Introduction
Osteosarcoma is a tumor of mesenchymal origin that
accounts for ~20% of all primary bone cancers. Osteosarcoma
develops primarily among children and adolescents and demonstrates
a low 5-year survival rate, a high amputation rate and a poor
post-operative function recovery rate (1,2). Long
non-coding RNAs (lncRNAs) are >200 nucleotides in length and are
non-protein-coding capacity transcripts. Previous studies have
demonstrated that lncRNAs regulate the expression of genes at
different levels through a variety of mechanisms, including
chromatin modification, transcription, splicing, translation and
post-transcriptional regulation (3,4). It has
been demonstrated that lncRNAs also serve important roles in the
biological processes of cancer cells, including cell proliferation,
invasion, differentiation and apoptosis (5–7).
Deregulated lncRNA expression has been observed in various cancers,
which suggests that lncRNAs may serve a vital regulatory function
in tumorigenesis and cancer progression (8,9).
The ANRIL lncRNA is transcribed as a 3.8 kb mRNA in
the opposite orientation of INK4b-ARF-INK4a gene cluster, which has
been identified as a genetic susceptibility locus associated with
coronary disease, intracranial aneurysm, type 2 diabetes and
various types of cancer (10,11). In
addition, ANRIL activates polycomb repressive complexes (PRC) 1 and
PRC2, to regulate the expression of INK4b-ARF-INK4a (12,13).
Zhang et al (14)
demonstrated that ANRIL expression was increased in gastric cancer
tissues, and was associated with tumor size and tumor, node,
metastasis (TNM) stage. Further studies have demonstrated that
ANRIL knockdown significantly inhibits the proliferation and
invasion of cancer cells in vitro and in vivo
(14). Huang et al (15) identified that the expression of
lncRNA ANRIL is increased in hepatocellular carcinoma, and
demonstrated that it serves a role in tumor proliferation and
metastasis. The results of these studies therefore suggest that
dysregulation of ANRIL may be a contributing factor in human cancer
progression. However, the functional role and underlying mechanisms
of action of ANRIL in osteosarcoma remain unknown.
The present study investigated the biological
functions of the ANRIL lncRNA in osteosarcoma development by
examining the expression of ANRIL in osteosarcoma tissues. In
vitro assays were subsequently performed to identify the
biological functions of ANRIL in an osteosarcoma cell line.
Materials and methods
Patients and tissue samples
Osteosarcoma tissues and their adjacent
non-cancerous tissues (ANCT) were obtained from 19 patients (10
males and 9 females; 15–35 years) pathologically diagnosed with
osteosarcoma that underwent resection of osteosarcoma at the First
Affiliated Hospital of China Medical University (Shenyang, China)
between July 2010 and July 2014. None of the patients received
therapy prior to surgery. Patients at stage IIB/III were included,
and patients with any other primary disease were excluded. All
tissues were immediately stored in liquid nitrogen and maintained
at −80°C. The associated clinical data (including sex, age, TNM
stage) was collected from each of the patient's medical records.
The current study was approved by the Ethics Committee of the First
Affiliated Hospital of China Medical University, and informed
consent was obtained from all participants.
Cell culture
The human osteosarcoma cell line U2OS (Shanghai
Gefan Biotechnology, Shanghai, China) was cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; PAN-Biotech GmbH,
Aidenbach, Germany) and 5% horse serum (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in 5% CO2.
Lentivirus-mediated RNA
interference
U2OS cells were transfected with lentivirus core
vector (hU6-MCS-CMV-RFP) carrying shRNA targeting human ANRIL
(si-ANRIL) and non-targeting control (si-NC),
5′-TTCTCCGAACGTGTCACGT-3′ (GeneChem Co., Ltd., Shanghai, China).
The sequence of the shRNA target for ANRIL was
5′-GGUCAUCUCAUUGCUCUAU-3′ (GeneChem Co., Ltd.), which has been
identified as an effective interference target sequence of ANRIL by
Kotake et al (12). U2OS
cells were infected at a multiplicity of infection of 20, and
incubated for 72 h prior to performing the subsequent assays.
Cell proliferation assays
Cell viability was measured using an MTT kit
(Nanjing KeyGen Biotech Co., Ltd., Jiangsu, China) according to the
manufacturer's instructions. A total of 3,000 transfected
cells/well were seeded in 96-well plates and incubated at 37°C in
5% CO2 for 1 day. Then, 50 ml MTT solution was added
into the medium for 4 h incubation at 37°C in 5% CO2,
then each well was replaced with 150 ml dimethyl sulfoxide. The
absorbance of each sample was recorded at 490 nm. Cell
proliferation was analyzed at 24, 48, 72 and 96 h.
The colony formation assay was performed by seeding
transfected cells into each well of a 6-well plate (500 cells/well)
containing RPMI-1640 medium supplemented with 10% FBS, and
incubating the cells for 2 weeks. The medium was refreshed every 4
days. Colonies were fixed with pure methanol for 15 min at room
temperature and stained with 0.1% crystal violet in PBS for 15 min
at room temperature. The total number of stained colonies was
counted. The experiments were performed in triplicate.
Cell migration and invasion
assays
At 48 h following transfection, U2OS cells were
harvested. The migration assay was performed by seeding
1×105 transfected cells in the upper chamber of a
Transwell insert (Corning Incorporated, Corning, NY, USA). The
invasion assay was performed by seeding 1×105
transfected cells in the upper chamber containing RPMI-1640 medium
without serum and coated with Matrigel (Corning Incorporated). The
inserts were placed in the lower chamber wells of a 24-well plate
containing Dulbecco's modified Eagle's medium supplemented with 10%
FBS. Following 24 h incubation at 37°C in 5% CO2, cells
remaining on the upper membrane were removed by scraping with a
cotton swab. The migrated or invaded cells on the lower membrane
were fixed with pure methanol for 10 min at room temperature and
stained with 0.1% crystal violet for 15 min at room temperature.
Cell numbers were counted under a Leica DMi8 inverted microscope
(Leica Microsystems GmbH, Wetzlar, Germany) at magnification, ×200.
Experiments were independently repeated in triplicate.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific
Inc.). cDNA synthesis was performed using the PrimeScript RT
Reagent kit with gDNA Eraser (cat. no. RR047A; Takara Biotechnology
Co., Ltd., Dalian, China). qPCR was performed with SYBR Premix Ex
Taq (Takara Biotechnology Co., Ltd.). The results were normalized
to the expression of GAPDH. The sequences of specific RNA primers
for ANRIL and GAPDH were as follows: ANRIL forward,
5′-CTGATTCAACAGCAGAGATCAAAGA-3′ and reverse,
5′-CACACCTAACAGTGATGCTTGAAC-3′; GAPDH forward,
5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′. RT-qPCR analysis and data collection
were performed using an Applied Biosystems 7900 Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: Initial denaturation
stage (95°C for 30 sec); PCR reaction stage (40 cycles of 95°C for
5 sec and 60°C for 34 sec); dissociation stage (95°C for 15 sec,
60°C for 1 min and 95°C for 15 sec). The relative expression of
ANRIL was calculated and normalized using the 2−∆∆Cq
method (16).
Statistical analysis
All statistical analyses were performed using SPSS
18.0 (SPSS, Inc., Chicago, IL, USA). Significant differences
between groups were estimated using a Student's t-test. P<0.05
was determined to indicate statistically significant
difference.
Results
ANRIL is upregulated in osteosarcoma
tissues
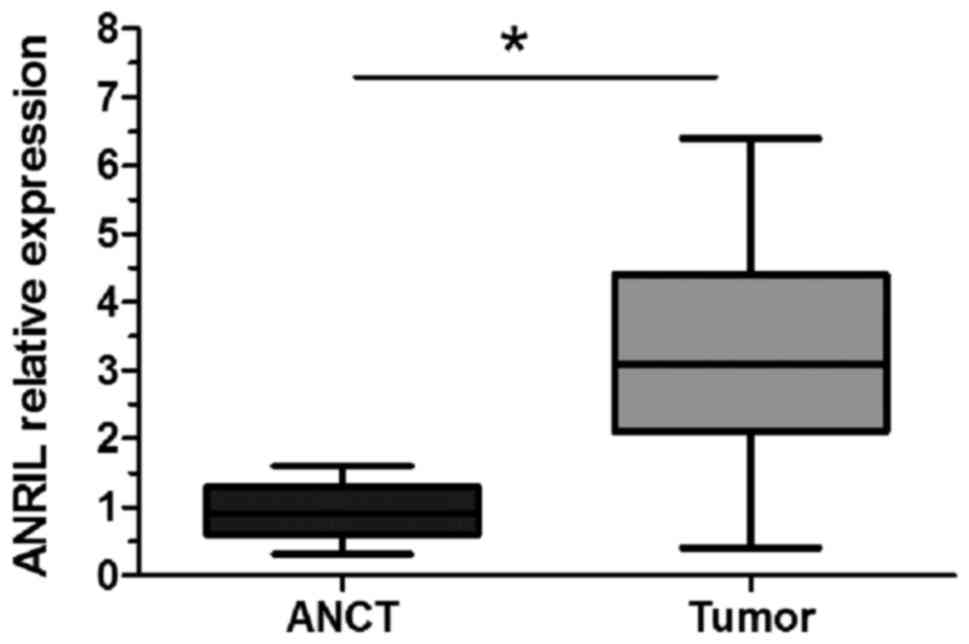
RT-qPCR was performed to detect the level of ANRIL
expression in 19 osteosarcoma tissues and ANCT samples. The results
demonstrated that ANRIL expression was significantly upregulated in
osteosarcoma tissues when compared with ANCT (P<0.05; Fig. 1).
Knockdown of ANRIL inhibits the
proliferation of osteosarcoma cells
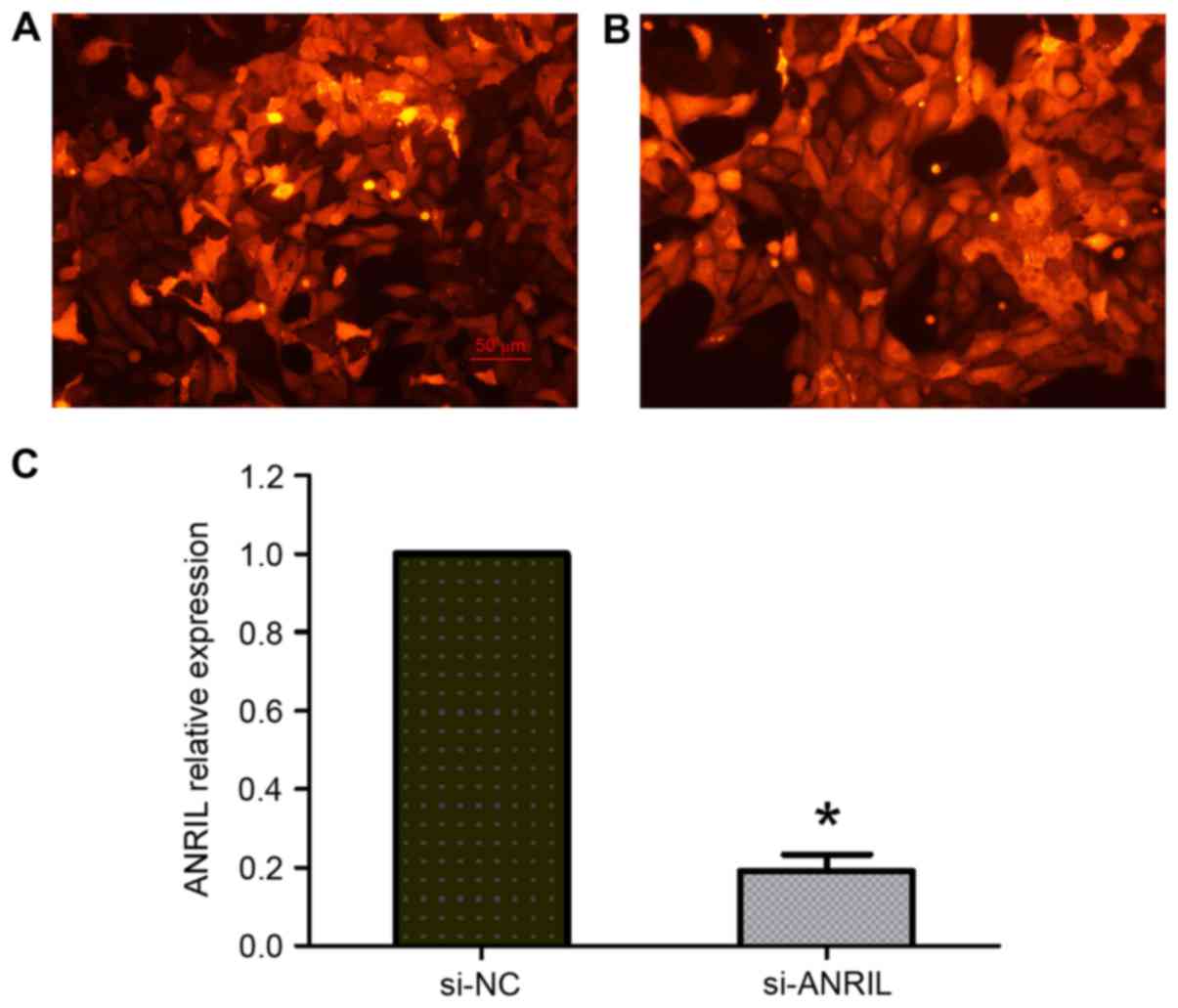
The function of ANRIL on the proliferation of
osteosarcoma cells was investigated by lentivirus-mediated
knockdown of ANRIL expression in U2OS cells. Transfected cells
labeled with red fluorescent protein were observed under a
fluorescence microscope, and the transfection efficiency was ~80%
(Fig. 2A and B). The RT-qPCR results
demonstrated that ANRIL expression was significantly decreased in
U2OS cells transfected with si-ANRIL compared with the si-NC group
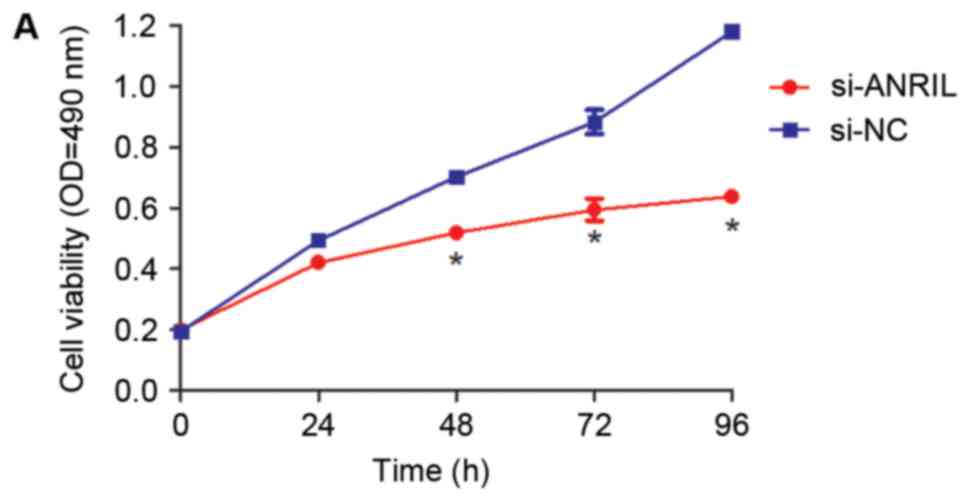
(P<0.05; Fig. 2C). The MTT assay
results demonstrated that knockdown of ANRIL expression
significantly decreased the proliferation of U2OS cells when
compared with the si-NC group at 48, 72 and 96 h following
transfection (P<0.05; Fig. 3A).
Similarly, the colony formation assay indicated that knockdown of
ANRIL significantly decreased the number of U2OS cell colonies
(P<0.05; Fig. 3B).
Downregulation of ANRIL inhibits the
invasion and migration of osteosarcoma cells
The effect of knockdown of ANRIL expression on
osteosarcoma cell invasion and migration was investigated using a
Transwell invasion and migration assay, respectively. The results
demonstrated that the invasion ability of U2OS osteosarcoma cells
transfected with si-ANRIL was significantly decreased compared with
the si-NC group (P<0.05; Fig. 3C and
D). Similar results were observed in the migration assay,
whereby U2OS cells transfected with si-ANRIL exhibited a
significantly lower level of migration when compared with the si-NC
group (P<0.05; Fig. 3C and D).
These results suggest that ANRIL may promote osteosarcoma cell
metastasis.
Discussion
Up to 70% of the human genome is transcribed into
RNA, and only ~2% of the genome is composed of protein-encoding
genes (1). Therefore, the human
genome contains higher non-coding information than coding
information, which subsequently leads to the expression of a
greater number of non-coding RNA (ncRNA) transcripts. NcRNAs are
commonly divided into small ncRNAs and lncRNAs. LncRNAs are >200
nucleotides in length, are non-coding and lack an open reading
frame.
LncRNAs are emerging as novel gene regulators that
are associated with various human diseases, including different
cancers (17–19). It has been demonstrated that the
dysregulation of lncRNAs may serve a role in tumorigenesis and
cancer progression (3). Gupta et
al (20) demonstrated that the
expression of hox transcript antisense RNA (HOTAIR) lncRNA was
increased in primary breast tumors, and may be a useful predictor
of subsequent metastasis and mortality. In addition, growth arrest
specific 5 has been identified to promote prostate cancer cell
apoptosis, and as the cells acquire castrate-resistance, its
expression decreases (8). Wang et
al (21) demonstrated that the
expression of the PlncRNA-1 lncRNA was significantly higher in
human esophageal squamous cell carcinoma, and was correlated with
lymph node metastasis and an advanced clinical stage. Knockdown of
PlncRNA-1 expression inhibited cell proliferation and increased
apoptosis in vitro (21).
Previous studies have also indicated that transcription factors,
c-myc and p53, activate HOTAIR transcription and regulate PVT-1
expression, respectively (22,23). The
results of these studies suggest that the aberrant expression of
lncRNAs may be associated with the progression of multiple tumor
types, and may be useful as a prognostic indicator. However, the
function of ANRIL in osteosarcoma remains unknown.
The ANRIL lncRNA is transcribed as a 3.8-kb mRNA in
the opposite orientation of the INK4b-ARF-INK4a gene cluster. The
INK4b-ARF-INK4a locus encodes three critical tumor suppressors,
p16INK4A, p14ARF and p15INK4B. Common disease genome wide
association studies have identified the ANRIL gene as a genetic
susceptibility locus associated with coronary disease, intracranial
aneurysm and type 2 diabetes. ANRIL interacts with p16INK4A, p14ARF
and p15INK4B. When ANRIL combines with chromobox 7 (CBX7) and SUZ12
polycomb repressive complex 2 subunit (SUZ12), PRC1 and PRC2, which
serve a role in chromatin modification, are activated,
respectively. ANRIL promotes the interaction between SUZ12 (a
component of PRC2) and the p15INK4B locus, which inhibits its
expression, thus leading to an increase in cellular proliferation
(12). Zhang et al (14) demonstrated that ANRIL lncRNA
expression was increased in gastric cancer tissues and was
associated with tumor size and TNM stage, and also have indicated
that ANRIL knockdown significantly inhibits proliferation and
invasion in vitro and in vivo, for instance in lung
cancer cells, gastric cancer cells and liver cancer cells (14,15,24). In
addition, knockdown of ANRIL upregulates the expression of microRNA
(miR)-99a/miR-449a in gastric cancer cell lines, SGC-7901 and
BGC-823 (14). Yap et al
(10) demonstrated that ANRIL and
CBX7 are upregulated in prostate cancer tissues, and high
expression of ANRIL and CBX7 inhibits INK4A transcription. In
addition, Lin et al (24)
demonstrated that ANRIL expression was increased in human non-small
cell lung cancer, and the aberrant expression of ANRIL was
correlated with tumor size and TNM stage. These results indicate
that the dysregulation of ANRIL may serve a role in the progression
of a variety of human cancers. However, the functional role and
underlying mechanism of ANRIL in osteosarcoma remains unknown.
In the present study, the expression of ANRIL was
observed to be upregulated in osteosarcoma tissues when compared
with ANCT, which suggests that ANRIL may serve an important role in
osteosarcoma development and progression. To investigate the
underlying mechanisms of the ANRIL lncRNA in osteosarcoma
progression further, the expression of ANRIL was reduced in U2OS
osteosarcoma cells by lentivirus-mediated RNA interference. RT-qPCR
analysis was subsequently performed to determine the expression of
ANRIL in U2OS osteosarcoma cells following transfection with
si-ANRIL or si-NC. MTT, colony formation and transwell assays were
used to analyze the proliferation, migration and invasion capacity
of transfected U2OS osteosarcoma cells. The results of the current
study demonstrated that ANRIL knockdown significantly decreased the
proliferation, migration and invasion of osteosarcoma cells in
vitro. This indicates the importance of ANRIL in the cellular
biology and oncogenesis of osteosarcoma cells.
In conclusion, ANRIL was observed to be upregulated
in human osteosarcoma tissues. In addition, knockdown of ANRIL
reduced osteosarcoma cell proliferation, invasion and migration
in vitro. The current study identified ANRIL expression as a
potential novel diagnostic marker and therapeutic target of
osteosarcoma; however, further study is required in order to
identify the exact pathway through which ANRIL influences
osteosarcoma, and how this pathway works.
Acknowledgements
The current study was supported by the Natural
Science Foundation of Science and Technology Department of Liaoning
Province (grant no. 201302106) and the Science and Technology
Department of Shenyang City (grant no. F14-231-1-48).
References
|
1
|
Huang J, Ni J, Liu K, Yu Y, Xie M, Kang R,
Vernon P, Cao L and Tang D: HMGB1 promotes drug resistance in
osteosarcoma. Cancer Res. 72:230–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clark JC, Dass CR and Choong PF: A review
of clinical and molecular prognostic factors in osteosarcoma. J
Cancer Res Clin Oncol. 134:281–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ginger MR, Shore AN, Contreras A, Rijnkels
M, Miller J, Gonzalez-Rimbau MF and Rosen JM: A noncoding RNA is a
potential marker of cell fate during mammary gland development.
Proc Natl Acad Sci USA. 103:5781–5786. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yacqub-Usman K, Pickard MR and Williams
GT: Reciprocal regulation of GAS5 lncRNA levels and mTOR inhibitor
action in prostate cancer cells. Prostate. 75:693–705. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, et al: Decreased expression of
the long non-coding RNA FENDRR is associated with poor prognosis in
gastric cancer and FENDRR regulates gastric cancer cell metastasis
by affecting fibronectin1 expression. J Hematol Oncol. 7:632014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yap KL, Li S, Muñoz-Cabello AM, Raguz S,
Zeng L, Mujtaba S, Gil J, Walsh MJ and Zhou MM: Molecular interplay
of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by
polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell.
38:662–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pasmant E, Laurendeau I, Heron D, Vidaud
M, Vidaud D and Bieche I: Characterization of a germ-line deletion,
including the entire INK4/ARF locus, in a melanoma-neural system
tumor family: Identification of ANRIL, an antisense noncoding RNA
whose expression coclusters with ARF. Cancer Res. 67:3963–3969.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aguilo F, Zhou MM and Walsh MJ: Long
noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a
expression. Cancer Res. 71:5365–5369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang EB, Kong R, Yin DD, You LH, Sun M,
Han L, Xu TP, Xia R, Yang JS, De W and Chen Jf: Long noncoding RNA
ANRIL indicates a poor prognosis of gastric cancer and promotes
tumor growth by epigenetically silencing of miR-99a/miR-449a.
Oncotarget. 5:2276–2292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang MD, Chen WM, Qi FZ, Xia R, Sun M, Xu
TP, Yin L, Zhang EB, De W and Shu YQ: Long non-coding RNA ANRIL is
upregulated in hepatocellular carcinoma and regulates cell
apoptosis by epigenetic silencing of KLF2. J Hematol Oncol.
8:502015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang CM, Wu QQ, Li SQ, Chen FJ, Tuo L, Xie
HW, Tong YS, Ji L, Zhou GZ, Cao G, et al: Upregulation of the long
non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma
cell proliferation and correlates with advanced clinical stage. Dig
Dis Sci. 59:591–597. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma MZ, Li CX, Zhang Y, Weng MZ, Zhang MD,
Qin YY, Gong W and Quan ZW: Long non-coding RNA HOTAIR, a c-Myc
activated driver of malignancy, negatively regulates miRNA-130a in
gallbladder cancer. Mol Cancer. 13:1562014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barsotti AM, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: p53-Dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin L, Gu ZT, Chen WH and Cao KJ:
Increased expression of the long non-coding RNA ANRIL promotes lung
cancer cell metastasis and correlates with poor prognosis. Diagn
Pathol. 10:142015. View Article : Google Scholar : PubMed/NCBI
|