Introduction
Hepatocellular carcinoma (HCC) is a common liver
cancer; a previous report demonstrated that it is the third most
frequent cause of cancer-related mortality worldwide (1). The incidence of HCC has increased
markedly in previous years, due to the increasing incidence of
viral infections, including hepatitis B and C, in addition to other
causes of cirrhosis, such as alcohol abuse (2–4).
Therapeutic strategies for this malignancy include surgical hepatic
resection or liver transplantation. However, the overall 5-year
survival rate remains only ~5% (5).
It is currently understood that HCC is an epithelial tumor with its
origin from mature hepatocytes or stem cells (6). Hepatocarcinogenesis has been considered
a multistep process involving different genetic alterations that
ultimately lead to malignant transformation (7). Therefore, an improved understanding of
the molecular pathways involved in the development and progression
of HCC may identify novel therapeutic strategies to improve the
treatment of this malignancy.
Previously, advances have been made in the research
and identification of regulatory RNA families, particularly those
of long non-coding RNAs (lncRNAs), which are currently defined as
transcripts of >200 nucleotides without evident protein coding
capacity (8,9). In previous research, lncRNAs have
served as key components of the address code, allowing genes,
chromosomes and protein complexes to be trafficked to their
destinations subject to activation or deactivation (8). Mounting evidence has established the
association between lncRNAs and tumorigenesis (10). HOTAIR, for example, is one common
lncRNA that has been reported to reprogram the chromatin state to
promote cancer metastasis (11).
Another non-protein coding lncRNA, growth arrest specific 5, has
been reported to regulate breast cancer cell apoptosis (12). In addition, lncRNAs have also emerged
as potential diagnostic or prognostic biomarkers for a number of
human malignancies, including lung, breast and colon cancer
(13–15).
Notably, lncRNAs comprise natural antisense
transcripts and long intergenic non-coding RNAs (lincRNAs).
LincRNAs, originating from the intergenic regions of certain genes,
function as competitive endogenous RNAs and sequester miRNAs (hence
the term ‘miRNA sponges’) (16), and
target chromatin modification complexes or RNA-binding proteins to
alter gene expression (17).
Previous evidence has revealed that lincRNAs have notable roles in
multiple stages of cancer progression (18). The first identified lincRNA, HOTAIR,
has been demonstrated to promote metastasis in breast cancer
(11), and has a prognostic role and
exhibits oncogenic activity in pancreatic cancer (19). Another lincRNA, integrin subunit β1
(ITGB1; linc-ITGB1), has recently been a focus of investigation. It
was demonstrated that linc-ITGB1 was aberrantly expressed in breast
and gallbladder cancer. A knockdown of linc-ITGB1 by short hairpin
RNA (shRNA) significantly inhibited cell migration and invasion
abilities in breast and gallbladder cancer (20,21).
These data suggest that linc-ITGB1 may be critically involved in
the progression of human cancer. However, the implication of
linc-ITGB1 in other types of solid tumors, such as HCC, remains
largely unknown.
The current study aimed to investigate the role of
linc-ITGB1 in cell proliferation and migration in HCC cells. A
specific shRNA against linc-ITGB1 (shlinc-ITGB1) was synthesized
and employed to deplete the expression of linc-ITGB1 in HCC cells.
The effects of linc-ITGB1 knockdown on cell proliferation and
migration were examined using Cell Counting Kit-8 and Transwell
assays, respectively. The results of the present study suggest that
linc-ITGB1 has a pro-oncogenic activity in HCC. Thus, the findings
of the current study may provide a novel insight into the diagnosis
and treatment of HCC.
Materials and methods
Patient samples
A total of 30 patients (45±5 years old; male:
female, 18:12) with liver cancer were included in the present
study. The patients had never received any radiotherapy or
chemotherapy. Liver cancer tissues and their adjacent non-cancerous
normal tissues were collected from Weifang People's Hospital
(Weifang, China). The tissues were frozen with liquid nitrogen
immediately. The current study was approved by the Ethical
Committee of Weifang People's Hospital. Full informed consent to
participate in the study was obtained from all patients.
Cell culture and shRNA infection
The human liver cancer cell lines, MHCC97-L,
MHCC97-H and HCCLM3, were purchased from the Shanghai Institute of
Cell Biology, (Shanghai, China) and cultured in Dulbecco's modified
Eagle's medium (DMEM, Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.). All cell culture was
maintained at 37°C and 5% CO2 in a humidified incubator.
The specific shRNA against human Linc-ITGB1 was designed and
chemically synthesized by Invitrogen (Thermo Fisher Scientific,
Inc.). The sequence was as follows:
5′-GCAGCTGTTTCCAGAATATTGCTCGAGCAATATTCTGGAAACAGCTGC-3′ and control
sequence:
5′-GCGGAGGGTTTGAAAGAATATCTCGAGATATTCTTTCAAACCCTCCGCTTTTTT-3′. Using
this shRNA, construction of a lentivirus-delivered short hairpin
RNA against Linc-ITGB1 was completed by Shanghai GenePharma, Co.,
Ltd. (Shanghai, China). For lentivirus infection, HCCLM3 cells were
seeded into a 6-well plate (5×104 cells per well) and
infected with the lentivirus at a multiplicity of infection of 50.
Following 96 h of incubation at 37°C, fluorescence microscopy was
used to examine the infection efficiency by calculating the
percentage of red fluorescence protein-positive cells. For nuclear
analysis, staining was performed using
4′,6-diamidino-2-phenylindole dye (Beyotime Institute of
Biotechnology, Dalian, China) for 10 min at room temperature at a
dilution of 1:1,000.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Whole RNA from human tissues and cultured liver
cancer cells were extracted using TRIzol reagent (Takara
Biotechnology, Co., Ltd., Dalian, China) following the
manufacturer's protocol. The concentration and quality of each
sample were measured by reading the absorbance at 260 and 280 nm
with a Nanodrop 2000 (Thermo Fisher Scientific, Inc.). RNA was
purified using the TRIzol reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol and then PrimeScript RT
Master Mix (Perfect Real Time; Takara Biotechnology, Co., Ltd.) was
used to obtain the first-strand cDNAs. PCR was performed in an ABI
PRISM 7900 Sequence detection system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the SYBR Premix Ex Taq kit
(Takara Biotechnology, Co., Ltd.). The thermocycling conditions
were as follows: 95°C for 5 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 30 sec. The primers of each gene included were
as follows: Lnc-ITGB1, forward 5′-CCTCTCAGCCTCCAGCGTTG-3′ and
reverse 5′-TGCTCTTGCTCACTCACACTCC-3′; and GAPDH (internal control),
forward 5′-ATGTCTTTCCGTGTTCCTACTGT-3′ and reverse
5′-TTTCCCTCAGACTCCTCCTTG-3′. All quantitative data were normalized
to GAPDH using the 2−∆∆Cq method (22).
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) assay (Beyotime
Institute of Biotechnology) was performed to evaluate the
proliferation ability of liver cancer cell line HCCLM3 with and
without administration of specific shRNAs against Linc-ITGB1
(shLinc-ITGB1). In total, 2×103 HCCLM3 cells were seeded
into 96-well plates and cultured for 12 h. Cells were then treated
with either the control shRNA or shLinc-ITGB1 (20 nM) and incubated
at 37°C for another 72 h. Cell viability was determined within five
consecutive days using MTT solution (Beyotime Institute of
Biotechnology). On each monitored day, 2 mg/ml of MTT reagent was
added into each well prior to another 4 h incubation at 37°C.
Following this incubation, culture medium was discarded and 200 µl
of dimethyl sulfoxide was added to each well. The plates were then
agitated for 5 min and the optical density was obtained at an
absorbance of 570 nm with a micoplate reader (Tecan Group Ltd.,
Männedorf, Switzerland).
Colony formation assay
HCCLM3 liver cancer cells (2,000 cells/well) were
pretreated with specific shRNA against Linc-ITGB1, resuspended in 2
ml of 0.4% agarose solution and DMEM/F12 or F12 culture medium
(Gibco; Thermo Fisher Scientific, Inc.) and overlaid onto a 0.8%
bottom agar layer in 6-well plates. The plates were incubated at
37°C for 3 to 6 weeks and colonies were calculated by counting the
number of colonies >80 µm in diameter. All separate experiments
were repeated at least three times in triplicate to obtain the mean
and standard deviations on the basis of the colony counts.
Cell cycle determination by flow
cytometry
The culture medium was discarded from a sample of
4×105 HCCLM3 liver cancer cells. Cells were washed with
phosphate-buffered saline (PBS) twice prior to digestion with a
Trypsin/EDTA solution. Following this, cells were incubated at 37°C
in an incubator containing 5% CO2 until they were
detached from the surface of the plate (~1 min). The cells
collected were scattered with PBS and subsequently gathered by
low-speed centrifugation (840 × g for 5 min at 4°C). Cells were
mixed with 70% ice-cold alcohol and stored at −20°C overnight. The
mixture was then centrifuged (840 × g for 5 min at 4°C) and the
alcohol solution was removed to obtain the cell pellet. Following
resuspension in PBS, 0.1% Triton-100, 4 mg/ml of RNase A (Beyotime
Institute of Biotechnology) and 2 mg/ml of propidium iodide (PI)
were added into the cells. Cells were then incubated at room
temperature in the dark for 30 min, a 35-micron nylon mesh was
included to filter the cell suspension. A flow cytometry instrument
(Beckman Coulter, Inc., Brea, CA, USA) and cell cycle analysis
software (Modfit Version 4.0; Verity Software House, Topsham, ME,
USA) were used to analyze the cell cycle distribution in HCCLM3
liver cancer cells, according to the manufacturers' protocols.
Transwell assay
Migration and invasion assays were performed in
triplicate with a chemotaxis chamber (8-µm pore size; Corning Inc.,
Corning, NY, USA). HCCLM3 cells were administered shRNAs 48 h prior
to harvest and resuspended in DMEM culture medium without any FBS.
A total of 1×104 cells (~200 µl) were seeded into the
upper chambers and lower chambers were filled with 600 µl DMEM
media supplied with 10% FBS. Following 12 h of free migration, the
membrane was fixed with cold 100% methanol (15 min, 4°C) and
stained with crystal violet for 5 min. Cells were quantified by
using a light microscope (magnification, ×200) to count cells that
had migrated through the membrane. For the invasion assay, the
membrane of the chamber was coated with a Matrigel solution
(Corning Inc.) 6 h prior to the experiment in a 37°C incubator.
Cells were allowed to invade for 12 h prior to staining by crystal
violet. All procedures were repeated in triplicate.
Statistical analysis
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. All results are presented as the
mean + standard deviation. Student's t-test was performed to
determine whether differences between groups were significant. Each
experiment was repeated at least three times in triplicate.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Linc-ITGB1 is highly expressed in HCC
tissues and in the invasive HCC cell line
Initially, the expression of linc-ITGB1 in clinical
HCC tissues was assessed by RT-qPCR. It was demonstrated that the
relative transcription level of linc-ITGB1 in cancerous tissues
were significantly increased (4-fold) compared with their paired
adjacent non-cancerous tissues (Fig.
1A; P<0.05). Furthermore, three increasingly invasive HCC
cell lines (MHCC97-L, MHCC97-H and HCCLM3) were cultured. It was
demonstrated that the relative transcription level of linc-ITGB1
was the highest in the most invasive HCCLM3 cells, while the least
expression of linc-ITGB1 was observed in the least invasive cell
line, MHCC97-L (Fig. 1B), indicating
that a high expression of linc-ITGB1 was associated with
invasiveness in HCC. These observations suggest that linc-ITGB1 is
overexpressed in HCC tissues.
Specific shRNA against linc-ITGB1 is
effective to knockdown the expression of linc-ITGB1 in HCCLM3
cells
Specific shRNAs against linc-ITGB1 (shLinc-ITGB1)
were synthesized to deplete the expression of linc-ITGB1 in HCCLM3
cells. The HCCLM3 cell line was selected because it exhibited the
highest expression of linc-ITGB1 (Fig.
1B). Following transfection of shLinc-ITGB1, it was observed
that the majority of cells (90%) exhibited red fluorescence
(Fig. 2A), indicating a high
efficiency of transfection. Total RNA was then isolated and it was
further detected that the transcript level of linc-ITGB1 was
significantly depleted by up to 60% in shLinc-ITGB1-transfected
cells (P<0.05), while control shRNA-transfected cells
demonstrated almost no change in linc-ITGB1 expression (Fig. 2B). These data suggest that
synthesized shRNA is effective at depleting the expression of
linc-ITGB1.
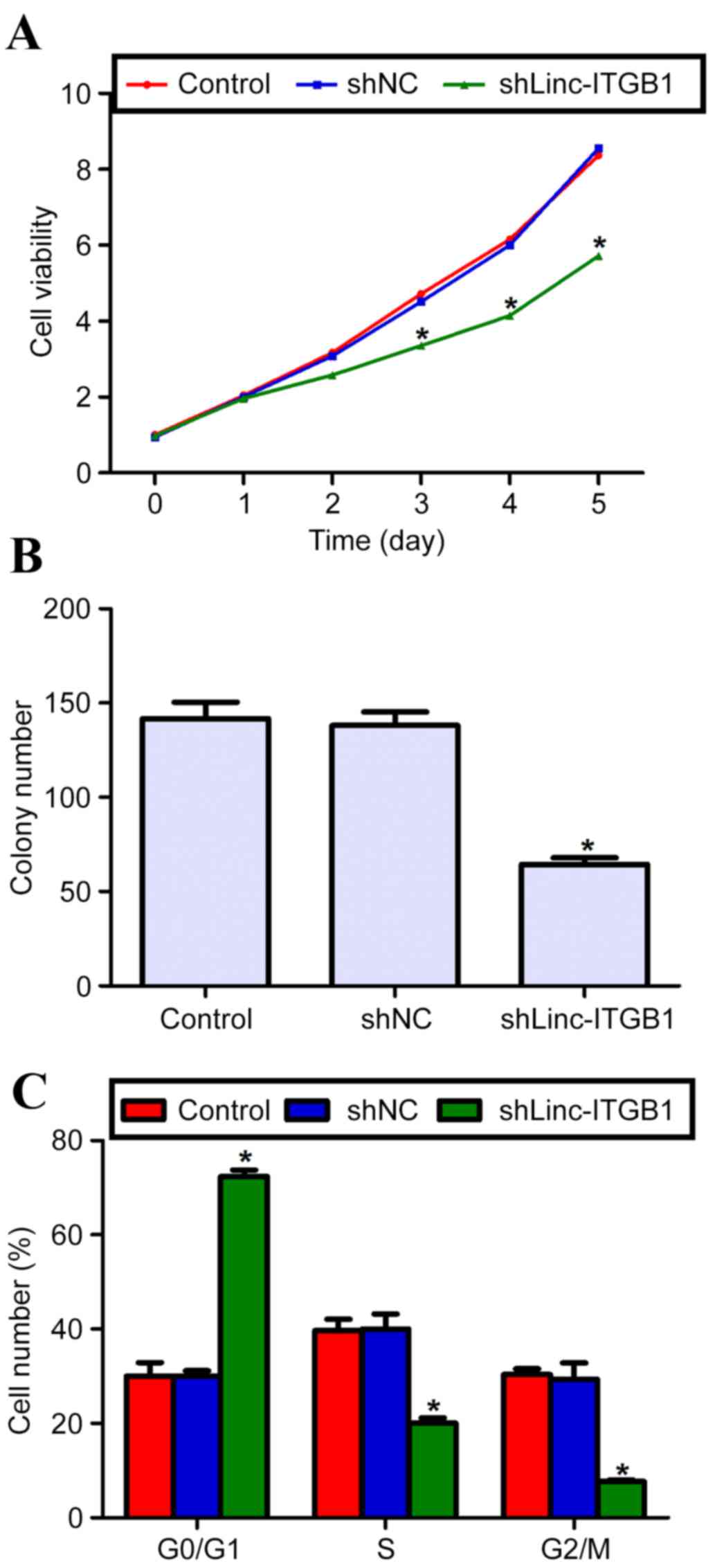
Knockdown of linc-ITGB1 inhibits cell
proliferation and colony formation in vitro
Using shLinc-ITGB1, the effects of linc-ITGB1
knockdown on cell growth in vitro were assessed. Following
the CCK-8 assay, there was no significant difference in cell
proliferation observed among the three groups for the first 2 days.
However, cell proliferation in the shLinc-ITGB1-transfected HCCLM3
cells was significantly reduced from the third day (Fig. 3A; P<0.05). Cell viability was
significantly inhibited in the shLinc-ITGB1 group when compared
with either control or scramble shRNA (shNC) group (Fig. 3A; P<0.05). Colony formation
ability was assessed by counting the colonies and it was
demonstrated that the control and shNC groups exhibited a similar
colony formation ability, by forming an equal amount of 140
colonies. However, in the shLinc-ITGB1 group, only 62 colonies were
counted, which is a significant 57.4% decrease compared with its
counterpart (Fig. 3B; P<0.05).
These data suggest that knockdown of linc-ITGB1 significantly
eliminates the cell proliferative and colony forming abilities of
the highly invasive HCCLM3 cells.
Knockdown of linc-ITGB1 arrests cell
cycle in G0/G1 phase in HCCLM3 cells
To explain the inhibited cell proliferation by
knockdown of linc-ITGB1, cell cycle progression was analyzed. The
percentage of cells in each phase was quantified. It was
demonstrated that control and shNC cell groups had a similar
percentage of cells in each phase (G0/G1, S and G2/M phase;
Fig. 3C). However, in the
shLinc-ITGB1 group, cells in the G0/G1 phase were significantly
increased, with the cell percentage as high as 72% in this phase
(compared with ~30% in the control group; Fig. 3A; P<0.05). Accordingly, the
percentage of cells in S or G2/M phase was significantly decreased
in the shLinc-ITGB1 group (Fig. 3C;
P<0.05). These data suggest that cell cycle progression is
arrested by a knockdown of linc-ITGB1 in HCCLM3 cells.
Knockdown of linc-ITGB1 inhibits cell
migration and invasion in HCCLM3 cells
In the three cultured cell lines, HCCLM3 cells
exhibited the highest invasive ability. To assess the effects of
linc-ITGB1 knockdown on cell migration and invasion, HCCLM3 cells
were further cultured and transfected with shLinc-ITGB1. Following
12 h of free migration, cells were stained with crystal violet.
Images of three randomly selected fields were captured (Fig. 4A). Transmigrated cells were counted
and the cell number of each group is presented in Fig. 4B. In the migration assay, ~500 cells
migrated to the lower surface of the chamber in the control or shNC
group. However, only an average of 214 linc-ITGB1-depleted cells
successfully migrated, a significant 57.2% decrease compared with
control cells (P<0.05). Similarly, in the invasion assay, ~200
cells invaded through the Matrigel in the control or shNC group.
However, only ~70 cells invaded through the Matrigel coat to the
lower surface of the chamber (Fig.
4B; P<0.05). These observations indicate that knockdown of
linc-ITGB1 significantly inhibits cell migration and invasion
abilities in HCCLM3 cells (P<0.05).
Discussion
HCC is a prevalent primary malignancy characterized
by aggressive invasion, early metastasis and resistance to existing
chemotherapeutics (1). Despite an
increase in the 5-year survival rate, certain patients suffering
from HCC remain threatened by poor prognosis due to the aggressive
growth and metastasis of HCC (23,24).
Thus, a further understanding of the molecular mechanisms
underlying the progression and metastases of HCC may improve the
current therapy (25).
lncRNA has emerged as a key component of the address
code allowing genes, chromosomes and protein complexes to be
trafficked to their destinations and be subjected to activation or
deactivation (8). Mounting evidence
has established the implication of lncRNAs in tumorigenesis
(10). LincRNA are a class of lncRNA
that are currently receiving wide attention from researchers
worldwide. The first identified lincRNA, HOTAIR, has been
demonstrated to promote metastasis in breast cancer (11), serve a progrostic role and exhibit
oncogenic activity in pancreatic cancer (19). Notably, linc-ITGB1 has been
demonstrated to promote cell migration and invasion in breast and
gallbladder cancer (20,21). The present study initially confirmed
that expression of linc-ITGB1was significantly elevated in HCC
tissues compared with their matched adjacent non-cancerous tissues.
The overexpression of linc-ITGB1 in HCC tissues was consistent with
previous studies that reported the aberrantly increased expression
of linc-ITGB1 in breast and gallbladder cancer (20,21).
Furthermore, in the present study it was observed that the
expression of linc-ITGB1 was positively correlated with cell
invasiveness, as indicated by the finding that the most invasive
HCCLM3 exhibited the highest transcript level of linc-ITGB1. This
observation indicated that linc-ITGB1 may be associated with
aggressive invasion in HCC.
Therefore, the present study modified the expression
of linc-ITGB1 using a specific shRNA in HCCLM3 cells. It was
observed that knockdown of linc-ITGB1 significantly inhibited tumor
cell growth in the proliferation assay and colony formation assay.
The proliferation assay using CCK-8 is useful for assessing cell
viability in an adhesion condition, while colony formation assay is
a convenient tool to assess cell growth ability in
anchorage-independent condition and is closely associated to the
in vivo situation (26,27).
Suppression of cell proliferation by shLinc-ITGB1 in both adhesion
presence and anchorage-independent conditions suggests that
linc-ITGB1 has a critically positive role in promoting tumor growth
in HCC. Furthermore, it was observed that depletion of linc-ITGB1
also decreased cell migration and invasion abilities. The name
linc-ITGB1 suggests that this intergenic lincRNA is potentially
co-regulated with B1-integrin, which is also a pro-migration gene
(20). Promotion of cell migration
and invasion by linc-ITGB1 has already been observed in breast and
gallbladder cancer models (20,21).
Thus, it would not be noteworthy that knockdown of linc-ITGB1 also
suppressed cell migration and invasion in HCCLM3 cells. The results
of the present study suggest that linc-ITGB1 exhibits a critical
role in cell migration and invasion, potentially in extensive
cancer types, including breast, bladder and liver cancer. The
present study is, to the best of our knowledge, the first to
identify linc-ITGB1 as a critical mediator of cell proliferation,
migration and invasion in HCC cells.
The mechanism(s) by which linc-ITGB1 promotes cell
proliferation, migration and invasion in HCCLM3 cells remain
unknown. The data of the current study indicates that a knockdown
of linc-ITGB1 arrested cell cycle progression in the G0/G1 phase.
Considering that this cell cycle arrest, specifically in the G0/G1
phase, links HCCLM3 cells to apoptosis (28,29).
Deregulation of cell cycle progression is a hallmark of tumor
growth (30). It is possible that
linc-ITGB1 contributes to cell proliferation by controlling cell
cycle progression and inducing cell apoptosis. This hypothesis was
supported by a study that reported the deregulation of cell cycle
key regulators by knockdown of linc-ITGB1 (21). In addition, the mechanisms of how
linc-ITGB1 mediates cell migration and invasion remains largely
unknown. Currently, the available data indicates that the
epithelial-to-mesenchymal transition (EMT) process is involved in
linc-ITGB1-mediated cell migration and invasion in breast and
gallbladder cancer (20,21). However, details on the regulation of
EMT by linc-ITGB1 are yet to be elucidated. Furthermore, the name
of linc-ITGB1 suggests that the gene B1-integrin may be
co-regulated by linc-ITGB1. How these regulatory networks
collectively contribute to linc-ITGB1-mediated aggressive
activities remains to be elucidated in HCC.
In conclusion, the present study has, to the best of
our knowledge, been the first to introduce a novel lncRNA
linc-ITGB1, which promotes cell proliferation, migration and
invasion in HCC. The data of the current study suggests that a
knockdown of linc-ITGB1 may be a promising therapeutic strategy to
treat HCC. However, the underlying detailed mechanisms remains to
be elucidated.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu YH, Ai X, Liu FY, Liang HF, Zhang BX
and Chen XP: c-Jun N-terminal kinase inhibitor favors transforming
growth factor-β to antagonize hepatitis B virus X proteininduced
cell growth promotion in hepatocellular carcinoma. Mol Med Rep.
13:1345–1352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fong ZV and Tanabe KK: The clinical
management of hepatocellular carcinoma in the United States,
Europe, and Asia: A comprehensive and evidence-based comparison and
review. Cancer. 120:2824–2838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
van Malenstein H, van Pelt J and Verslype
C: Molecular classification of hepatocellular carcinoma anno 2011.
Eur J Cancer. 47:1789–1797. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Minicis S, Marzioni M, Benedetti A and
Svegliati-Baroni G: New insights in hepatocellular carcinoma: From
bench to bedside. Ann Transl Med. 1:152013.PubMed/NCBI
|
|
8
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Q, Huang J, Zhou N, Zhang Z, Zhang A,
Lu Z, Wu F and Mo YY: LncRNA loc285194 is a p53-regulated tumor
suppressor. Nucleic Acids Res. 41:4976–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsai MC, Spitale RC and Chang HY: Long
Intergenic Noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gloss B, Moran-Jones K, Lin V, Gonzalez M,
Scurry J, Hacker NF, Sutherland RL, Clark SJ and Samimi G: ZNF300P1
Encodes a lincRNA that regulates cell polarity and is
epigenetically silenced in type II epithelial ovarian cancer. Mol
Cancer. 13:32014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Zhang Y, Lv W, Lu J, Mu J, Liu Y
and Dong P: Long non-coding RNA Linc-ITGB1 knockdown inhibits cell
migration and invasion in GBC-SD/M and GBC-SD gallbladder cancer
cell lines. Chem Biol Drug Des. 86:1064–1071. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan M, Zhang L, Li G, Xiao S, Dai J and
Cen X: Long non-coding RNA linc-ITGB1 promotes cell migration and
invasion in human breast cancer. Biotechnol Appl Biochem. 64:5–13.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge Z, Zhang B, Bu X, Wang Y, Xiang L and
Tan J: Molecular mechanism of activating protein-4 regulated growth
of hepatocellular carcinoma. Tumor Biol. 35:12441–12447. 2014.
View Article : Google Scholar
|
|
24
|
Wang J, Su H, Han X and Xu K: Inhibition
of fibroblast growth factor receptor signaling impairs metastasis
of hepatocellular carcinoma. Tumor Biol. 35:11005–11011. 2014.
View Article : Google Scholar
|
|
25
|
Tian GY, Zang SF, Wang L, Luo Y, Shi JP
and Lou GQ: Isocitrate dehydrogenase 2 suppresses the invasion of
hepatocellular carcinoma cells via matrix metalloproteinase 9. Cell
Physiol Biochem. 37:2405–2414. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang LH: Molecular signaling regulating
anchorage-independent growth of cancer cells. Mt Sinai J Med.
71:361–367. 2004.PubMed/NCBI
|
|
27
|
Thullberg M and Strömblad S:
Anchorage-independent cytokinesis as part of oncogenic
transformation? Cell Cycle. 7:984–988. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu XY, Xia P, Yu M, Nie XC, Yang X, Xing
YN, Liu YP, Takano Y and Zheng HC: The roles of REIC gene and its
encoding product in gastric carcinoma. Cell Cycle. 11:1414–1431.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Cheung KF, Ma X, Tian L, Zhao J, Go
MY, Shen B, Cheng AS, Ying J, Tao Q, et al: Epigenetic inactivation
of paired box gene 5, a novel tumor suppressor gene, through direct
upregulation of p53 is associated with prognosis in gastric cancer
patients. Oncogene. 31:3419–3430. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|