based on network topological
centralities. In order to achieve this, recruitment and
preprocessing of the gene expression profile was initially
conducted, and differentially expressed genes (DEGs) in CAD
patients were identified before and after applying sevoflurane or
propofol, respectively. Next, pathway analysis of the DEGs was
performed using the Kyoto Encyclopedia of Genes and Genomes
database. A co-expression network was constructed by weighted gene
co-expression network analysis (WGCNA), and pathway-related modules
were mined. Finally, significant pathway-related modules were
identified by conducting analysis on the topological centralities
of the co-expression network, in order to further understand the
underlying mechanisms of these anesthetics on the CAD patients
during the CABG process according to systematically analyzing the
pathway-related modules of the co-expression network.
Materials and methods
Data recruitment and
preprocessing
The gene expression profile of E-GEOD-4386 was
obtained from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/). E-GEOD-4386
existed on the A-AFFY-44-Affymetrix GeneChip Human Genome U133 Plus
2.0 Platform. The data were obtained from patients that had
undergone off-pump CABG surgery, and they were allocated either to
receive the anesthetic gas sevoflurane or the intravenous
anesthetic propofol. The samples were then divided into two groups:
Baseline sevoflurane (n=10)-sevoflurane (n=10) and baseline
propofol (n=10)-propofol (n=10) (9).
Furthermore, the microarray data and annotation files were
downloaded for further analysis.
Background-corrected signal intensities were
determined using the Micro Array Suite 5.0 (MAS 5.0) software
(Affymetrix, Inc., Santa Clara, CA, USA) (18). The normalization of datasets obtained
from the ArrayExpress database was performed using a robust
multichip average method (19) and
quantile based algorithm (20).
Meanwhile, the gene expression value was transformed to a
comparable level. Additionally, a gene-filter package was used to
screen the data. Each probe was mapped to one gene, and the probe
was discarded if it did not match any genes. Furthermore, the
expression value averaged over probes was used as the gene
expression value if the gene had multiple probes, and 20,102 genes
were obtained overall.
Identification of DEGs
The linear model for microarray data (LIMMA)
package, a core component of Bioconductor, is an R-based
open-source software development project in statistical genomics
(21). A core capability is the use
of linear models to assess differential expression in the context
of multifactor designed experiments. In the present study, for
genes with scores greater than an adjustable threshold, empirical
Bayes method that was implemented in the package (22) was used to identify DEGs in the
sevoflurane and propofol groups, respectively. Furthermore, the
false discovery rate was used to proofread the P-values. Values of
[log Fold Change (FC)] >2.0 and P<0.01 were selected as the
cut-off criteria.
Functional enrichment analysis of
DEGs
KEGG pathway database is a recognized and
comprehensive database including all types of biochemical pathways
(23). In the present study, the
KEGG database was applied to investigate the enrichment analysis of
the nodes in order to find the biochemical pathways of DEGs that
were involved in patients that had undergone off-pump CABG surgery
before and after applying sevoflurane or propofol. The Database for
Annotation, Visualization and Integrated Discovery (DAVID)
(24) was used to perform the KEGG
pathway enrichment analysis with the P<0.05 and gene count
>5.
Co-expression network analysis
Identifying differential co-expression by
WGCNA
Gene co-expression networks, which represent a major
application of correlation network methodology, are instrumental
for describing the pair-wise relationships among gene transcripts
and facilitate the understanding of their function and
identification of their key players (25,26).
WGCNA, as a statistical approach based on correlations, has been
widely used to analyze transcriptional profiles, and has proved to
be an informative approach for the functional annotation of
uncharacterized genes (27). A
coefficient of variation (CV=µ/σ) filtering was applied to remove
genes that were constitutively expressed, unexpressed or vary only
modestly across experimental treatments or conditions (28). In this study, a CV cutoff value of
0.6 was selected to obtain co-expression interactions.
Co-expression network construction
Cytoscape provides an environment for the
visualization and analysis of networks and associated annotations
(29). The primary audience for
Cytoscape is the biological community, and it supports a number of
standard use cases for analyzing and visualizing biological data
(30). In the present study, the
co-expression network was constructed using Cytoscape version
3.1.0. Meanwhile, the expression values of each node were mapped to
the co-expression network, where different colors represent the
differences in the expression value of the nodes.
Pathway-related module mining and topological
analysis
In the present study, pathway-related modules were
extracted from a co-expression network in order to investigate
significant genes and modules that played key roles in patients
undergoing off-pump CABG surgery before and after applying
sevoflurane or propofol. To achieve this, firstly, genes in each
significant pathway of the two groups were explored and mapped into
the co-expression network. Next, pathway genes in the network and
their adjacent genes were captured to form a sub-network, which
were also called pathway-related modules. Finally, module
topological analysis (the mean degree centrality of genes in the
corresponding module) was conducted to evaluate significant
pathway-related modules.
Results
Identifying DEGs
After having preprocessed the profile, the empirical
Bayes method (F test) that was implemented in the LIMMA package was
used to identify DEGs in the sevoflurane and propofol group. Under
this condition, when the threshold values of [log(FC)]>2.0 and
P<0.01 were set, a total of 269 DEGs were obtained in the
sevoflurane group and a total of 129 DEGs in the propofol
group.
KEGG pathway analysis of the DEGs
Based on human genomes, DAVID for KEGG pathway
enrichment analysis was performed to further investigate the
biological functions of the DEGs. When the threshold of P-value was
set to 0.05, eight significant pathways in the sevoflurane
(Table I) and seven in the propofol
(Table II) groups were
obtained.
 | Table I.KEGG pathway of DEGs in the
sevoflurane group. |
Table I.
KEGG pathway of DEGs in the
sevoflurane group.
| ID | Term | Count | P-value |
|---|
| hsa04621 | NOD-like receptor
signaling pathway | 11 |
7.39×10−8 |
| hsa04060 | Cytokine-cytokine
receptor interaction | 17 |
7.92×10−6 |
| hsa04610 | Complement and
coagulation cascades | 9 |
2.06×10−5 |
| hsa05219 | Bladder cancer | 5 |
5.68×10−3 |
| hsa04010 | MAPK signaling
pathway | 12 |
5.92×10−3 |
| hsa04115 | p53 signaling
pathway | 6 |
6.13×10−3 |
| hsa04062 | Chemokine signaling
pathway | 9 |
1.50×10−2 |
| hsa04630 | Jak-STAT signaling
pathway | 8 |
1.72×10−2 |
 | Table II.KEGG pathway of DEGs in the propofol
group. |
Table II.
KEGG pathway of DEGs in the propofol
group.
| ID | Term | Count | P-value |
|---|
| hsa04621 | NOD-like receptor
signaling pathway | 7 |
4.68×10−5 |
| hsa04060 | Cytokine-cytokine
receptor interaction | 11 |
4.14×10−4 |
| hsa04010 | MAPK signaling
pathway | 10 |
2.00×10−3 |
| hsa04115 | p53 signaling
pathway | 5 |
5.98×10−3 |
| hsa05219 | Bladder cancer | 4 |
1.02×10−2 |
| hsa05120 | Epithelial cell
signaling in Helicobacter pylori infection | 4 |
3.67×10−2 |
| hsa04610 | Complement and
coagulation cascades | 4 |
3.80×10−2 |
It was evident that nucleotide-binding
oligomerization domain (NOD)-like receptor signaling pathway,
cytokine-cytokine receptor interaction, complement and coagulation
cascades, mitogen-activated protein kinase and p53 signaling
pathways were enriched in both groups. While the chemokine and
Janus kinase/signal transducers and activators of transcription
signaling pathways were only enriched in the sevoflurane group,
epithelial cell signaling in Helicobacter pylori infection
was only enriched in the propofol group.
Co-expression network construction and
topological analysis
After having identified the DEGs, co-expression
analysis was conducted on these 269 DEGs in the sevoflurane group
and 129 DEGs in the propofol group using the WGCNA method. By
setting a threshold CV cutoff value of 0.6, 813 (180 DEGs) and
1,216 (119 DEGs) co-expression interactions were obtained in two
groups. Furthermore, two co-expression networks were obtained via a
conducting network with the co-expression interactions in Cytoscape
Version 3.1.0. Additionally, as we mapped the expression values of
each node to the co-expression network separately, two networks
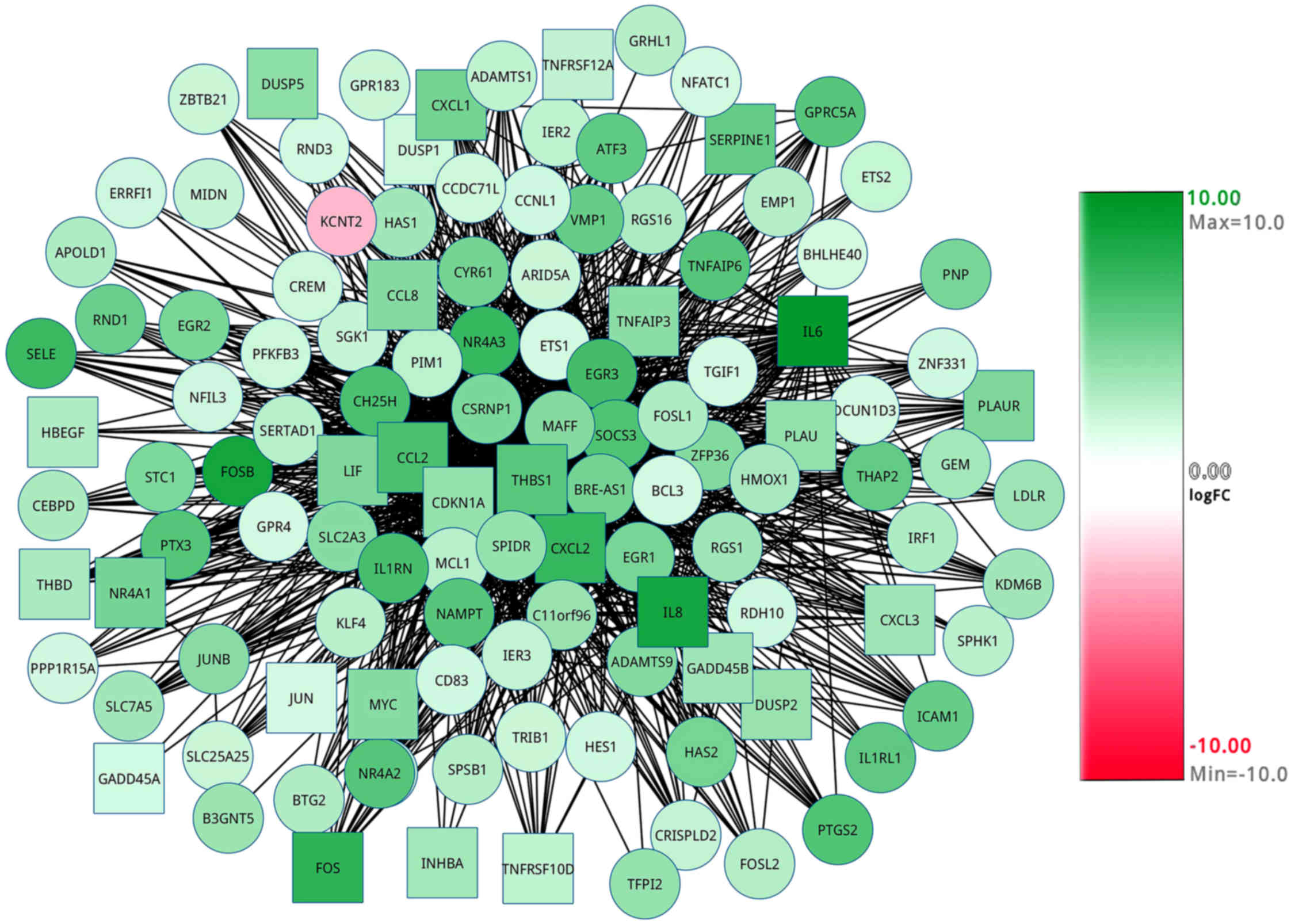
with expression values were obtained (Figs. 1 and 2). It was evident that all of the pathway
genes were in green ([log(FC)]>2.0), which meant that these
genes were all upregulated.
Pathway-related module mining and
topological analysis
After having separately investigated genes in each
significant pathway of the two groups, and having mapped them into
the co-expression network, eight and seven pathway-related modules
were obtained in the sevoflurane group (Figs. 3 and 4) and in the propofol group (Figs. 5 and 6), respectively. To further investigate the
biological functions of these modules, degree centrality analysis
was performed, the details of which are shown in Table III (sevoflurane group) and Table IV (propofol group). The mean degree
of modules of complement and coagulation cascades related, p53
signaling related, NOD-like receptor signaling related and
cytokine-cytokine receptor interaction related were >20 in both
of the groups. Moreover, the complement and coagulation cascades
pathway-related module revealed the highest mean degree in both
groups, which in the sevoflurane group had a mean degree of 56.67,
and in the propofol group a mean degree of 71.88. However, the
chemokine signaling pathway-related module only existed in the
sevoflurane group with a mean degree of 36.92, and epithelial cell
signaling in H. pylori infection pathway-related module only
existed in the propofol group with a mean degree of 35.98.
 | Table III.Mean degree centrality of
pathway-related modules in the sevoflurane group. |
Table III.
Mean degree centrality of
pathway-related modules in the sevoflurane group.
| Pathway-term | Degree |
|---|
| Complement and
coagulation cascades | 56.67 |
| p53 signaling
pathway | 55.69 |
| Chemokine signaling
pathway | 36.92 |
| NOD-like receptor
signaling pathway | 33.31 |
| Cytokine-cytokine
receptor interaction | 22.29 |
| MAPK signaling
pathway | 15.67 |
| Bladder cancer | 15.64 |
| Jak-STAT signaling
pathway | 15.52 |
 | Table IV.Mean degree centrality of
pathway-related modules in the propofol group. |
Table IV.
Mean degree centrality of
pathway-related modules in the propofol group.
| Pathway Term | Degree |
|---|
| Complement and
coagulation cascades | 71.88 |
| MAPK signaling
pathway | 53.46 |
| Epithelial cell
signaling in Helicobacter pylori infection | 35.98 |
| NOD-like receptor
signaling pathway | 25.01 |
| Cytokine-cytokine
receptor interaction | 24.79 |
| p53 signaling
pathway | 20.77 |
| Bladder cancer | 20.76 |
By conducting analysis on the frequency of genes
contained in the pathway-related modules, it was identified that
there were several genes that presented in more than one module in
both of the groups, as presented in Table V (sevoflurane group) and Table VI (propofol group). Furthermore, it
was evident that in the sevoflurane group, genes IL8, CXCL2, CCL2,
IL6, IL1B, CXCL1, CCL11 and MYC had a frequency >3 and in the
propofol group, genes IL8 and CXCL1 had a frequency >3. In
addition, genes IL8, CXCL2, CCL2, CXCL1 and CCL11 were all enriched
in the chemokine signaling pathway-related module of the
sevoflurane group, and genes IL8, CXCL1 were all enriched in
epithelial cell signaling in H. pylori infection
pathway-related module of the propofol group.
 | Table V.Frequency of genes that are in the
pathway-related modulus of the sevoflurane group. |
Table V.
Frequency of genes that are in the
pathway-related modulus of the sevoflurane group.
| Symbol | No. |
|---|
| IL8 | 4 |
| CXCL2 | 3 |
| CCL2 | 3 |
| IL6 | 3 |
| IL1B | 3 |
| CXCL1 | 3 |
| CCL11 | 3 |
| MYC | 3 |
| CCL8 | 2 |
| CXCL3 | 2 |
| CCL20 | 2 |
| CSF3 | 2 |
| LIF | 2 |
| SERPINE1 | 2 |
| CDKN1A | 2 |
| THBS1 | 2 |
| GADD45A | 2 |
| GADD45B | 2 |
| BIRC3 | 1 |
| TNFAIP3 | 1 |
| INHBA | 1 |
| TNFRSF12A | 1 |
| TNFRSF10D | 1 |
| PLAUR | 1 |
| SOCS2 | 1 |
| CISH | 1 |
| THBD | 1 |
| PLAU | 1 |
| DUSP1 | 1 |
| NR4A1 | 1 |
| FOS | 1 |
| DUSP2 | 1 |
| JUN | 1 |
| DUSP5 | 1 |
| CDKN1A | 1 |
| CXCL2 | 1 |
| SOCS3 | 1 |
| PIM1 | 1 |
 | Table VI.Frequency of genes that are in the
pathway-related modulus of the propofol group. |
Table VI.
Frequency of genes that are in the
pathway-related modulus of the propofol group.
| Symbol | No. |
|---|
| IL8 | 4 |
| CXCL1 | 3 |
| IL5 | 2 |
| CXCL2 | 2 |
| CCL2 | 2 |
| CCL8 | 2 |
| GADD45A | 2 |
| GADD45B | 2 |
| MYC | 2 |
| DUSP2 | 2 |
| JUN | 2 |
| SERPINE1 | 2 |
| THBS1 | 2 |
| CDKN1A | 2 |
| TNFAIP3 | 1 |
| LIF | 1 |
| TNFRSF10D | 1 |
| INHBA | 1 |
| TNFRSF12A | 1 |
| CXCL3 | 1 |
| DUSP5 | 1 |
| DUSP1 | 1 |
| NR4A1 | 1 |
| FOS | 1 |
| HBEGF | 1 |
| THBD | 1 |
| PLAU | 1 |
| PLAUR | 1 |
Discussion
In the present study, an analysis on the gene
profiles of patients who had undergone off-pump CABG surgery before
and after applying sevoflurane or propofol was conducted based on a
pathway-related module associated co-expression network. Since the
co-expression network that separately mapped all of the genes
present in the significant pathways was analyzed, it was shown that
the mean degree of several modules (complement and coagulation
cascades pathway-related module, p53 signaling pathway-related
module, NOD-like receptor signaling pathway-related module and
cytokine-cytokine receptor interaction pathway-related module) were
>20 in both of the groups, and the mean degree of complement and
coagulation cascades pathway-related module in both of the groups
were the highest. However, a chemokine signaling pathway-related
module only existed in the sevoflurane group with a mean degree of
36.92, and epithelial cell signaling in H. pylori infection
pathway-related module only existed in the propofol group with a
mean degree of 35.98.
The complement and coagulation systems were
described as separate cascades and as descendants of a common
ancestral pathway. Both proteolytic cascades were composed of
serine proteases with common structural characteristics, including
highly conserved catalytic sites of histidine, aspartate and serine
(31,32). Furthermore, both systems belonged to
a complex inflammatory network (33)
and exhibited some similar characteristics with regard to the
specialized functions of their inhibitors and activators (34). It was indicated that cardiac surgery
with cardiopulmonary bypass gives rise to a systemic inflammatory
reaction, caused by the extracorporeal circuit and surgical trauma
(35), which generated activation of
the complement, fibrinolytic, kallikrein and coagulation cascades,
activation of leukocytes and endothelial cells with expression of
adhesion molecules and the release of inflammatory mediators, such
as cytokines (36). Pathways of
cytokine-cytokine receptor interaction were at the top of an
enriched pathway list in an CAD gene KEGG pathway analysis
(37).
Cytokines represent a diverse group of molecules
that transmit intercellular signals. These signals may be paracrine
or autocrine. Both of these situations could occur simultaneously
(38). In addition, injured
endothelial cells produce cytokines (including interleukin) that
stimulate the expression of adhesion proteins selectins and cell
adhesion molecules including vascular cell adhesion and
intercellular adhesion molecules on the endothelial surface
(39). It had been concluded that
among apparently healthy women and men, elevated levels of IL8 are
associated with an increased risk of CAD (40). There has also been increasing
recognition that in various pathological conditions, CB1 receptor
activation by endocannabinoids may promote activation of signaling
pathways promoting cell death (41).
The chemokines are a family of low-molecular-weight
proteins involved in leukocyte activation and migration (42). Significant advances have been made in
understanding the role of chemokines and their receptors in
cardiovascular diseases (43) tumor
growth as well as metastasis (44)
and inflammatory diseases (45). It
has been indicated that the molecular mechanisms responsible for
monocyte accumulation in plaque of atherosclerosis are likely to
include chemokines and their receptors, as these molecules were
major regulators of specific leukocyte trafficking (46). In the present study, the chemokine
signaling pathway-related module only existed in the sevoflurane
group, which meant that this anesthetic might give the patients
with more protection in hematopoiesis, angiogenesis, metastasis and
tumor rejection or inflammatory diseases. However, it has also been
reported that epithelial cell signaling in H. pylori
infection was mainly associated with peptic ulceration, chronic
gastritis and more rarely with gastric adenocarcinoma (47).
In conclusion, in the present study, complement and
coagulation cascade related modules were successfully identified to
be significant in both groups of sevoflurane and propofol, which
meant that during the CABG, these anesthetics might activate the
complement and coagulation systems so as to exert some
cardioprotective properties. While the chemokine signaling
pathway-related module only existed in the sevoflurane group, which
meant that this anesthetic might provide the patients with more
protection in hematopoiesis, angiogenesis, metastasis and tumor
rejection or inflammatory diseases.
However, there remained certain limitations in the
present study that must be taken into account. First of all, the
sample size was not large enough. Additionally, an experimental
verification analysis should be conducted in order to verify the
results obtained by the bioinformatics method used in the present
study. Although disadvantages existed, it is believed that this
method and the results offered investigators valuable resources for
better understanding the underlying mechanisms of sevoflurane and
propofol of the CAD patients who undergo CABG on the pathway
level.
Acknowledgements
The present study was supported by the Department of
Anesthesiology of Qilu Hospital of Shandong University, the
Departments of Anesthesiology and Neurosurgery of the Affiliated
Hospital of Binzhou Medical College. The authors would like to
thank all the members of the research group. Meanwhile, the authors
are grateful to the Ji'nan Evidence Based Medicine
Science-Technology Center, who provided technical support during
the data processing and analysis.
References
|
1
|
Mendis S, Puska P and Norrving B: Global
atlas on cardiovascular disease prevention and control. 2011.
|
|
2
|
Wong ND: Epidemiological studies of CHD
and the evolution of preventive cardiology. Nat Rev Cardiol.
11:276–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scott NB, Turfrey DJ, Ray DA, Nzewi O,
Sutcliffe NP, Lal AB, Norrie J, Nagels WJ and Ramayya GP: A
prospective randomized study of the potential benefits of thoracic
epidural anesthesia and analgesia in patients undergoing coronary
artery bypass grafting. Anesth Analg. 93:528–535. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heindl B, Reichle FM, Zahler S, Conzen PF
and Becker BF: Sevoflurane and isoflurane protect the reperfused
guinea pig heart by reducing postischemic adhesion of
polymorphonuclear neutrophils. Anesthesiology. 91:521–530. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jakobsen CJ, Berg H, Hindsholm KB, Faddy N
and Sloth E: The influence of propofol versus sevoflurane
anesthesia on outcome in 10,535 cardiac surgical procedures. J
Cardiothorac Vasc Anesth. 21:664–671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jakobsen CJ, Berg H, Hindsholm KB, Faddy N
and Sloth E: The influence of propofol versus sevoflurane
anesthesia on outcome in 10,535 cardiac surgical procedures. J
Cardiothorac Vasc Anesth. 21:664–671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karwacki Z, Kowiański P, Moryś J,
Dziewiatkowski J, Kaczmarek E and Suchorzewska J: Effect of
sevoflurane on intracranial pressure and cardiovascular function in
rabbits with experimental intracerebral haematoma. Med Sci Monit.
7:212–217. 2001.PubMed/NCBI
|
|
8
|
Conzen PF, Fischer S, Detter C and Peter
K: Sevoflurane provides greater protection of the myocardium than
propofol in patients undergoing off-pump coronary artery bypass
surgery. Anesthesiology. 99:826–833. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lucchinetti E, Hofer C, Bestmann L,
Hersberger M, Feng J, Zhu M, Furrer L, Schaub MC, Tavakoli R,
Genoni M, et al: Gene regulatory control of myocardial energy
metabolism predicts postoperative cardiac function in patients
undergoing off-pump coronary artery bypass graft surgery:
Inhalational versus intravenous anesthetics. Anesthesiology.
106:444–457. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bergmann S, Ihmels J and Barkai N:
Similarities and differences in genome-wide expression data of six
organisms. PLoS Biol. 2:E92004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Choi JK, Yu U, Yoo OJ and Kim S:
Differential coexpression analysis using microarray data and its
application to human cancer. Bioinformatics. 21:4348–4355. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Basso K, Margolin AA, Stolovitzky G, Klein
U, Dalla-Favera R and Califano A: Reverse engineering of regulatory
networks in human B cells. Nat Genet. 37:382–390. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang B and Horvath S: A general framework
for weighted gene co-expression network analysis. Stat Appl Genet
Mol Biol. 4:Article 172005. View Article : Google Scholar
|
|
15
|
Mumford JA, Horvath S, Oldham MC,
Langfelder P, Geschwind DH and Poldrack RA: Detecting network
modules in fMRI time series: A weighted network analysis approach.
Neuroimage. 52:1465–1476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ravasz E, Somera AL, Mongru DA, Oltvai ZN
and Barabasi AL: Hierarchical organization of modularity in
metabolic networks. Science. 297:1551–1555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davidson EH, McClay DR and Hood L:
Regulatory gene networks and the properties of the developmental
process. Proc Natl Acad Sci USA. 100:pp. 1475–1480. 2003;
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pepper SD, Saunders EK, Edwards LE, Wilson
CL and Miller CJ: The utility of MAS5 expression summary and
detection call algorithms. BMC Bioinformatics. 8:2732007.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ma L, Robinson LN and Towle HC: ChREBP*
Mlx is the principal mediator of glucose-induced gene expression in
the liver. J Biol Chem. 281:28721–28730. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rifai N and Ridker PM: Proposed
cardiovascular risk assessment algorithm using high-sensitivity
C-reactive protein and lipid screening. Clin Chem. 47:28–30.
2001.PubMed/NCBI
|
|
21
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ritchie ME, Silver J, Oshlack A, Holmes M,
Diyagama D, Holloway A and Smyth GK: A comparison of background
correction methods for two-colour microarrays. Bioinformatics.
23:2700–2707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord G, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carey VJ, Gentry J, Whalen E and Gentleman
R: Network structures and algorithms in Bioconductor.
Bioinformatics. 21:135–136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cokus S, Rose S, Haynor D, Grønbech-Jensen
N and Pellegrini M: Modelling the network of cell cycle
transcription factors in the yeast Saccharomyces cerevisiae. BMC
Bioinformatics. 7:3812006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Iancu OD, Darakjian P, Walter NA,
Malmanger B, Oberbeck D, Belknap J, McWeeney S and Hitzemann R:
Genetic diversity and striatal gene networks: Focus on the
heterogeneous stock-collaborative cross (HS-CC) mouse. Bmc
Genomics. 11:5852010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Childs KL, Davidson RM and Buell CR: Gene
coexpression network analysis as a source of functional annotation
for rice genes. PLoS One. 6:e221962011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krem MM and Di Cera E: Evolution of enzyme
cascades from embryonic development to blood coagulation. Trends
Biochem Sci. 27:67–74. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Esmon CT: The impact of the inflammatory
response on coagulation. Thromb Res. 114:321–327. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rittirsch D, Flierl MA and Ward PA:
Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 8:776–787.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Amara U: Molecular intercommunication
between the complement and coagulation systems. J Immunology.
185:5628–5636. 2010. View Article : Google Scholar
|
|
35
|
Levy JH and Tanaka KA: Inflammatory
response to cardiopulmonary bypass. Ann Thorac Surg. 75:S715–S720.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wehlin L, Vedin J, Vaage J and Lundahl J:
Activation of complement and leukocyte receptors during on-and off
pump coronary artery bypass surgery. Eur J Cardiothorac Surg.
25:35–42. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu H, Liu W, Liao Y, Cheng L, Liu Q, Ren
X, Shi L, Tu X, Wang QK and Guo AY: CADgene: A comprehensive
database for coronary artery disease genes. Nucleic Acids Res.
39(Database issue): D991–D996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Leonard WJ and Lin JX: Cytokine receptor
signaling pathways. J Allergy Clin Immunol. 105:877–888. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Auer J, Weber T, Berent R, Lassnig E, Lamm
G and Eber B: Genetic polymorphisms in cytokine and adhesion
molecule genes in coronary artery disease. Am J Pharmacogenomics.
3:317–328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Boekholdt SM, Peters RJ, Hack CE, Day NE,
Luben R, Bingham SA, Wareham NJ, Reitsma PH and Khaw KT: IL-8
plasma concentrations and the risk of future coronary artery
disease in apparently healthy men and women: The EPIC-Norfolk
prospective population study. Arterioscler Thromb Vasc Biol.
24:1503–1508. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dalton GD, Bass CE, Van Horn CG and
Howlett AC: Signal transduction via cannabinoid receptors. CNS
Neurol Disord Drug Targets. 8:422–431. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mellado M, Rodríguez-Frade JM, Mañes S and
Martínez AC: Chemokine signaling and functional responses: The role
of receptor dimerization and TK pathway activation. Annu Rev
Immunol. 19:397–421. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang JM, Shen W and Su S: Chemokines and
their role in cardiovascular diseases. Trends Cardiovasc Med.
8:169–174. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rossi D and Zlotnik A: The biology of
chemokines and their receptors. Annu Rev Immunol. 18:217–242. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Murdoch C and Finn A: Chemokine receptors
and their role in inflammation and infectious diseases. Blood.
95:3032–3043. 2000.PubMed/NCBI
|
|
46
|
Moatti D, Faure S, Fumeron F, Amara Mel-W,
Seknadji P, McDermott DH, Debré P, Aumont MC, Murphy PM, de Prost D
and Combadière C: Polymorphism in the fractalkine receptor CX3CR1
as a genetic risk factor for coronary artery disease. Blood.
97:1925–1928. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Crabtree JE and Naumann M: Epithelial Cell
Signaling in Helicobacter pylori Infection. Curr Sig Transduc Ther.
1:53–65. 2006. View Article : Google Scholar
|