Introduction
Bone regeneration is a cascade of complex biological
events of bone induction and translation, and is required to
optimize skeletal repair and restore skeletal function (1,2). It has
been shown to be involved in numerous conditions, including
skeletal reconstruction of large bone defects created by trauma,
orthopedic surgery and osteotomy (3). The most common form of bone
regeneration is fracture healing in the clinical setting (4). Thus, understanding the mechanism of
fracture healing and accelerating the overall regeneration process
will help to improve the healing outcome and alleviate the pain
felt be patients to some extent.
Fracture consolidation can be influenced by
mechanical factors, and mechanical stability is confirmed to be a
key factor for determining the healing outcome of bone regeneration
(5,6). It has also been demonstrated that the
optimal fixation rigidity, which is neither too stiff nor too
gentle, is associated with the fastest growth of the bone (7). Ranganathan et al (8) also reported that an appropriate
fixation stability was necessary to promote timely fracture
healing. In addition, the early fracture hematoma has been shown to
have potency and potential in fracture healing and has become a
subject of attention (9). An in
vitro study has indicated that fracture hematoma contains
multilineage mesenchymal progenitor cells able to differentiate
into a chondrogenic or osteogenic cell type and thus is important
in bone healing (10). Furthermore,
fracture hematoma has been shown to contain growth factor vascular
endothelial growth factor, which is crucial in angiogenesis and
fracture healing (11,12). Sarahrudi et al (13) demonstrated that fracture hematoma had
significantly higher concentrations of transforming growth factor,
β 1 (Tgfβ1) than the peripheral serum of patients. Therefore, the
early fracture hematoma may be important in the early healing
period and may determine the healing outcome. Furthermore, gene
expression in the early fracture hematoma is also reported to be
influenced by fixation stability (14). However, it so far remains
incompletely understood what molecules are involved in the early
fracture hematoma associated with fixation stability, as well as
their roles during fracture healing.
In a previous study, GSE53256 microarray data was
utilized to analyze the interactive effects of age and mechanical
stability on bone defect healing using an early transcriptional
analysis (14). In the present
study, the GSE53256 microarray data was downloaded and a
bioinformatics approach was applied in order to identify the
differentially expressed genes (DEGs) in fracture hematoma tissues
from old rats with rigid fixation in comparison with fracture
hematoma tissues from old rats with semi-rigid fixation. In
addition, functional enrichment analysis and protein-protein
interaction (PPI) network analysis were performed. The aim was to
identify the potential key genes influenced by fixation stability
in early fracture hematoma, as well as to elucidate their roles in
the process of fracture healing.
Materials and methods
Affymetrix microarray data
The gene expression profile of GSE53256 deposited by
Ode et al (14) was obtained
from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database; the
profile was performed on the platform of GPL1335 [Rat 230_2]
Affymetrix Rat Genome 230 2.0 Array (Affymetrix, Santa Clara, CA,
USA). A total of 12 fracture hematoma tissues from four groups of
Sprague-Dawley rats with a 1.5-mm osteotomy gap in the femora with
varying age (12 vs. 52 weeks) and fixator stiffness (rigid vs.
semi-rigid fixation; n=3 per group) were used for the development
of this microarray data. However, in this study, only the
expression data from the six fracture hematoma tissues of the old
(52-week-old) rats after 3 days of an osteotomy and with rigid or
semi-rigid fixation were used for the analysis.
Data preprocessing and DEG
screening
The raw expression data were first background
corrected and quantile normalized by the robust multiarray average
(15) method with application of the
Affy package in R/Bioconductor. Next, the DEGs in fracture hematoma
tissues from old rats with rigid fixation compared with fracture
hematoma tissues from old rats with semi-rigid fixation were
identified using limma (16) package
in R/Bioconductor (http://www.bioconductor.org/packages/release/bioc/html/limma.html),
where a fold-change ≥1.5 and P<0.05 were defined as the cutoff
value. Additionally, hierarchical clustering analysis of the DEGs
was performed and visualized using the pheatmap package in R
(17).
Functional enrichment analysis
Gene Ontology (GO; http://www.geneontology.org) (18) is widely applied for the biological
unification of large-scale gene lists, which are mainly classified
into three categories, namely biological process (BP), molecular
function and cellular component. In the present study, GO-BP
enrichment analysis for DEGs was performed using the BiNGO
(http://www.psb.ugent.be/cbd/papers/BiNGO/) (19) plugin and was then visualized using
Cytoscape software (20). In order
to reduce false positives, multiple testing correction was
performed using the Beniamini-Hochberg method (21), and the P-value was then adjusted as
the false discovery rate (FDR). Finally, significant enrichment
threshold of GO-BP terms was set as FDR<0.05.
PPI network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING; http://string-db.org/) (22) database collects experimental and
predicted information associated with the interactions of protein
pairs in a given cell context via calculating the combined score of
PPIs. The higher the combined score, the more reliable the PPIs
are. In the present study, PPIs with a combined score >0.4 were
considered to be significant. Thus, the DEGs were mapped into PPIs
and a PPI network was then constructed based on the information of
the STRING database.
Results
DEG analysis
Following data processing, the data was normalized
for subsequent analysis. A total of 265 DEGs were obtained using
the limma package in fracture hematoma tissues from old rats with
rigid fixation compared with fracture hematoma tissues from old
rats with semi-rigid fixation, and included 158 upregulated and 107
downregulated genes. For example, chemokine (C-X-C motif) ligand 12
(Cxcl12), chemokine (C-C motif) ligand 9 (Ccl9) and matrix
metallopeptidase 9 (mmp9) were upregulated, while Tgfβ1, serpin
peptidase inhibitor, clade E (nexin, plasminogen activator
inhibitor type 1), member 1 (serpine1) and angiotensin II receptor,
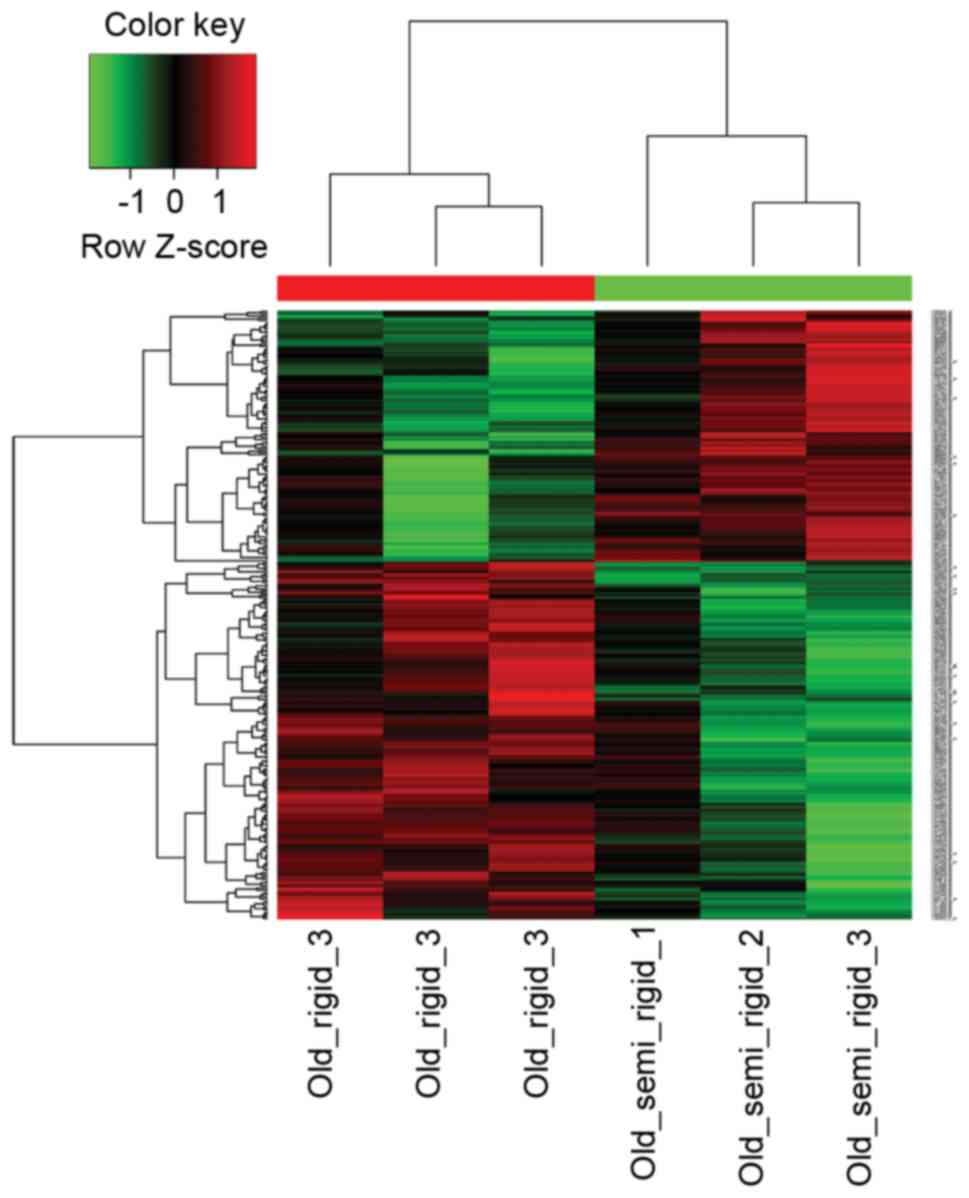
type 1a (Agtr1a) were downregulated. The heat map plot of DEGs
derived from hierarchical clustering analysis is shown in Fig. 1.
GO enrichment analysis
In order to understand the function of these DEGs,
GO enrichment analysis was performed for DEGs. Table I displays the top 20 most significant
GO-BPs, and the most overrepresented GO terms of these DEGs were
found to be associated with the extracellular region, positive
regulation of locomotion, response to external stimulus, positive
regulation of cell migration and response to wounding.
 | Table I.Top 20 most significant pathways. |
Table I.
Top 20 most significant pathways.
| GO-ID | Description | Corrected
P-value |
|---|
| 44421 | Extracellular region
part |
2.49×10−12 |
| 40017 | Positive regulation
of locomotion |
5.06×10−9 |
| 9605 | Response to external
stimulus |
5.06×10−10 |
| 30335 | Positive regulation
of cell migration |
1.02×10−9 |
| 9611 | Response to
wounding |
1.04×10−9 |
| 5576 | Extracellular
region |
1.53×10−9 |
| 51272 | Positive regulation
of cellular component movement |
1.80×10−9 |
| 42221 | Response to chemical
stimulus |
1.85×10−9 |
| 32879 | Regulation of
localization |
3.69×10−9 |
| 5615 | Extracellular
space |
6.92×10−9 |
| 6954 | Inflammatory
response |
1.25×10−8 |
| 32502 | Developmental
process |
2.14×10−8 |
| 30334 | Regulation of cell
migration |
2.27×10−8 |
| 42330 | Taxis |
2.27×10−8 |
| 6935 | Chemotaxis |
2.27×10−8 |
| 7275 | Multicellular
organismal development |
2.40×10−8 |
| 31012 | Extracellular
matrix |
3.70×10−8 |
| 40011 | Locomotion |
4.00×10−8 |
| 48731 | System
development |
5.95×10−8 |
| 8009 | Chemokine
activity |
7.92×10−8 |
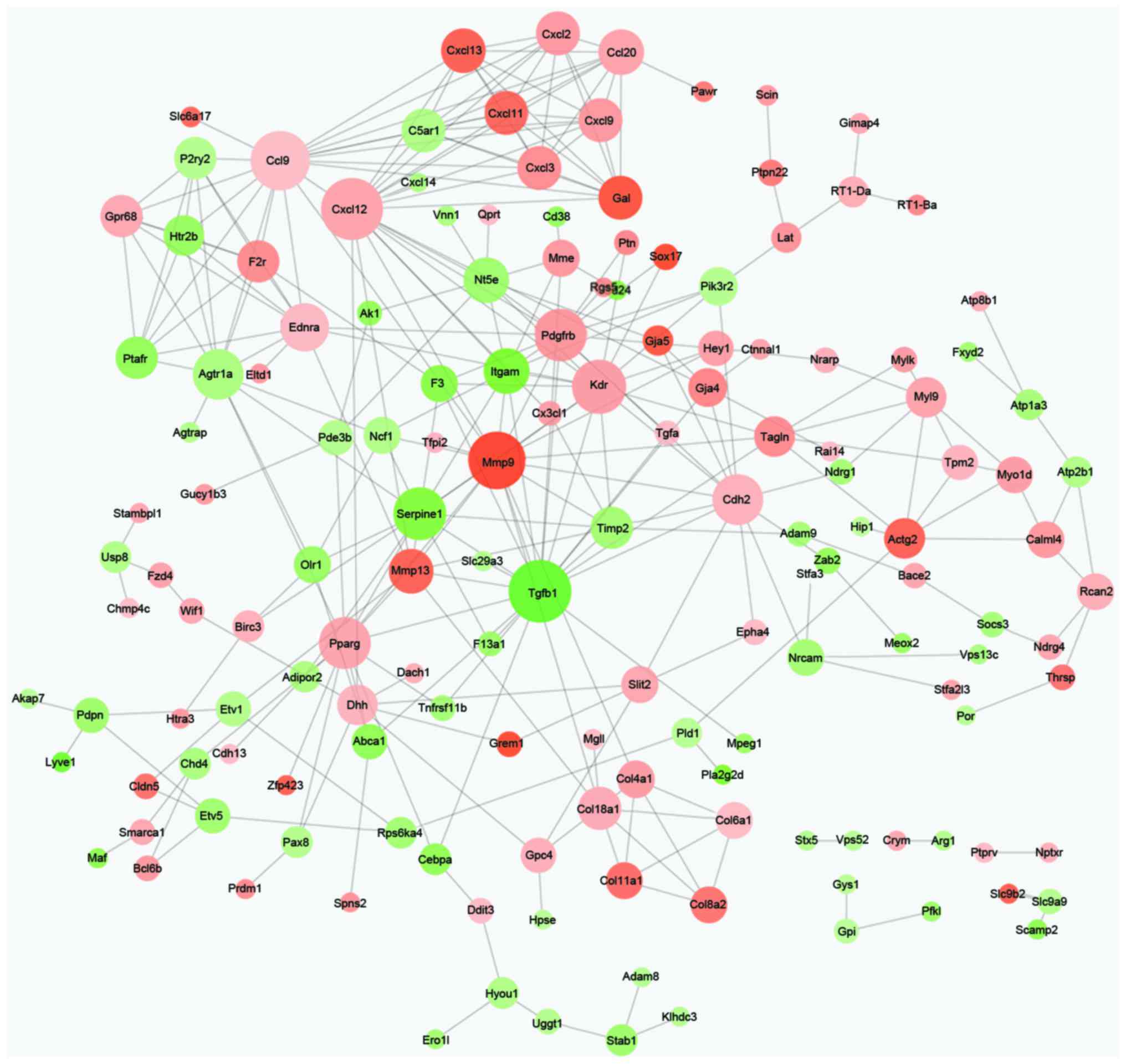
PPI network analysis
Based on the information of the STRING database, the
PPI network of DEGs was constructed with 153 nodes and 302 edges
(Fig. 2). Additionally, the node
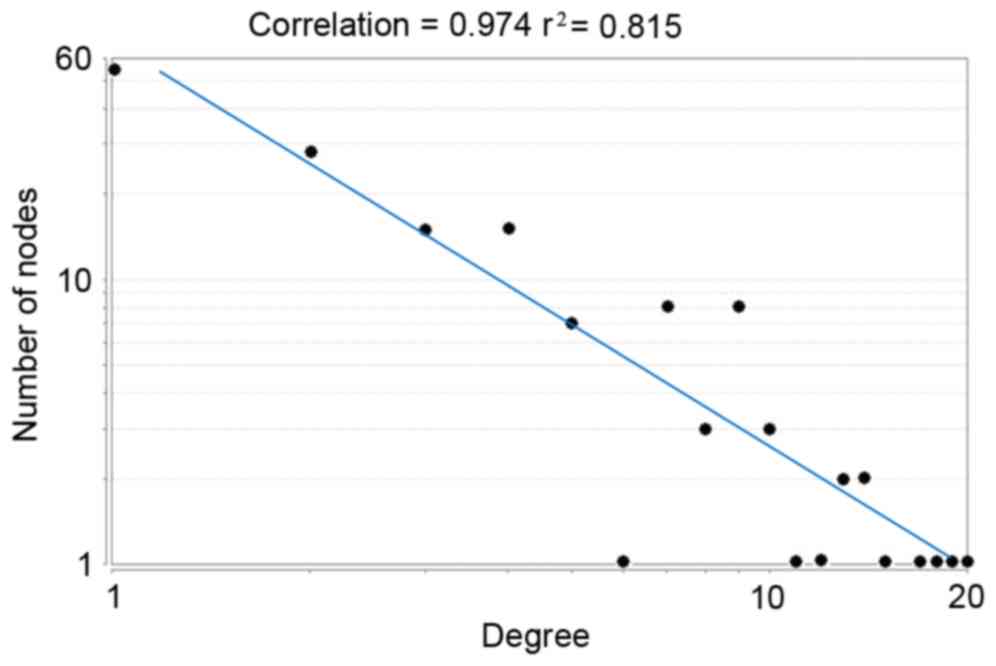
degree distribution displayed that this PPI network was a
scale-free network (Fig. 3). The
biggest characteristic of the scale-free network was that a small
number of nodes had higher degrees while the majority of the nodes
had lower degrees, indicating that nodes with higher degrees may be
important in network connectivity. Therefore, the nodes Tgfβ1
(degree, 20), Cxcl12 (degree, 19), Ccl9 (degree, 18), mmp9 (degree,
17), kinase insert domain receptor (degree, 15), platelet derived
growth factor receptor, β polypeptide (degree, 14), serpine1
(degree, 14), cadherin 2 (degree, 13), peroxisome
proliferator-activated receptor γ (degree, 13) and Agtr1a (degree,
12) were hub proteins in the PPI network (Table II).
 | Table II.Genes with degree ≥10. |
Table II.
Genes with degree ≥10.
| Gene | Name | Log FC | Degree |
|---|
| Tgfβ1 | Transforming growth
factor, β 1 | −1.567666199 | 20 |
| Cxcl12 | Chemokine (C-X-C
motif) ligand 12 |
0.8016909 | 19 |
| Ccl9 | Chemokine (C-C
motif) ligand 9 |
0.588012021 | 18 |
| Mmp9 | Matrix
metallopeptidase 9 |
1.752023742 | 17 |
| Kdr | Kinase insert
domain receptor |
0.846463992 | 15 |
| Pdgfrb | Platelet derived
growth factor receptor, β polypeptide |
0.906423663 | 14 |
| Serpine1 | Serpin peptidase
inhibitor, clade E (nexin, plasminogen activator inhibitor type 1),
member 1 | −1.17741059 | 14 |
| Cdh2 | Cadherin 2 |
0.691787895 | 13 |
| Pparg | Peroxisome
proliferator-activated receptor γ |
0.78658078 | 13 |
| Agtr1a | Angiotensin II
receptor, type 1a | −0.7331974 | 12 |
| Ednra | Endothelin receptor
type A |
0.637602954 | 11 |
| Ccl20 | Chemokine (C-C
motif) ligand 20 |
0.807308765 | 10 |
| Itgam | Integrin, α M | −1.252174861 | 10 |
| Nt5e | 5′ nucleotidase,
ecto | −0.831568237 | 10 |
Discussion
The early phase of bone healing is likely to be
sensitive to the conditions of mechanical loading (23). In the present study, a bioinformatics
approach was used to identify key genes influenced in fixation
stability from fracture hematoma tissues harvested 3 days
post-osteotomy. Upregulated genes, including Cxcl12 and mmp9, and
downregulated genes, such as Tgfβ1 and serpine1, were identified as
hub nodes and appeared to be important in the process of fracture
healing associated with fixation stability.
The fracture healing process may be divided into
three stages: Acute inflammation, repair and remodeling (24). In the present study, upregulated
genes, including Cxcl12 and mmp9, were key inflammatory cytokines
and may be involved in the initial inflammatory response as the
first step of fracture healing. The chemokine Cxcl12 has been
identified to regulate the inflammatory response associated with
the healing process (25).
Furthermore, it is an important contributor to bone marrow MSC
homing and localization within the bone marrow (26). Grassi et al (27) also indicated that Cxcl12 was involved
in a number of inflammatory pathologies and was crucial in the
regulation of osteoclast differentiation and function. In addition,
mmp9 has been confirmed to have effects on skeletal cell
differentiation during fracture healing via regulation of the
inflammatory response and the inflammatory cell distribution
(28). Furthermore, Beamer et
al (12) demonstrated that mmp9
was implicated in the regulation of chondrogenic and osteogenic
cell differentiation during the early stages of bone repair.
Notably, Wang et al (29)
indicated that the mechanical environment affected the inflammatory
response and influenced skeletal cell differentiation during bone
repair via the regulation of mmp9. Therefore, the results of the
present study imply that fixation stability may induce an
inflammatory response during fracture healing via the regulation of
Cxcl12 and mmp9.
Tgfβ1 was identified as a hub node in the PPI
network with the highest degree. It is a member of the transforming
growth factor-β family and functions as a regulatory protein
involved in bone remodeling during the fracture healing process
(13). Furthermore, Tgfβ1 is thought
to induce the migration of bone mesenchymal stem cells and to
regulate the coordination of bone resorption and subsequent bone
formation (30). Additionally, it
has also been found to be involved in fracture healing through
regulation of the activation and differentiation of osteoblasts and
osteoclasts (31). Tgfβ1 induces the
production of extracellular bone matrix proteins, including
alkaline phosphatase, collagen, osteonectin, osteopontin and
proteoglycans (32,33), and can regulate different cell types
implicated in bone turnover and fracture healing (34). Therefore, Tgfβ1 may be important in
the regulation of bone remodeling during the fracture healing
process. However, in the present study, Tgfβ1 was downregulated in
fracture hematoma tissues from old rats with rigid fixation
compared with fracture hematoma tissues from old rats with
semi-rigid fixation, implying that rigid fixation may influence
bone remodeling during the fracture healing process by the
downregulation of Tgfβ1.
Furthermore, serpine1 was identified as another
downregulated hub node. Serpine1 [also known as plasminogen
activator inhibitor-1 (PAI-1)], as a component of the fibrinolytic
system, is the principal inhibitor of plasminogen activators
(35). It has been shown to have
various functions, including regulation of extracellular matrix
(ECM) degradation and cell migration (36). Tamura et al (37) confirmed that PAI-1 suppressed the
mRNA expression levels of runt-related transcription factor 2 and
type I collagen in primary osteoblasts from mouse calvaria,
suggesting that PAI-1 may suppress osteoblast differentiation
during the bone repair process. In addition, serpine1 has been
reported to be important in the regulation of fracture callus size,
cartilage formation and resorption during bone fracture repair
(38). Mao et al (24) also demonstrated that PAI-1 may be
involved in ECM remodeling and thus be crucial in the bone repair
process in patients with diabetes. It may thus be speculated that
serpine1 is important in bone fracture repair. In addition, Mao
et al (24) also demonstrated
that PAI-1 deficiency attenuated diabetic impaired bone repair in
patients with diabetes. In the present study, serpine1 was
downregulated in fracture hematoma tissues with rigid fixation when
compared with those with semi-rigid fixation, indicating that
suppression of serpine1 may promote bone repair following treatment
using rigid fixation.
In conclusion, the observations of the present study
indicate that fixation stability may influence the fracture healing
process, and that important DEGs in fracture hematoma, including
Cxcl12, mmp9, Tgfβ1 and serpine1 may be important in this process.
The present observations shed new light on the molecular mechanism
of fracture healing and have implications for future research.
However, the small sample size and lack of experimental validation
were limitations in the present study. To further validate the
results, it would be prudent to perform the same analysis on a
group of young rats and compare the two gene sets for common DEGs
upon fixation rigidity in young and old rats.
References
|
1
|
Einhorn TA: The cell and molecular biology
of fracture healing. Clin Orthop Relat Res. 355 Suppl:S7–S21. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dimitriou R, Tsiridis E and Giannoudis PV:
Current concepts of molecular aspects of bone healing. Injury.
36:1392–1404. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dimitriou R, Jones E, McGonagle D and
Giannoudis PV: Bone regeneration: Current concepts and future
directions. BMC Med. 9:662011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferguson C, Alpern E, Miclau T and Helms
JA: Does adult fracture repair recapitulate embryonic skeletal
formation? Mech Dev. 87:57–66. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Strube P, Sentuerk U, Riha T, Kaspar K,
Mueller M, Kasper G, Matziolis G, Duda GN and Perka C: Influence of
age and mechanical stability on bone defect healing: Age reverses
mechanical effects. Bone. 42:758–764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mehta M, Strube P, Peters A, Perka C,
Hutmacher D, Fratzl P and Duda GN: Influences of age and mechanical
stability on volume, microstructure, and mineralization of the
fracture callus during bone healing: Is osteoclast activity the key
to age-related impaired healing? Bone. 47:219–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Epari DR, Kassi JP, Schell H and Duda GN:
Timely fracture-healing requires optimization of axial fixation
stability. J Bone Joint Surg. 89:1575–1585. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ranganathan SI, Ferrari M and Decuzzi P:
Design maps for scaffold constructs in bone regeneration. Biomed
Microdevices. 15:1005–1013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kolar P, Schmidt-Bleek K, Schell H, Gaber
T, Toben D, Schmidmaier G, Perka C, Buttgereit F and Duda GN: The
early fracture hematoma and its potential role in fracture healing.
Tissue Eng Part B Rev. 16:427–434. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oe K, Miwa M, Sakai Y, Lee SY, Kuroda R
and Kurosaka M: An in vitro study demonstrating that haematomas
found at the site of human fractures contain progenitor cells with
multilineage capacity. J Bone Joint Surg Br. 89:133–138. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Street J, Winter D, Wang JH, Wakai A,
McGuinness A and Redmond HP: Is human fracture hematoma inherently
angiogenic? Clin Orthop Relat Res. 378:224–237. 2000. View Article : Google Scholar
|
|
12
|
Beamer B, Hettrich C and Lane J: Vascular
endothelial growth factor: An essential component of angiogenesis
and fracture healing. HSS J. 6:85–94. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sarahrudi K, Thomas A, Mousavi M, Kaiser
G, Köttstorfer J, Kecht M, Hajdu S and Aharinejad S: Elevated
transforming growth factor-beta 1 (TGF-β1) levels in human fracture
healing. Injury. 42:833–837. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ode A, Duda GN, Geissler S, Pauly S, Ode
JE, Perka C and Strube P: Interaction of age and mechanical
stability on bone defect healing: An early transcriptional analysis
of fracture hematoma in rat. PLoS One. 9:e1064622014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
17
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of Gene Ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B. 57:289–300. 1995.
|
|
22
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Klein P, Schell H, Streitparth F, Heller
M, Kassi JP, Kandziora F, Bragulla H, Haas NP and Duda GN: The
initial phase of fracture healing is specifically sensitive to
mechanical conditions. J Orthop Res. 21:662–669. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mao L, Kawao N, Tamura Y, Okumoto K, Okada
K, Yano M, Matsuo O and Kaji H: Plasminogen activator inhibitor-1
is involved in impaired bone repair associated with diabetes in
female mice. PLoS One. 9:e926862014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Galliera E, Corsi M and Banfi G: Platelet
rich plasma therapy: Inflammatory molecules involved in tissue
healing. J Biol Regul Homeost Agents. 26 2 Suppl 1:35S–42S.
2011.
|
|
26
|
Honczarenko M, Le Y, Swierkowski M, Ghiran
I, Glodek AM and Silberstein LE: Human bone marrow stromal cells
express a distinct set of biologically functional chemokine
receptors. Stem cells. 24:1030–1041. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grassi F, Cristino S, Toneguzzi S,
Piacentini A, Facchini A and Lisignoli G: CXCL12 chemokine
up-regulates bone resorption and MMP-9 release by human
osteoclasts: CXCL12 levels are increased in synovial and bone
tissue of rheumatoid arthritis patients. J Cell Physiol.
199:244–251. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pape HC, Marcucio R, Humphrey C, Colnot C,
Knobe M and Harvey EJ: Trauma-induced inflammation and fracture
healing. J Orthop Trauma. 24:522–525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Yu YY, Lieu S, Yang F, Lang J, Lu
C, Werb Z, Hu D, Miclau T, Marcucio R and Colnot C: MMP9 regulates
the cellular response to inflammation after skeletal injury. Bone.
52:111–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z,
Zhao L, Nagy TR, Peng X, Hu J, et al: TGF-beta1-induced migration
of bone mesenchymal stem cells couples bone resorption with
formation. Nat Med. 15:757–765. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Estai MA, Suhaimi F, Das S, Shuid AN,
Mohamed Z and Soelaiman IN: Expression of TGF-β1 in the blood
during fracture repair in an estrogen-deficient rat model. Clinics
(Sao Paulo). 66:2113–2119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
D'Amelio P, Cristofaro MA, Grimaldi A,
Ravazzoli M, Pluviano F, Grosso E, Pescarmona GP and Isaia GC: The
role of circulating bone cell precursors in fracture healing.
Calcif Tissue Int. 86:463–469. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sandblrg MM, Aro HT and Vuorio EI: Gene
expression during bone repair. Clin Orthop Relat Res. 1–312.
1993.PubMed/NCBI
|
|
34
|
Vo TN, Kasper FK and Mikos AG: Strategies
for controlled delivery of growth factors and cells for bone
regeneration. Adv Drug Deliv Rev. 64:1292–1309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gils A and Declerck PJ: Plasminogen
activators inhibitorsPlasminogen: Structure, Activation and
Regulation. Waisman DM: Springer; New York: pp. 47–66. 2003
|
|
36
|
Declerck PJ and Gils A: Three decades of
research on plasminogen activator inhibitor-1: A multifaceted
serpin. Semin Thromb Hemost. 39:356–364. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tamura Y, Kawao N, Okada K, Yano M,
Okumoto K, Matsuo O and Kaji H: Plasminogen activator inhibitor-1
is involved in streptozotocin-induced bone loss in female mice.
Diabetes. 62:3170–3179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rundle CH, Wang X, Wergedal JE, Mohan S
and Lau KH: Fracture healing in mice deficient in plasminogen
activator inhibitor-1. Calcif Tissue Int. 83:276–284. 2008.
View Article : Google Scholar : PubMed/NCBI
|