Introduction
Ventilator-associated pneumonia (VAP) is a common
complication among mechanical ventilated neonates in neonatal
intensive care unit (NICU). It has been reported that VAP is the
second most frequent nosocomial infection among patients in the
pediatric intensive care unit (PICU) (1), and is the leading cause of death among
ventilated patients (2). The
incidence of VAP in NICU is even higher than that in PICU (3). A previous meta-analysis on neonatal VAP
in China reported that the total weighted incidence and case
fatality rates of VAP in NICU were 42.8% (95% confidence interval;
39.8–45.9%) and 16.4% (95% confidence interval; 13.8–18.9%),
respectively (4). In spite of the
high mobility and mortality, studies addressing neonatal VAP are
limited in the literature (3). Thus,
VAP in NICU remains a serious public health issue that endangers
the lives of critically ill neonates.
Within 24 h following intubation, a polymicrobial
biofilm harboring potential pathogens forms on the inner surface of
the endotracheal tube (ET) (5–7).
Bacterial biofilm cells are able to detach from the surface of the
ET and travel to the lower airway due to gas flow during continuous
ventilation (5), thus promoting VAP
microbial persistence and consequently affecting patient prognosis
(7,8). Therefore, biofilms are crucial in the
pathogenesis of VAP and a better understanding of biofilms,
especially the biofilm flora, is required. However, studies on ET
biofilm flora among intubated neonates are scarce. Furthermore, due
to concerns for patient safety and the fact that reintubation
itself carries a significant risk of neonatal VAP (3), ETs are not able to be extubated during
intubation. The majority of ET samples are only available at the
time of extubation (9–11), which means that the microbial
signatures of ET biofilms prior to extubation are unknown.
Therefore, identifying substituent samples to investigate
microbiota in ET biofilms is beneficial to identify the floral risk
factors associated with VAP. Specimens, such as throat swabs and
tracheal aspirates, are able to be obtained at different times
during intubation. Furthermore, it has been reported that a vast
range of identical and potential pathogens are found in upper and
lower respiratory tract (12,13), and
ET is the conduit between them. This indicates a potential
consistence of microorganisms among throat swab, tracheal aspirate
and ET biofilms, which means that tracheal aspirates and throat
swabs may be the substituents of ET biofilms. A previous study
demonstrated that nose swabs, throat swabs and sputum samples were
able to predict the presence of several nosocomial pathogens in ET
biofilms among intubated adults (14). However, this remains to be
investigated in other populations, in particular intubated
neonates. Furthermore, the previous study only addressed several
nosocomial pathogens based on culture results (14). Culture dependent methods typically
fail to recover and grow all biofilm cells from implanted medical
devices, and therefore lack sensitivity for the detection of
microflora (10,15). Culture-independent methods based on
microbial 16S ribosomal RNA (rRNA) genes overcome the limitation of
culture, and allow a more comprehensive understanding of human
microbiome (16). Denaturing
gradient gel electrophoresis (DGGE) is a culture-independent method
that facilitates rapid analyses and comparisons of microbial
communities (17), and is simpler
and less expensive than other culture-independent methods. DGGE has
been widely applied in microbiological studies (9,17,18),
such as a recent study among intubated adults, which reported a
considerable biofilm compositional complexity based on DGGE
(9). However, to the best of our
knowledge similar studies among neonatal patients are scarce,
particularly those with VAP.
The present study utilized polymerase chain reaction
(PCR)-DGGE to characterize the microbial communities in ET
biofilms, throat swabs and tracheal aspirate samples of intubated
neonates with VAP. The aim of the present study was to test the
consistency of microbial communities among the three sources of
specimens based on a culture-independent approach, thereby
identifying alternatives for the surveillance of microbial profiles
and potential pathogens in ET biofilms during intubation of
intubated neonates.
Materials and methods
Patients
The study period ran from January to December 2014.
A total of 15 mechanically ventilated neonates with VAP at the
Children's Hospital of Chongqing Medical University (Chongqing,
China) were included in the present study. VAP was diagnosed
according to the accepted VAP criterion defined by the Centers for
Disease Control and Prevention of America (19,20). Of
the 15 intubated neonates with VAP included in the present study, 8
(53.3%) were male and 7 (46.7%) were female (Table I). The gestational age of the study
patients ranged from 28.6–41.3 weeks, including 10 (66.7%)
premature infants. A total of 8 (53.3%) of the study patients were
diagnosed with neonatal respiratory distress syndrome, followed by
asphyxia neonatorum (20.0%; Table
I). The length of intubation ranged from 2–10.9 days, with a
median of 5.8 days (Table I). Prior
to extubation, antibiotics (moxalactam) were administered among all
the study patients for a range of 1–13 days, with a median of 5.4
days (Table I).
 | Table I.Demographic and clinical
characteristics of the 15 intubated neonates with
ventilator-associated pneumonia in the present study. |
Table I.
Demographic and clinical
characteristics of the 15 intubated neonates with
ventilator-associated pneumonia in the present study.
| Patient ID | Sex | Gestation age
(weeks) | Weight (g) | Diagnosis | Intubation period
(days) | Antibiotic
administration (days) | Outcome | Culture
finding |
|---|
| 1 | Male | 32.6 | 2,160 | NRDS |
4.2 | 2 | Improved | Normal flora |
| 2 | Female | 31.0 | 1,680 | NRDS | 10.8 | 9 | Healed | Klebsiella
pneumoniae |
| 3 | Female | 33.7 | 1,650 | NRDS | 15.0 | 13 | Improved | Normal flora |
| 4 | Female | 38.6 | 2,750 | Asphyxia
neonatorum |
5.0 | 2 | Improved | Normal flora |
| 5 | Male | 28.6 | 1,325 | NRDS |
4.0 | 4 | Healed | Negative |
| 6 | Male | 29.7 | 1,540 | Asphyxia
neonatorum |
3.0 | 3 | Died | Acinetobacter
baumannii |
| 7 | Female | 37.0 | 4,270 | Asphyxia
neonatorum |
5.0 | 9 | Improved | Normal flora |
| 8 | Male | 35.9 | 2,800 | Pneumonia |
2.0 | 2 | Healed | Pseudomonas
aeruginosa |
| 9 | Male | 41.3 | 4,700 | Brain injury |
5.0 | 5 | Healed | Normal flora |
| 10 | Male | 39.3 | 3,430 | Pneumonia |
5.5 | 7 | Healed | Negative |
| 11 | Female | 32.1 | 1,260 | NRDS | 10.9 | 9 | Improved | Negative |
| 12 | Female | 36.1 | 3,250 | NRDS |
4.6 | 5 | Healed | Negative |
| 13 | Female | 34.1 | 1,950 | NRDS |
5.6 | 5 | Improved | Normal flora |
| 14 | Male | 31.7 | 2,300 | NRDS |
4.0 | 5 | Healed | Negative |
| 15 | Male | 40.6 | 3,080 | MAS |
2.0 | 1 | Healed | Negative |
Study samples
The ET was collected immediately following
extubation and stored in sterile PBS at 4°C. Vortex and sonication
were conducted to release biofilms from the inner surface of the
endotracheal tube as previously described (9). Prior to extubation, throat swabs and
tracheal aspirates were collected and processed as previously
described (14). The three types of
specimen from each patient were collected and processed on the same
day. Microbial DNA was extracted using a Mini BEST Bacterial
Genomic DNA Extraction Kit Ver 3.0 (Takara Biotechnology Co., Ltd.,
Dalian, China) according to the manufacturer's protocol. The study
protocol was reviewed and approved by the Medical Ethics Committee
of Chongqing Medical University (099/2014). Informed consent was
obtained from the parents or legal guardians of the study patients.
Demographic and clinical data were obtained from medical records
and anonymized information was used in the present study.
PCR-DGGE
The viable V3 region of 16S rRNA gene was amplified
using universal bacterial primers 357f with ‘GC’ clamp
(5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCAGGGGCCTACGGGAGGCAGCAG-3′) and
518r (5′-ATTACCGCGGCTGCTGG-3′). The PCR was conducted as previously
described (21). The reaction
included 6 µl microbial DNA as template DNA, 25 µl Premix Taq
Version 2.0 (TaKaRa Biotechnology Co., Ltd., Dalian, China), 0.5 µl
each primer and 18 µl sterile ddH2O. Template DNA of
sterile ddH2O was regarded as the negative control. The
reaction conditions were as follows: Initial denaturation at 94°C
for 5 min, 10 cycles of denaturation at 94°C for 30 sec, annealing
at 61 to 56°C (−0.5°C/cycle), and extension at 72°C for 1 min, 25
cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30
sec, and extension at 72°C for 1 min, and a final extension at 72°C
for 7 min (21). To evaluate the
accuracy of PCR, the size of amplicons was determined by 2% agarose
electrophoresis and visualized using Benchtop 3UV ultraviolet
transilluminator (Ultra-Violet Products, Ltd., Cambridge, UK). The
195 bp PCR products were processed for DGGE based on DCode
Universal Mutation Detection System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Following 16 h of electrophoresis at 85 V and
60°C, the DGGE gels were stained using SYBR-Green I (Bioteke
Corporation, Beijing, China) for 30 min and scanned using Benchtop
3UV ultraviolet transilluminator (UVP, Inc., Upland, CA, USA).
Visible bands in the DGGE gel were excised, washed twice with
sterile ddH2O, placed in 30 µl sterile ddH2O
and incubated at 4°C overnight to release DNA.
Cloning and sequencing
A second PCR targeting 16S rRNA gene V3 region was
conducted using primers 357f without the ‘GC’ clamp
(5′-CCTACGGGAGGCAGCAG-3′) and 518r as previously described
(21). The PCR products were
purified using Agarose Gel DNA Purification Kit Version 3.0 (Takara
Bio, Inc., Otsu, Japan). The purified DNA was cloned using the
PMD18-T Vector system (Takara Bio, Inc.) and transformed into
Escherichia coli DH5α competent cells (Takara Biotechnology Co.,
Ltd.). The resulting cells were cultured overnight at 37°C on
Luria-Bertani (LB) medium with ampicillin and positive clones were
selected for sequencing (Biotech Corporation, Shanghai, China) as
described previously (21).
Sequences were assigned and identified to phylogenetic species
using BLAST in NCBI Genbank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) with 97%
similarity. The PCR experiments performed were repeated twice.
Diversity and similarity analysis of
DGGE fingerprints
DGGE profiles were analyzed and compared using
Quantity One software (Version 4.6.2; Bio-Rad Laboratories, Inc.).
To evaluate the microbial diversity and richness, the
Shannon-Wiener index and Simpson's index for each sample were
calculated using Bio-Dap software (nhsbig.inhs.uiuc.edu/wes/populations.html). The
band number of each sample in DGGE images was also analyzed to
evaluate the microbial diversity. Cluster maps of DGGE fingerprints
were generated based on the Dice coefficient using the unweighted
pair group method with arithmetic averages (UPGMA) to assess the
similarity between samples in each DGGE image using Quantity-One
software.
Statistical analysis
To assess the consistency of microflora in
endotracheal tube biofilms with throat swab and tracheal aspirate,
microbial diversity and taxonomic composition were compared using
independent sample t-test, Chi-square test and Fisher's exact test
as appropriate. Sensitivities and specificities for alternative
samples (i.e., throat swab and tracheal aspirate samples) for the
detection of certain taxa in ET biofilms were also calculated based
on the microbiological findings in ET biofilms. All the statistical
analyses were performed using SPSS 18.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
A total of 8 (53.3%) of the study patients were
healed, clinical symptoms of 6 (40.0%) patients were improved and
the remaining patient succumbed to mortality. Clinical culture
results of tracheal aspirate (following extubation) demonstrated
that 60.0% of patients were culture-positive. Among those
culture-positive subjects, normal flora was the most frequently
identified, accounting for 66.7% (6/9; Table I).
Microbial diversity analysis in the
study samples
PCR-DGGE was successfully performed for all 15
throat swabs and ET biofilm samples; however 2 (13.3%) tracheal
aspirate samples failed to generate a positive PCR result. The DGGE
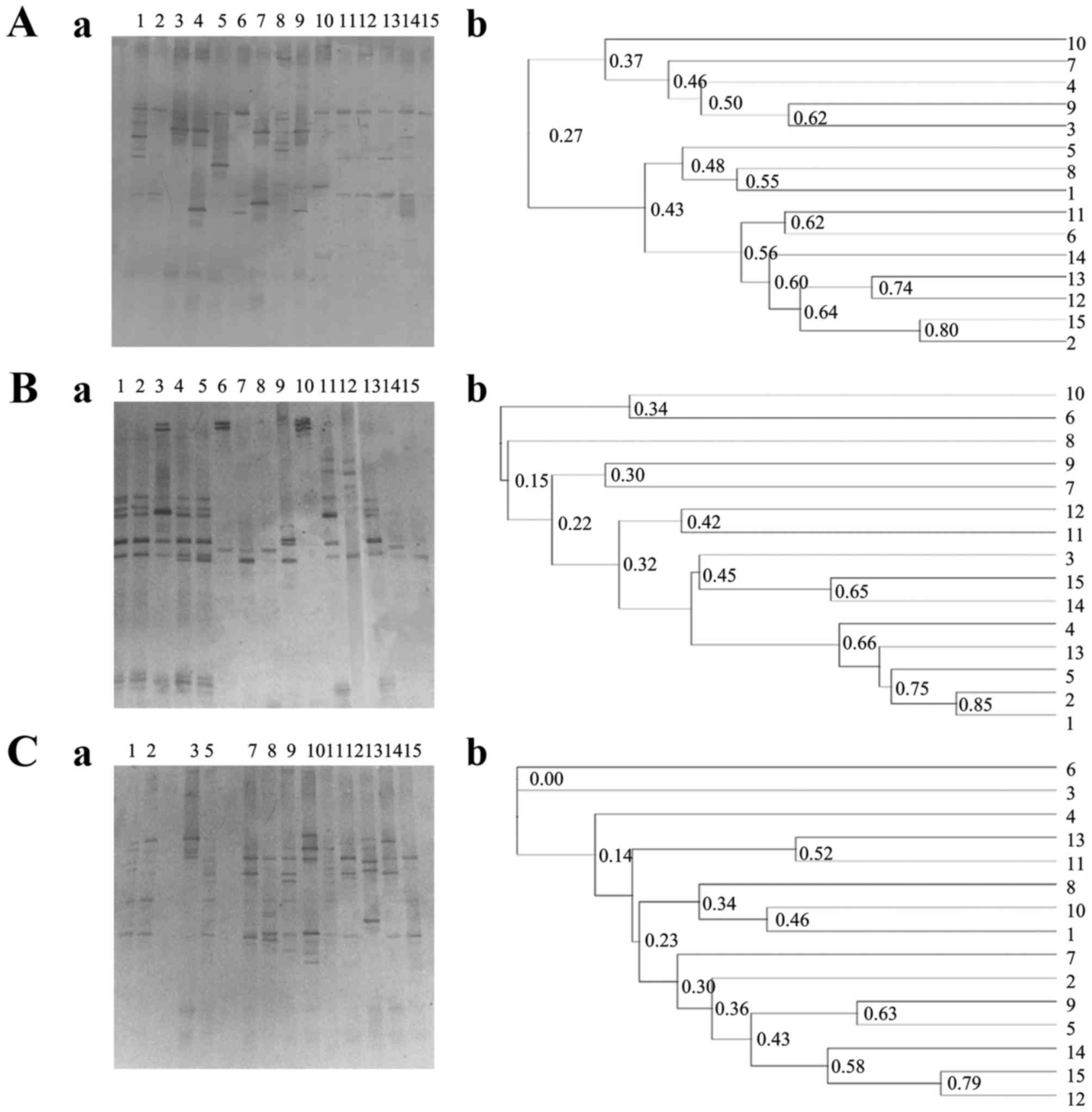
fingerprints for the three types of specimens are displayed in
Fig. 1. No significant difference
between the three sources of specimens was found in the Dice
coefficients in UPGMA based cluster maps (Fig. 1), with no significant differences in
the microbial similarity among the three types of samples.
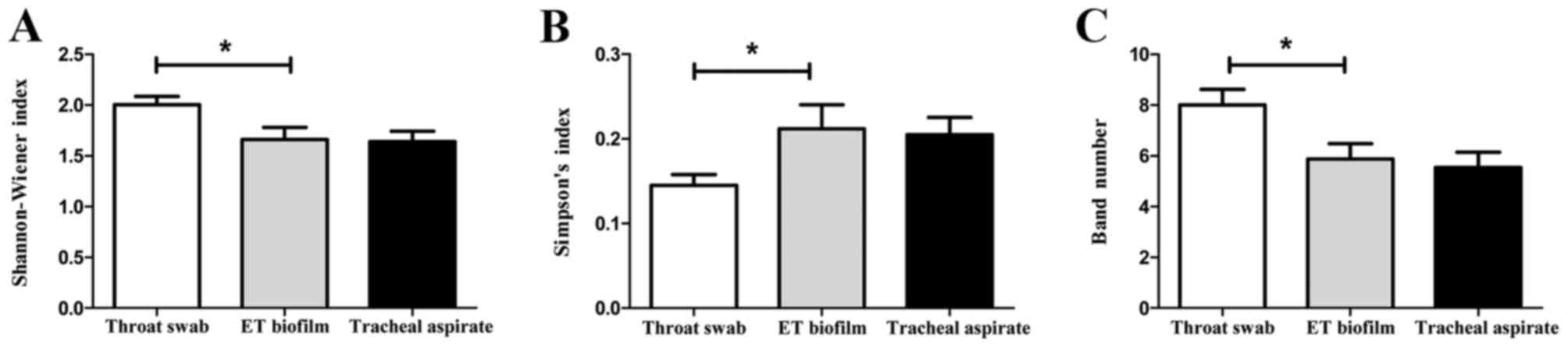
Shannon-Wiener index, Simpson's index and band number of tracheal
aspirate samples were similar to those of ET biofilms (Fig. 2), which indicates a similar level of
microbial diversity between the ET biofilm and tracheal aspirate
samples. However, significant differences were found between throat
swab and ET biofilm samples in the above three indices (P<0.05;
Fig. 2).
Taxonomic analysis of microbial
community in the study samples
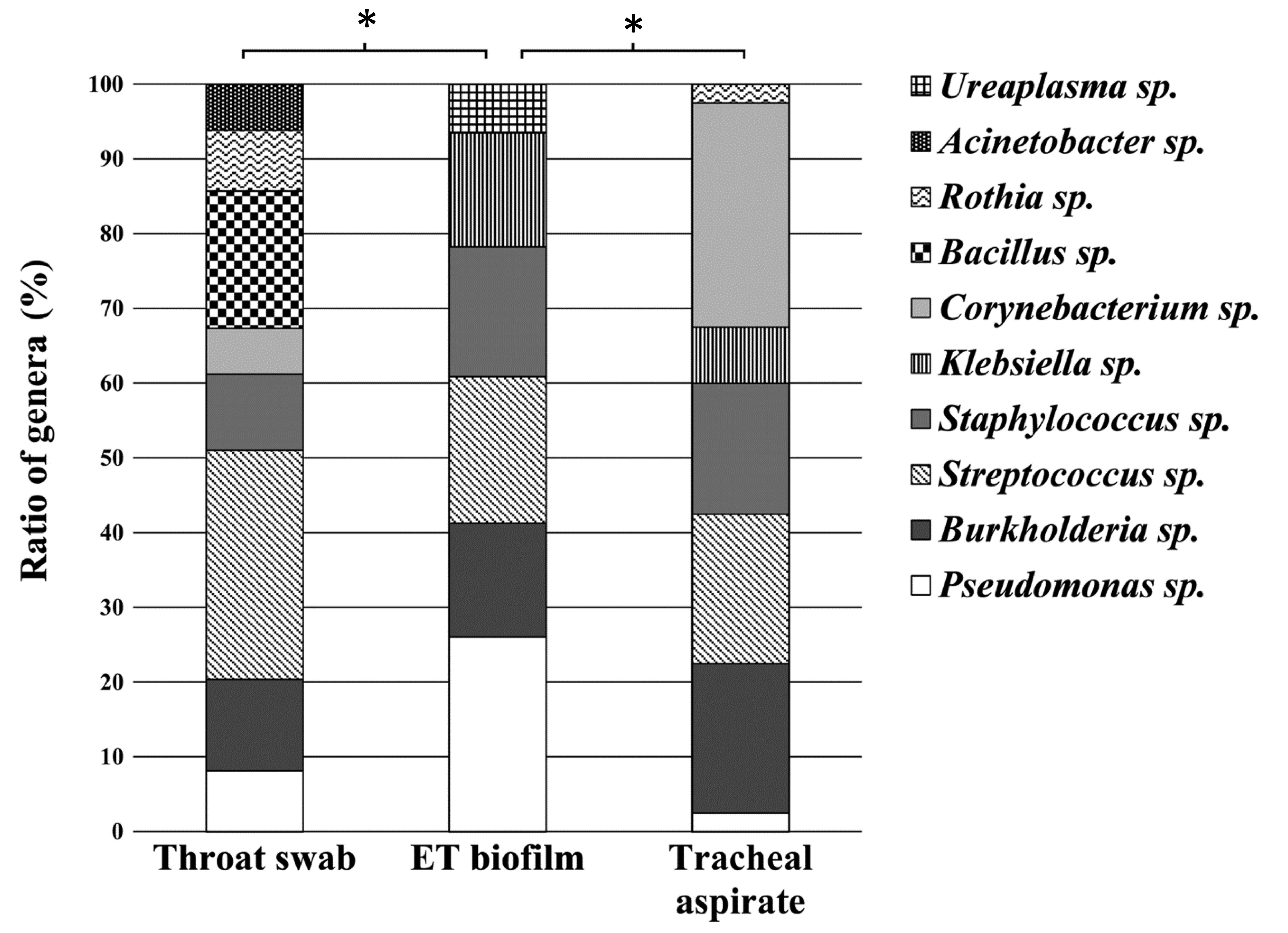
A total of 10 genera were identified in the study
samples (Table II). The overall
constituent ratio of microflora differed significantly in throat
swabs and tracheal aspirates compared with ET biofilm samples
(P<0.05; Fig. 3). Notably, four
genera (Pseudomonas sp., Staphylococcus sp.,
Streptococcus sp. and Burkholderia sp.) were shared by the
three different types of specimens. Klebsiella sp. was
identified in ET biofilm and tracheal aspirate. As a common
pathogen to cause VAP (4),
Klebsiella sp. was also included in the subsequent analysis.
To evaluate the consistency of the detection of identical taxa, the
detection rates of the five common genera in throat swab and
tracheal aspirate were compared with ET biofilms (Table III). The detection rates of
Staphylococcus sp. and Streptococcus sp. were similar
between ET biofilms and the corresponding tracheal aspirate samples
(Table III); however, a
significant difference was observed in the detection rate of
Pseudomonas sp. between ET biofilms and the corresponding
tracheal aspirate samples (P<0.05; Table III). The detection rates of
Pseudomonas sp. and Streptococcus sp. in throat swab
samples were significantly different from those in ET biofilm
samples (P<0.05; Table
III).
 | Table II.Sequencing results of denaturing
gradient gel electrophoresis bands. |
Table II.
Sequencing results of denaturing
gradient gel electrophoresis bands.
| NCBI BLAST
result | Accession
number | Identity (%) |
|---|
| Acinetobacter
sp. | KR088390.1 | 99 |
| Bacillus
sp. | AJ000648.1 | 99 |
| Burkholderia
sp. | AB011288.1 | 98 |
| Corynebacterium
sp. | AB012207.1 | 98 |
| Klebsiella
sp. | KP645384.1 | 100 |
| Pseudomonas
sp. | HQ268805.1 | 99 |
| Rothia
sp. | KC632201.1 | 99 |
| Staphylococcus
sp. | KT277495.1 | 100 |
| Streptococcus
sp. | KM225751.1 | 100 |
| Ureaplasma
sp. | AF073457.1 | 100 |
 | Table III.Detection rates of the five common
genera among throat swab, ET biofilm and tracheal aspirate
specimens respectively in the present study. |
Table III.
Detection rates of the five common
genera among throat swab, ET biofilm and tracheal aspirate
specimens respectively in the present study.
| Genus | Throat swab,
detection rate (%) | ET biofilm,
detection rate (%) |
P-valuea | Tracheal aspirate,
detection rate (%) |
P-valueb |
|---|
|
Staphylococcus | 5/15
(33.3) | 8/15
(53.3) |
0.27 | 7/13
(53.8) |
0.98 |
|
Klebsiella | 0/15 (0.0) | 7/15
(46.7) |
0.06c | 3/13
(23.1) |
0.25c |
|
Pseudomonas | 4/15
(26.7) | 12/15 (80.0) | <0.01 | 1/13
(7.7) | <0.01 |
|
Burkholderia | 6/15
(40.0) | 7/15
(46.7) |
0.71 | 8/13
(61.5) |
0.43 |
|
Streptococcus | 15/15 (100.0) | 9/15
(60.0) |
0.02c | 8/13
(61.5) |
0.93 |
Sensitivity and specificity for the
detection of ET biofilm flora
To assess the possibility of using throat swabs,
tracheal aspirates and a combination of throat swab and tracheal
aspirate samples to predict microbial components in ET biofilms,
their sensitivities and specificities were calculated to detect the
five genera in ET biofilms (Table
IV). For the detection of Staphylococcus sp. in ET
biofilms, the sensitivity for tracheal aspirate samples was 85.7%,
which was higher than that for throat swab samples (37.5%; Table IV). Furthermore, the sensitivity for
the combination of tracheal aspirate and throat swab samples for
Staphylococcus sp. detection in ET biofilms was 100%
(Table IV). Although the
sensitivity of throat swab samples for detecting Pseudomonas
sp. in ET biofilms was 33.3%, the specificity was 100%
(Table IV). The sensitivities to
detect Klebsiella sp. were low in all the three groups
(i.e., throat swab, tracheal aspirate and combined throat swab and
tracheal aspirate groups; Table
IV). The sensitivities to detect Streptococcus sp. in
all the three groups were high; however the corresponding
specificities were low (Table
IV).
 | Table IV.Sensitivity and specificity to detect
the five major genera in endotracheal tube biofilms for the
corresponding throat swab, tracheal aspirate and the combination of
throat swab and tracheal aspirate samples, respectively. |
Table IV.
Sensitivity and specificity to detect
the five major genera in endotracheal tube biofilms for the
corresponding throat swab, tracheal aspirate and the combination of
throat swab and tracheal aspirate samples, respectively.
|
| Sensitivity
(%) | Specificity
(%) |
|---|
|
|
|
|
|---|
| Genus | T | A | T+A | T | A | T+A |
|---|
|
Staphylococcus | 37.5
(3/8) | 85.7
(6/7) | 100.0 (8/8) | 71.4
(5/7) | 83.3
(6/6) | 71.4
(5/7) |
|
Klebsiella |
0.0 (0/7) | 28.6
(2/7) | 28.6
(2/7) | 100.0 (8/8) | 87.5
(7/8) | 87.5
(7/8) |
|
Pseudomonas |
33.3 (4/12) |
9.1
(1/11) |
33.3 (4/12) | 100.0 (3/3) | 100.0 (2/2) | 100.0 (3/3) |
|
Burkholderia | 57.1
(4/7) | 50.0
(3/6) | 85.7
(6/7) | 75.0
(6/8) | 28.6
(2/7) | 37.5
(3/8) |
|
Streptococcus | 100.0 (9/9) | 62.5
(5/8) | 100.0 (9/9) |
0.0 (0/6) | 40.0
(2/5) |
0.0 (0/6) |
Discussion
ET biofilm is an important source in facilitating
bacterial contamination of the lower airway (10,22) and
so is critical for VAP surveillance. To the best of our knowledge,
this is the first study to address the consistency of microbial
signatures in ET biofilms and the corresponding throat swab and
tracheal aspirate samples using a culture-independent approach
among newborns. There were four major findings of the present
study. Firstly, the microbial richness and diversity of ET biofilms
were similar to tracheal aspirate samples, however significantly
different from throat swab samples. Secondly, the taxa composition
varied significantly among the three sources of specimens. Thirdly,
tracheal aspirate samples performed well in the detection of
Staphylococcus sp. in ET biofilms, with a sensitivity of
85.7% and a specificity of 83.3%. Finally, the detection of
Pseudomonas sp. in throat swabs helped its identification in
ET biofilms (sensitivity 33.3% and specificity 100%).
A previous study has demonstrated that decreased
microbial diversity may be associated with VAP among intubated
neonates (21), indicating that it
is an important measurement for VAP surveillance. In the present
study, the findings of similar microbial diversity shared by
tracheal aspirate and ET biofilm samples suggests that the
microbial diversity profile of ET biofilms may be evaluated using
the corresponding tracheal aspirate samples instead. However,
throat swab samples harbored a significantly higher microbial
diversity and richness than ET biofilms, which indicates that they
are not suitable as a substitute to represent the microbial
diversity and richness of ET biofilm samples.
In the present study, the presence of
Staphylococcus sp. was found to be similarly distributed in
tracheal aspirate samples and ET biofilms. Furthermore, the
sensitivity and specificity to detect Staphylococcus sp. in
ET biofilms by tracheal aspirate samples were high (85.7 and 83.3%,
respectively, Table IV). It is well
known that common VAP pathogen Staphylococcus aureus and
nosocomial pathogen Staphylococcus epidermidis are important
members of Staphylococcus sp., especially Staphylococcus
aureus (4). A review based on
the results of the SENTRY Antimicrobial Surveillance Program
(1997–2008, 31,436 cases) reported that Staphylococcus
aureus was the most common causative pathogen (28%) in VAP
(22). Therefore, the detection of
Staphylococcus sp. is important and may be useful for VAP
diagnosis and treatment. The findings of the present study suggest
that the detection of Staphylococcus sp. in tracheal
aspirate may predict its presence in ET biofilms among intubated
neonates.
Previous studies have reported a high concordance of
bacterial culture results between tracheal aspirate samples and ET
biofilms (8,23). For example, in a previous study
conducted among adults undergoing mechanical ventilation in Spain,
identical microorganisms with ET biofilms were isolated in 56% of
tracheal aspirate samples (8).
However, based on a culture-independent approach, the results of
the present study demonstrated that the overall microbial
composition was different in tracheal aspirates compared with ET
biofilm specimens in neonates with VAP, with a significant
difference in the presence of Pseudomonas sp. between
tracheal aspirate and ET biofilm samples (Table III). This suggests that microflora
in tracheal aspirate samples may not entirely represent all the
microbial components correctly in ET biofilms. Pseudomonas
sp. is considered to be a common and important nosocomial
pathogen to cause VAP (4). The
findings of the present study suggest that the presence of
Pseudomonas sp. in tracheal aspirate has little predictive
value of its presence in ET biofilms. This may possibly be due to
the distinct difference in responses to systemic antibiotics
between planktonic and biofilm microorganisms. Bacteria in tracheal
aspirates are present as a planktonic form, which are exposed to
antibiotics and more likely to be killed. However, bacteria in ET
biofilms are considered to be drug-resistant and free from systemic
antibiotics (8,24), which may lead to the disparity in the
levels of Pseudomonas sp. in the two different types of
samples. The similar constituent ratio of Staphylococcus sp.
between tracheal aspirate and ET biofilms suggests the presence of
drug-resistant Staphylococcus sp. strains in tracheal
aspirate in the study samples. This is consistent with several
epidemiological studies conducted in China demonstrating the
relatively high drug-resistant rate of Staphylococcus sp.
strains in NICU (4,25).
Unlike healthy patients (26), gram-negative bacilli such as
Pseudomonas aeruginosa are typically observed in the
oropharynx of critically ill hospitalized patients (22,27).
These bacilli are gram-negative and are considered to be associated
with the onset of nosocomial pneumonia (28). Although a statistical difference was
observed in the constitute ratio of Pseudomonas sp. between
throat swab and ET biofilm samples in the present study, it was
also noted that of the 4 patients with Pseudomonas sp.
present in throat swab, Pseudomonas sp. was also identified
in the corresponding ET biofilms, with a specificity of 100%. This
suggests that throat swabs may have a potential role in
facilitating the detection of Pseudomonas sp. in ET
biofilms. Feldman et al (23)
reported the time sequence of bacterial colonization in VAP
patients, and demonstrated that potential pathogens first colonized
the oropharynx and then the lower respiratory tract. Therefore,
there may be a high consistency of bacteria between oropharynx and
ET biofilms in the early stages of VAP. To confirm this, the study
isolates were classified into early-onset VAP (intubated for <5
days) and late-onset VAP groups (intubated for ≥5 days), and found
the sensitivity for throat swab samples to detect Pseudomonas
sp. in ET biofilms was higher among early-onset VAP (60.0%)
subjects than the late-onset subjects (14.3%). Therefore, the
detection of Pseudomonas sp. in throat swabs may be helpful
to predict its presence in ET biofilms of intubated neonates,
particularly for early-onset VAP patients.
Significant differences were observed in the
detection of Streptococcus sp. between throat swab and ET
biofilm samples in the present study (P<0.05). These results
suggest that throat swab samples are not useful for the
identification of Streptococcus sp in ET biofilms. As the
common colonizers in the oropharynx, Streptococcus sp. is
dominant in the oropharynx among infants (29). The results of the present study
confirmed this, with the detection rate of Streptococcus sp.
being 100.0% in throat swab samples. Simultaneously, the prevalence
of Streptococcus sp. in ET biofilms was found to be 60.0%,
which suggests Streptococcus sp. in ET biofilms may
originate from the oropharynx. It has been reported that
Streptococcus sp. enhanced biofilm formation of other
nosocomial pathogens such as Pseudomonas aeruginosa
(30), which is associated with the
progression of VAP. The results of the present study indicate the
possible source of Streptococcus sp. in ET biofilms among
intubated neonates and suggest the need to develop strategies
against Streptococcus sp. in the oropharynx to prevent VAP
among intubated neonates.
It has been reported that combined culture results
of throat swab, nose swab and sputum increases the sensitivity to
detect nosocomial pathogens in ET biofilms compared with results
from a single sample in intubated adults (14). In the present study, the performance
of a combination of throat swabs and tracheal aspirates was
assessed to identify potential pathogens in ET biofilms (Table IV). The combination of samples
indeed raised the sensitivity, however, decreased the specificity,
which impedes its further application.
The present study has some limitations. Firstly, the
samples size of the present study is small; however, it provides
the preliminary microbial data for neonatal VAP based on the
comprehensive culture-independent method. These findings need to be
confirmed and generalized using a larger sample size of intubated
neonates. Secondly, due to the limited resolution of the 16S rRNA
gene V3 region, the resulting sequences are only able to be
identified and assigned to the genus level. As a result, further
taxonomic information at species level is not provided.
In conclusion, the present study extended the
knowledge base of the microbial characteristics of intubated
neonates with VAP. The results suggest that microbial
investigations in throat swab and tracheal aspirate samples are
beneficial to investigate microbial profile and identify potential
pathogens in ET biofilms among intubated neonates, and suggest that
there are potential applications of using substituent samples to
investigate microbial signatures in ET biofilms for VAP
surveillance in NICU.
Acknowledgements
The authors of the present study would like to thank
Mrs. Xianhong Zhang in the Department of Neonatology, Children's
Hospital of Chongqing Medical University for her excellent
assistance in sample collection. This work was supported by the
National Natural Science Foundation of China (grant no. 81370744),
the Doctoral Degree Funding from Chinese Ministry of Education
(grant no. 20135503110009), the State Key Clinic Discipline Project
(grant no. 2011-873), the Subproject of National Science &
Technology Pillar Program during the 12th Five-year Plan Period in
China (grant no. 2012BAI04B05), and the Clinical Research Program
of Children's Hospital of Chongqing Medical University (grant no.
lcyj2014-11).
Glossary
Abbreviations
Abbreviations:
|
DGGE
|
denaturing gradient gel
electrophoresis
|
|
ET
|
endotracheal tube
|
|
NICU
|
neonatal intensive care unit
|
|
PICU
|
Pediatric intensive care unit
|
|
UPGMA
|
unweighted pair group method with
arithmetic averages
|
|
VAP
|
ventilator-associated pneumonia
|
References
|
1
|
National Nosocomial Infections
Surveillance System, . National Nosocomial Infections Surveillance
(NNIS) System Report, data summary from January 1992 through June
2004, issued October 2004. Am J Infect Control. 32:470–485. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aly H, Badawy M, El-Kholy A, Nabil R and
Mohamed A: Randomized, controlled trial on tracheal colonization of
ventilated infants: Can gravity prevent ventilator-associated
pneumonia? Pediatrics. 122:770–774. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tan B, Zhang F, Zhang X, Huang YL, Gao YS,
Liu X, Li YL and Qiu JF: Risk factors for ventilator-associated
pneumonia in the neonatal intensive care unit: A meta-analysis of
observational studies. Eur J Pediatr. 173:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan B, Xian-Yang X, Zhang X, Peng-Zhou X,
Wang P, Xue J, Ling-Huang Y, Li-Li Y and Fu-Qiu J: Epidemiology of
pathogens and drug resistance of ventilator-associated pneumonia in
Chinese neonatal intensive care units: A meta-analysis. Am J Infect
Control. 42:902–910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Inglis T, Millar M, Jones J and Robinson
D: Tracheal tube biofilm as a source of bacterial colonization of
the lung. J Clin Microbiol. 27:2014–2018. 1989.PubMed/NCBI
|
|
6
|
Adair C, Gorman S, Feron B, Byers L, Jones
D, Goldsmith C, Moore J, Kerr J, Curran M, Hogg G, et al:
Implications of endotracheal tube biofilm for ventilator-associated
pneumonia. Intensive Care Med. 25:1072–1076. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sottile FD, Marrie TJ, Prough DS, Hobgood
CD, Gower DJ, Webb LX, Costerton JW and Gristina AG: Nosocomial
pulmonary infection: Possible etiologic significance of bacterial
adhesion to endotracheal tubes. Crit Care Med. 14:265–270. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gil-Perotin S, Ramirez P, Marti V,
Sahuquillo JM, Gonzalez E, Calleja I, Menendez R and Bonastre J:
Implications of endotracheal tube biofilm in ventilator-associated
pneumonia response: A state of concept. Crit Care. 16:R932012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cairns S, Thomas JG, Hooper SJ, Wise MP,
Frost PJ, Wilson MJ, Lewis MA and Williams DW: Molecular analysis
of microbial communities in endotracheal tube biofilms. PLoS One.
6:e147592011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vandecandelaere I, Matthijs N, Van
Nieuwerburgh F, Deforce D, Vosters P, De Bus L, Nelis HJ, Depuydt P
and Coenye T: Assessment of microbial diversity in biofilms
recovered from endotracheal tubes using culture dependent and
independent approaches. PLoS One. 7:e384012012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Perkins SD, Woeltje KF and Angenent LT:
Endotracheal tube biofilm inoculation of oral flora and subsequent
colonization of opportunistic pathogens. Int J Med Microbiol.
300:503–511. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Charlson ES, Bittinger K, Haas AR,
Fitzgerald AS, Frank I, Yadav A, Bushman FD and Collman RG:
Topographical continuity of bacterial populations in the healthy
human respiratory tract. Am J Respir Crit Care Med. 184:957–963.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pneumatikos IA, Dragoumanis CK and Bouros
DE: Ventilator-associated pneumonia or endotracheal tube-associated
pneumonia? An approach to the pathogenesis and preventive
strategies emphasizing the importance of endotracheal tube.
Anesthesiology. 110:673–680. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vandecandelaere I, Matthijs N, Nelis HJ,
Depuydt P and Coenye T: The presence of antibiotic-resistant
nosocomial pathogens in endotracheal tube biofilms and
corresponding surveillance cultures. Pathog Dis. 69:142–148. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Costerton JW, Post JC, Ehrlich GD, Hu FZ,
Kreft R, Nistico L, Kathju S, Stoodley P, Hall-Stoodley L, Maale G,
et al: New methods for the detection of orthopedic and other
biofilm infections. FEMS Immunol Med Microbiol. 61:133–140. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Hoenig JD, Malin KJ, Qamar S,
Petrof EO, Sun J, Antonopoulos DA, Chang EB and Claud EC: 16S rRNA
gene-based analysis of fecal microbiota from preterm infants with
and without necrotizing enterocolitis. ISME J. 3:944–954. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu H, Qian G, Ren Z, Zhang C, Zhang H, Xu
W, Ye P, Yang Y and Li L: Alterations of Bacteroides sp., Neisseria
sp., Actinomyces sp. and Streptococcus sp. populations in the
oropharyngeal microbiome are associated with liver cirrhosis and
pneumonia. BMC Infect Dis. 15:2392015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu D, Yu J, Li L, Ai Q, Feng J, Song C
and Li H: Bacterial community structure associated with elective
cesarean section versus vaginal delivery in Chinese newborns. J
Pediatr Gastroenterol Nutr. 60:240–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Centers for Disease Control and
Prevention: Criteria for defining nosocomial pneumonia. http://www cdc
gov/ncidod/hip/NNIS/members/pneumonia/Final/PneumoCriteriaV1pdfFebruary
5–2009
|
|
20
|
Cernada M, Brugada M, Golombek S and Vento
M: Ventilator-associated pneumonia in neonatal patients: An update.
Neonatology. 105:98–107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu W, Yu J, Ai Q, Liu D, Song C and Li L:
Increased constituent ratios of Klebsiella sp., Acinetobacter sp.
and Streptococcus sp. and a decrease in microflora diversity may be
indicators of ventilator-associated pneumonia: A prospective study
in the respiratory tracts of neonates. PLoS One. 9:e875042014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jones RN: Microbial etiologies of
hospital-acquired bacterial pneumonia and ventilator-associated
bacterial pneumonia. Clin Infect Dis. 51 Suppl 1:S81–S87. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Feldman C, Kassel M, Cantrell J, Kaka S,
Morar R, Mahomed AG and Philips J: The presence and sequence of
endotracheal tube colonization in patients undergoing mechanical
ventilation. Eur Respir J. 13:546–551. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wolcott RD and Ehrlich GD: Biofilms and
chronic infections. Jama. 299:2682–2684. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu XL, Zhao L, Yang JC, Chen X and Wu XH:
Etiology and high risk factors of neonatal ventilator-associated
pneumonia. Zhongguo Dang Dai Er Ke Za Zhi. 9:549–552. 2007.(In
Chinese). PubMed/NCBI
|
|
26
|
Safdar N, Crnich CJ and Maki DG: The
pathogenesis of ventilator-associated pneumonia: Its relevance to
developing effective strategies for prevention. Respir Care.
50:725–741. 2005.PubMed/NCBI
|
|
27
|
Brennan MT, Bahrani-Mougeot F, Fox PC,
Kennedy TP, Hopkins S, Boucher RC and Lockhart PB: The role of oral
microbial colonization in ventilator-associated pneumonia. Oral
Surg Oral Med Oral Pathol Oral Radiol Endod. 98:665–672. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johanson WG Jr, Pierce AK, Sanford JP and
Thomas GD: Nosocomial respiratory infections with gram-negative
bacilli: The significance of colonization of the respiratory tract.
Ann Intern Med. 77:701–706. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rotimi V and Duerden B: The development of
the bacterial flora in normal neonates. J Med Microbiol. 14:51–62.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song S, Du L, Yu J, Ai Q, Pan Y, Fu Y and
Wang Z: Does Streptococcus mitis, a neonatal oropharyngeal
bacterium, influence the pathogenicity of Pseudomonas aeruginosa?
Microbes Infect. 17:710–716. 2015. View Article : Google Scholar : PubMed/NCBI
|