Introduction
Primary hypertension is a common chronic disease and
typically causes other health complications, including coronary
artery disease, stroke, renal failure and heart failure, and poses
a great threat to human health (1–3). At
present, ~300 million patients with hypertension, and 10 million
patients are diagnosed annually in China (4). However, the mechanisms underlying the
development of primary hypertension remain unclear. Over the last
decade, the incidence of primary hypertension and percentage of
young patients with primary hypertension in China has increased
(5), which has drawn the attention
of a number of researchers.
Vascular injury, including vascular endothelial
injury, smooth muscle cell proliferation and vascular remodeling,
is the basic pathological change of hypertension diseases (6–8). A
previous study demonstrated that vascular endothelial injury is
important in the development of hypertension (9). Hypertension induces vascular
endothelial dysfunction, disrupts the balance of nitric oxide (NO)
and secretion of endothelin from vascular endothelial cells and
increases the contraction of arteries, leading to pulmonary
arterial hypertension characterized by a persistent increase in
pulmonary artery pressure (10). In
a hypertensive microenvironment, the apoptosis of microvascular
endothelial cells is increased, resulting in a reduction of
pathways in the capillary network that allow gas exchange between
tissues and elevation of peripheral circulation pressure, which
ultimately leads to high blood pressure (11). However, the molecular mechanism
underlying hypertension-induced vascular endothelial injury is not
well understood and warrants further study.
MicroRNAs (miRNA) are a class of highly conserved,
non-coding RNAs of 18–22 nucleotides in length, which predominantly
serve as translational repressors by binding to complementary
sequences in the 3′ untranslated region (UTR) of their target mRNAs
(12). A recent study demonstrated
that the miRNA expression pattern in the peripheral blood of
patients with hypertension was altered, which may be used
clinically for the early diagnosis of hypertension and the
prognosis of hypertension-induced complications (13). In addition, miRNAs may regulate
hypertension-induced complications and vascular endothelial injury
(14). It has been demonstrated that
miR-34a is ectopically expressed in the peripheral blood of
patients with hypertension (15),
suggesting that this miRNA is associated with the development of
hypertension. However, the specific mechanism remains unclear. The
current study aimed to investigate the expression of miRNA
(miR)-34a in the peripheral blood of patients with hypertension and
its role in regulating vascular endothelial injury.
Materials and methods
Clinical data and peripheral blood
collection
Peripheral blood from 50 patients with primary
hypertension and 28 healthy volunteers was collected between
December 2013 and October 2014 from the Shandong Provincial
Hospital Affiliated to Shandong University (Jinan, China). The
blood samples were stored on ice for <20 min until further
processing. A total of 24 male and 26 female patients were included
in the study. The average age of patients was 61.5 years (ranging
between 51 and 82) and the median age was 62 years. Patients with a
duration of hypertension >5 years were enrolled. Patients with
combination of other chronic underlying diseases (such as diabetes)
or long history of medication were excluded. Written informed
consent was obtained from all patients prior to the study, and the
study was approved by the Ethics Review Board of Shandong
Provincial Hospital Affiliated to Shandong University.
Patients were divided into three phases, based on
the phase of hypertension observed (16). Phase I (n=21), systolic pressure is
140–159 mmHg or diastolic blood pressure is 90–99 mmHg; Phase II
(n=16), systolic pressure is 160–179 mmHg or diastolic blood
pressure is 100–109 mmHg; Phase III (n=13), systolic pressure
>180 mmHg or diastolic blood pressure >100 mmHg.
miRNA transfection of human umbilical
vein endothelial cells (HUVECs)
HUVECs were purchased from ScienCell Research
Laboratories, Inc. (Carlsbad, CA, USA; cat. no. 8000) and
maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.). Cells were seeded into 24-well plates at a
concentration of 1×105 cells/well. When the HUVECs
reached 70–90% confluence, the cells were transfected with 2.5 µl
miR-34a inhibitor (targeting sequence;
5′-CAATCAGCAAGTATACTGCCCT-3′, 25 pmol/µl) or scramble-miR (negative
control, NC; both from Guangzhou RiboBio Co., Ltd., Guangzhou,
China) using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
Cells were cultured 48 h post-transfection at 37°C with 5%
CO2 until collection for subsequent experiments.
RNA extraction and reverse
transcription quantitative polymerase chain reaction (RT-qPCR)
assay
Total RNA was extracted from cells using
TRIzol® isolation reagent (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Following
verification of RNA integrity by gel electrophoresis and
quantification using a UV spectrophotometer at 260 and 280 nm, 0.5
µg total RNA was reverse transcribed using an miScript II RT kit
(Qiagen, Hilden, Germany). The expression of small nuclear U6 was
used as an internal control. qPCR was performed using a KAPA
SYBR® FAST qPCR kit (Kapa Biosystems, Inc., Wilmington,
MA, USA) at 95°C for 10 min and 40 cycles of 95°C for 1 min and
60°C for 30 sec. The forward primer used for miR-34a amplification
was 5′-CAGTGTCTTAGCTGGTTG-3′, and the reverse primer was provided
within the kit. Primers used for amplification of U6 were as
follows: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. Relative expression levels were
determined using the ΔΔCq method (17).
Cell Counting Kit-8 (CCK-8) assay
To evaluate the effect of miR-34a on the
proliferation of HUVECs, cells were collected 48 h after
transfection and washed twice with phosphate-buffered saline (PBS).
The cells were then suspended in fresh DMEM containing 10% CCK-8
solution (Biyuntian Biotech Co., Ltd, Shanghai, China) and
incubated for 1 h at 37°C. The absorbance of each well was measured
with an ELx800 Microplate Reader (Biotek Instruments, Inc.,
Winooski, VT, USA) set at 450 nm.
Flow cytometric analysis of cell
apoptosis
At 48 h after transfection, 106 cells
were harvested 48 h after transfection by 0.25% trypsin digestion
at 37°C for 3 min, washed twice with cold PBS and stained with an
Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit
I (BD Biosciences, San Jose, CA, USA) according to the
manufacturer's instructions. Cells were immediately analyzed by BD
FACSVerse™ flow cytometer (BD Biosciences) with Modfit software
(version 1.0.1; Verity Software House, Inc., Topsham, ME, USA).
Flow cytometry analysis of cell
cycle
At 48 h after transfection, 1×106 cells
were washed twice with cold PBS and stained with a BD Cycletest
Plus DNA Reagent (cat. no. 340242; BD Biosciences), according to
the manufacturer's instructions. Cells were analyzed by flow
cytometry using a BD FACSVerse™ (BD Biosciences) with Modfit
software, version 2.0.
Migration assay
A total of 1×105 HUVECs in 200 µl
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) without FBS were
seeded into the top chamber of an 8-µm pore filter Transwell
chamber (Corning Incorporated, Corning, NY, USA) inserted in
24-well plates. RPMI-1640 plus 10% FBS (500 µl) was added to the
bottom chamber, and the cells were incubated at 37°C and 5%
CO2. After 24 h, the cells that did not pass through the
chambers were removed with a cotton swab, while the cells located
on the lower side of the chamber were fixed with 4%
paraformaldehyde at room temperature for 10 min, stained with
Giemsa and counted using an Olympus BX51/61 microscope (Olympus
Corporation, Tokyo, Japan) at a magnification of ×200. Migration
data were collected by counting the migrated cells in 5 randomly
selected fields.
Western blot analysis
Following transfection with miR-34a inhibitor or
scramble-miR, cells were collected and resuspended in
radioimmunoprecipitation assay lysis buffer with 1%
phenylmethanesulfonyl fluoride (Beyotime Institute of
Biotechnology, Beijing, China) at room temperature for 30 min to
extract total protein. Each sample was centrifuged at 12,000 × g
for 10 min at 4°C. The enhanced BCA protein assay kit (cat. no.
P0009; Beyotime Institute of Biotechnology) including bovine serum
albumin as the standard was used to measure the concentration of
total protein. A total of 20 µg protein per lane was separated by
12% SDS-PAGE and detected with rabbit anti-TGIF2 antibody (1:1,000;
11522-1-AP) based on results of the bioinformatics prediction and
mouse anti-GAPDH antibody (1:5,000; 60004-1-Ig) at 4°C overnight.
Antibodies were purchased from ProteinTech Group, Inc. (Wuhan
Sanying Biotechnology, Wuhan, China). Rabbit anti-mouse IgG H&L
conjugated to horseradish peroxidase (HRP; cat. no. ab6728;
1:5,000) and Goat anti-rabbit IgG H&L conjugated to HRP (cat.
no. ab6721; 1:5,000; Abcam, Cambridge, UK) were used as secondary
antibodies and incubated at room temperature for 2 h. Immunolabeled
bands were detected by BeyoECL Plus (Beyotime, Beijing, China; cat.
no. P0018). Each western blot analysis was replicated 3 times.
Bioinformatic prediction
The targets of miR-34a were predicted by Targetscan
7.1 (www.targetscan.org). Species set as
human and miR-34a was entered into the microRNA name field and
searched.
Dual-luciferase reporter gene
assay
According to the results of the bioinformatics
prediction, a conservative miR-34a binding sequence complimentary
to the 3′ UTR of Tgif2 mRNA with the smallest P-value (P<0.001)
was selected. Luciferase reporter plasmids were generated by
insertion of wild-type (5′-GGGUUUUCUAUGGAUCACUGCCA-3′) or mutant
binding sequences (5′-GGGUUUUCUAUGGAUAAGUACAA-3′) of TGIF2 into the
multiple cloning site (SpeI and HindIII) of a
pMIR-REPORT™ luciferase plasmid downstream of the luciferase
reporter gene, provided by Hanbio Biotechnology, Co., Ltd. HEK293T
cells (105) (ScienCell Research Laboratories, Inc.) were
cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.) for 24 h. Cell were transfected
with 1 µg constructed luciferase reporters and 100 nM miR-34a
mimics (5′-TGGCAGTGTCTTAGCTGGTTGT-3′) or NC RNA (Hanbio
Biotechnology Co., Ltd.). A total of 10 ng pMIR-REPORT™ β-gal
control plasmid (Beyotime Institute of Biotechnology) was
transfected as an internal control to determine transfection
efficiency. Luminescence was measured 24 h after transfection using
a dual-luciferase detection kit (cat. no. RG027; Beyotime Institute
of Biotechnology), according to the manufacturer's instructions.
Measurements of luminescence were performed with a luminometer
(Glomax® 20/20; Promega Corporation, Madison, WI,
USA).
Statistical analysis
The statistical significance of data was determined
with paired t-tests using SPSS 16.0 software (SPSS, Inc., Chicago,
IL, USA). All data were presented as the mean ± standard deviation
of 3 independent experiments, and P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-34a expression in the peripheral
blood of patients with primary hypertension
Initially, the potential dysregulation of miR-34a in
patients with hypertension was investigated. Using RT-qPCR
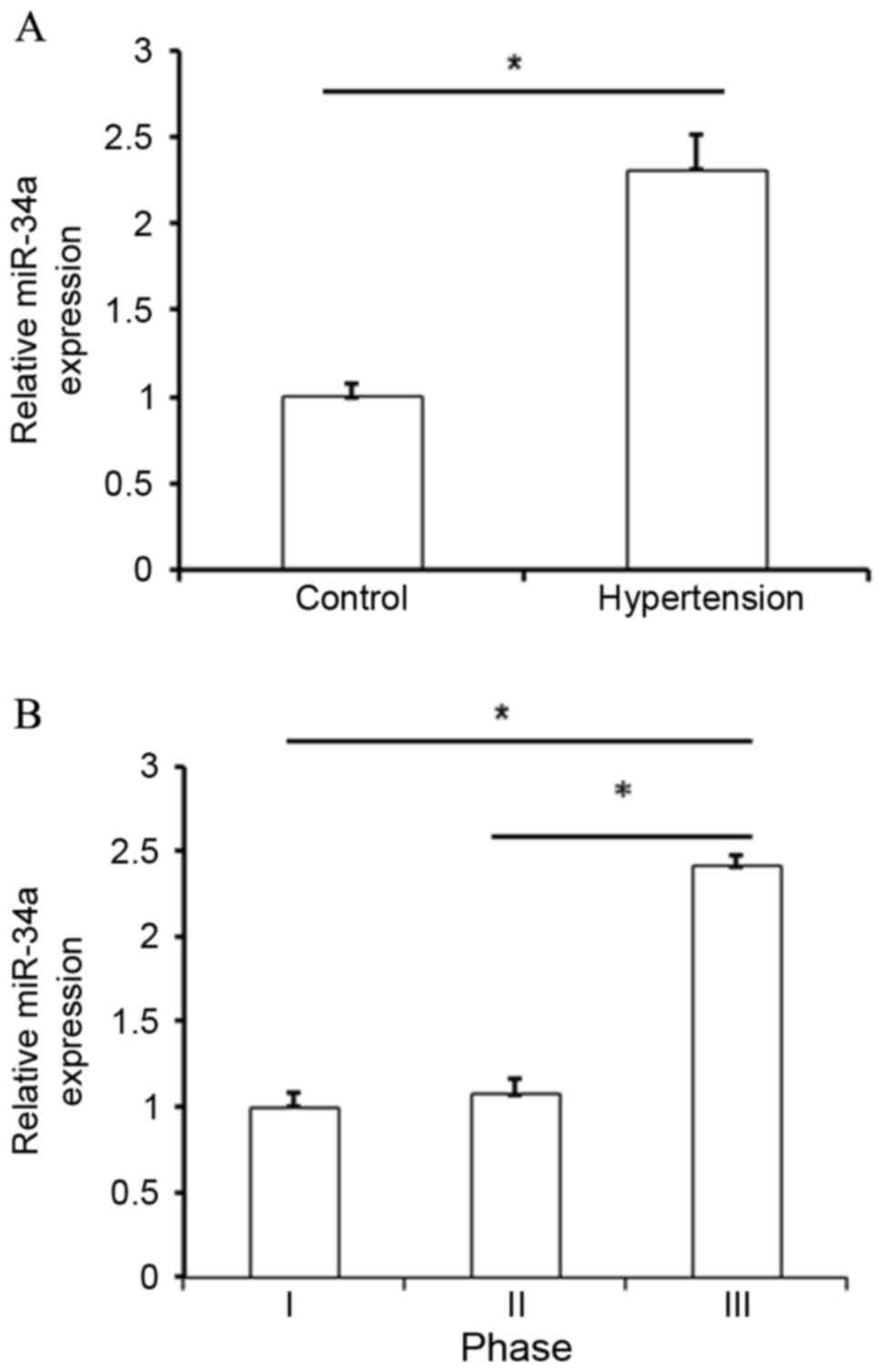
analysis, upregulation of miR-34a was identified in hypertensive
patients when compared with normal subjects (P<0.05; Fig. 1A). By analyzing miR-34a expression in
various groups based on clinical pathological features, it was also
observed that miR-34a was significantly upregulated in patients
with phase III hypertension when compared with patients presenting
with phase I and II hypertension (P<0.05; Fig. 1B and Table
I). These results indicated that upregulation of miR-34a was
correlated with the development of hypertension.
 | Table I.Clinical data of patients categorized
into different phases of hypertension. |
Table I.
Clinical data of patients categorized
into different phases of hypertension.
| Phase | Number | Mean age, years | Male/Female |
|---|
| I | 21 | 54.5 | 8/13 |
| II | 16 | 68.6 | 12/4 |
| III | 13 | 57.4 | 4/9 |
Effect of miR-34a on the proliferation
of HUVECs
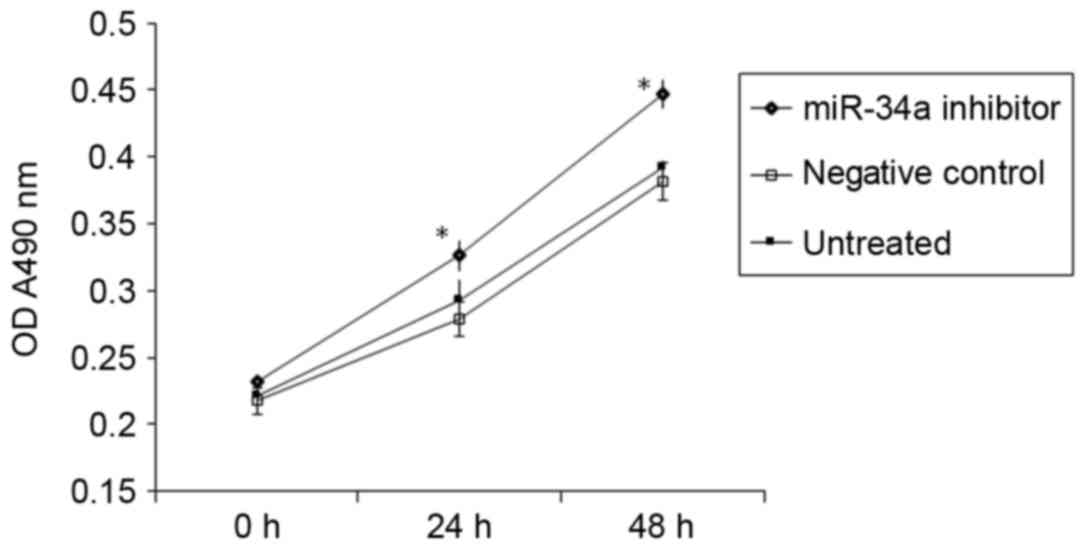
A CCK-8 assay demonstrated that transfection with
miR-34a inhibitor significantly promoted the proliferation of
HUVECs in vitro (P<0.05 vs. NC; Fig. 2). This suggests that increased
miR-34a expression in the peripheral blood of patients with
hypertension may inhibit the proliferation of vascular endothelial
cells.
miR-34a suppresses the migration of
HUVECs
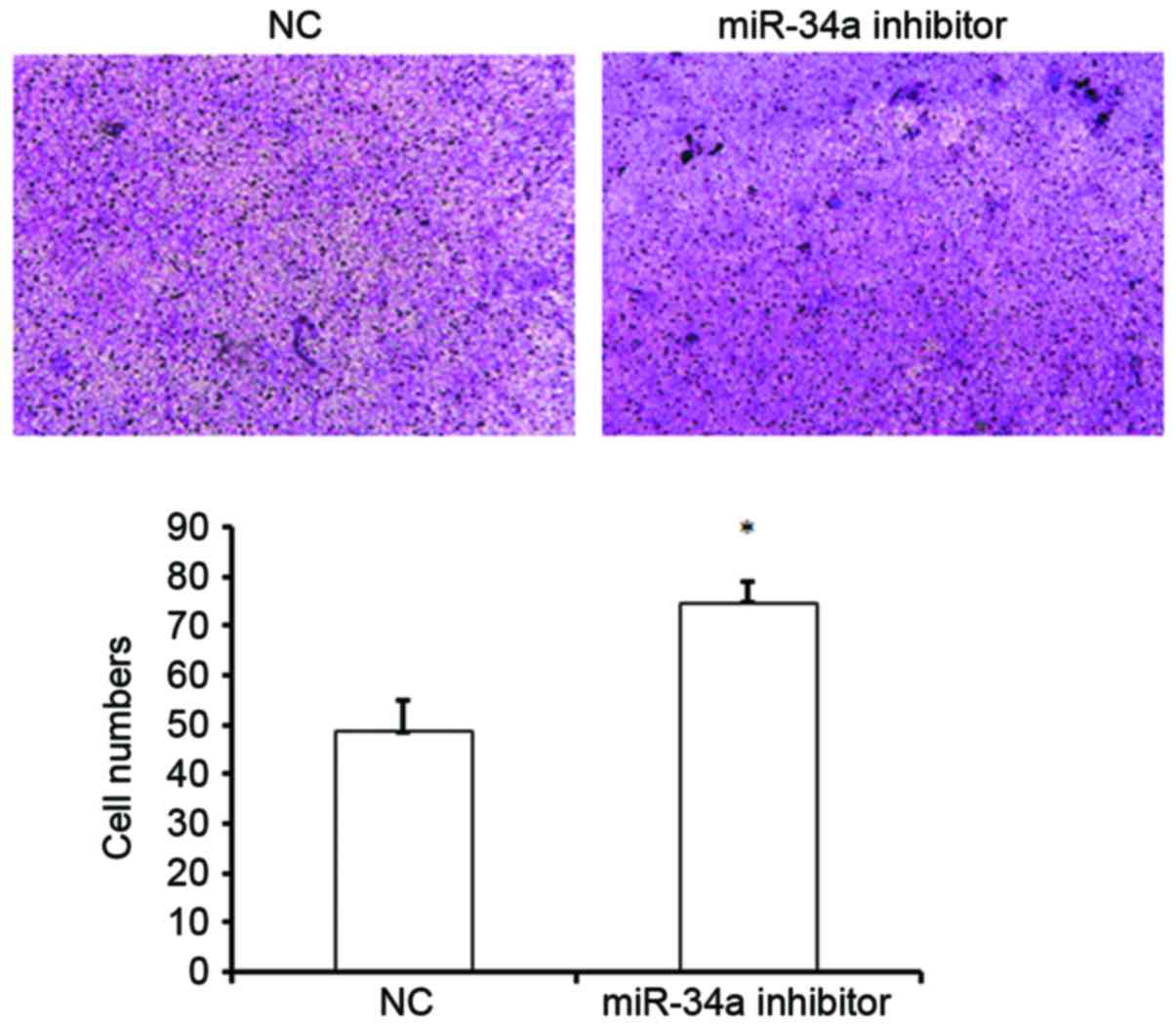
The effect of miR-34a on cell migration was
subsequently evaluated. A Transwell assay revealed that inhibition
of miR-34a expression significantly increased the migration of
HUVECs, as demonstrated by an increased number of cells that passed
through the chambers compared with the NC group (74.5±4.30 vs.
48.5±6.3; P<0.05; Fig. 3). This
result suggests that miR-34a may suppress the migration of HUVECs,
and thus may inhibit the migration of vascular endothelial cells to
sites of injury in vivo, leading to the inhibition of
vascular injury repair.
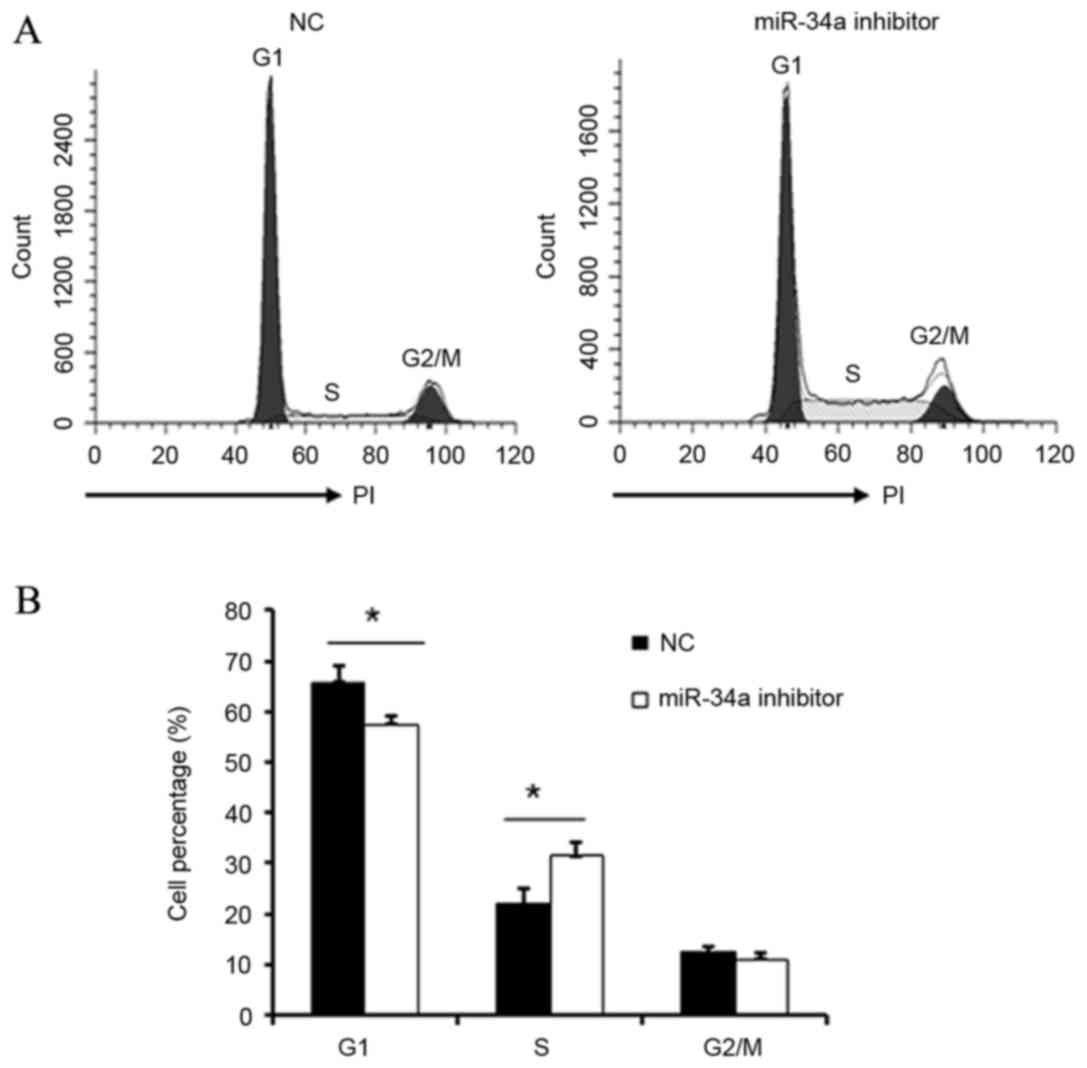
Effect of miR-34a on cell cycle
distribution
Following transfection of HUVECs with miR-34a
inhibitor, cell cycle distribution was evaluated by flow cytometry.
The G1/S transition was significantly promoted in cells transfected
with miR-34a inhibitor when compared with NC cells (P<0.05;
Fig. 4). This data indicates that
miR-34a may inhibit the proliferation and repair of vascular
endothelial cells by regulation of the G1/S transition.
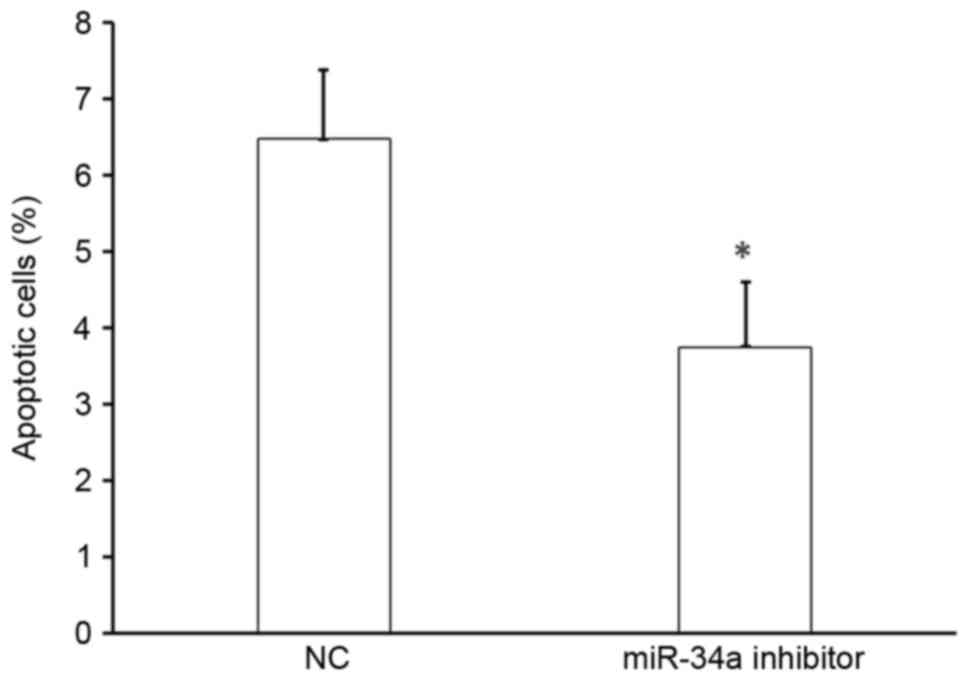
Effect of miR-34a on the apoptosis of
HUVECs
To determine the effect of miR-34a on cell
apoptosis, HUVECs were transfected with miR-34a inhibitor or
scramble-miR (NC), stained with Annexin V-FITC and propidium
iodide, and analyzed by flow cytometry. There was a significant
decrease in the number of apoptotic cells following transfection
with miR-34a inhibitor compared with that following NC transfection
(P<0.05; Fig. 5). Thus, miR-34a
promotes the apoptosis of HUVECs and miR-34a was upregulated in
patients with hypertension, suggesting that miR-34a may accelerate
vascular endothelial injury.
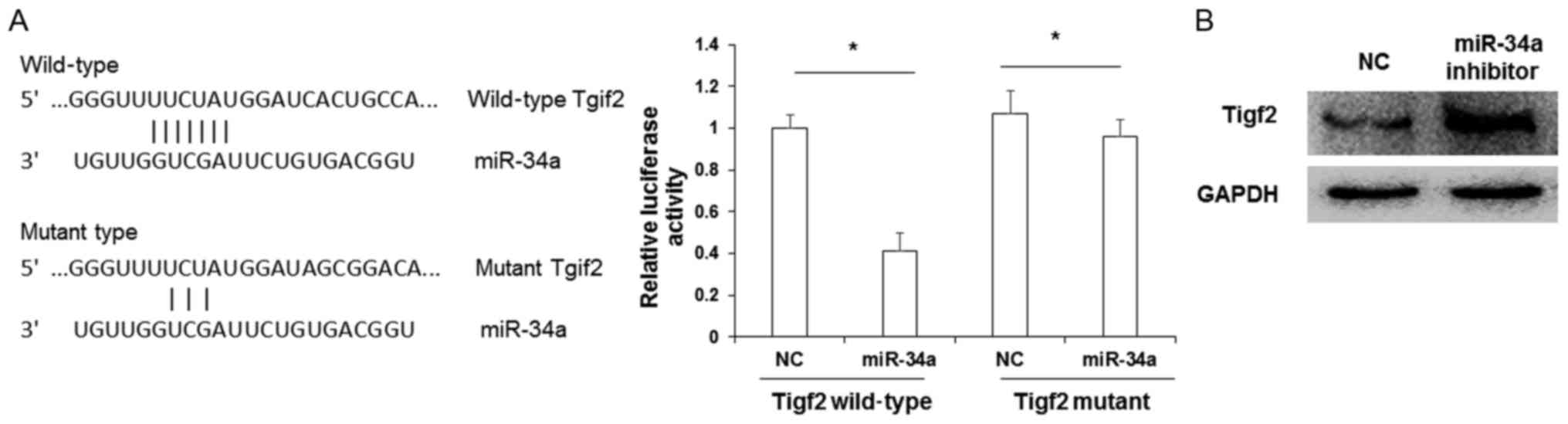
Tigf2 is a target of miR-34a
Next, the downstream targets of miR-34a were
investigated. As predicted by computational screening, the 3′ UTR
of Tigf2 was identified to contain multiple potential binding sites
for miR-34a. To determine whether miR-34a directly targeted Tigf2,
reporter gene assays were performed. The binding sequence of
miR-34a to Tigf2 is presented in Fig.
6A. Cotransfection of a Tigf2 wild-type 3′ UTR construct and
miR-34a mimic in HEK293T cells lead to a significant reduction in
relative luciferase activity (P<0.05 vs. Tigf2 wild-type 3′ UTR
+ NC transfectants). By contrast, relative luciferase activity
following cotransfection with Tigf2 mutant 3′ UTR and miR-34a mimic
was not significantly altered when compared with Tigf2 mutant 3′
UTR + NC transfectants. To verify that TIGF2 was a target of
miR-34a, a western blot analysis was performed. It was observed
that the protein expression of TIGF2 was markedly upregulated
following transfection with miR-34a inhibitor when compared with NC
cells (Fig. 6B). These data indicate
that the biological function of miR-34a may be correlated with the
level of TIGF2 expression.
Discussion
In the present study, miR-34a was significantly
upregulated in the peripheral blood of patients with hypertension,
and its expression was correlated with the clinical phase of
hypertension. In vitro assays also demonstrated that miR-34a
suppressed the proliferation and migration and promoted the
apoptosis of HUVECs. These results indicate that miR-34a may
accelerate vascular endothelial injury in the development of
hypertension.
Hypertension is a complex and common disease
resulting from the interaction of environmental and genetic
factors, and the molecular mechanism underlying the development of
hypertension remains unclear (18).
Recent studies demonstrated that numerous miRNAs were involved in
the development of hypertension by mediating vascular remodeling
and injury of the heart, kidney and other organs (19,20). For
instance, miR-122 induced endothelial NO metabolic disorder and
disrupted diastolic and contractile function of the vascular
endothelium by targeting solute carrier family 7 member 1, leading
to the development of primary hypertension (21). Furthermore, miR-204 may be involved
in vascular remodeling by regulating the proliferation and
apoptosis of vascular smooth muscle cells (22). In addition, a number of other miRNAs
have been associated with hypertension, including miR-296-5p,
let-7e, miR-15b and miR-185, and their roles in regulating the
development of hypertension require further investigation (23). The results of the present study
demonstrated that miR-34a was significantly upregulated in patients
with phase III hypertension compared with patients presenting with
phase I and II hypertension, thus indicating that miR-34a may be
closely associated with hypertension.
The vascular endothelium, which envelops circulating
blood in a continuous monolayer of squamous cells, serves key
functions in the regulation of blood flow and exchange of water and
small molecules (24). Additionally,
it has been observed that continuous hypertension may induce
vascular endothelial injury and vascular endothelial dysfunction
(25). Numerous miRNAs have been
implicated in hypertension-induced vascular endothelial injury
(26). miR-34a is a recently
identified miRNA that has been associated with tumor development,
vascular injury, cell proliferation and apoptosis (27). Furthermore, overexpression of miR-34a
may inhibit the proliferation and migration of various tumor cells,
including lung, colon and gastric cancer cells (28,29).
Overexpression of miR-34a may also induce the apoptosis of human
brain glioblastoma cells (30). In
particular, miR-34a has been closely associated with pulmonary
arterial hypertension, cell proliferation and apoptosis in patients
with chronic obstructive pulmonary disease (31). The present study demonstrated that
miR-34a may suppress the proliferation and migration and promote
the apoptosis of HUVECs, indicating a correlation between miR-34a
upregulation and vascular endothelial injury.
Using bioinformatics prediction methods, Tgif2 was
identified as a potential target of miR-34a. Tgif2 is a member of
the three-amino-acid loop super family, and the TGIF2 protein
encoded by Tgif2 mRNA may bind to Smad to inhibit the transforming
growth factor-β signaling pathway (32). Additionally, members of the TGIF
family have been documented to promote cell proliferation and
differentiation and inhibit cell apoptosis (33). However, there is no reports in the
role of TGIF in hypertension. The present study revealed that
miR-34a may directly bind to the 3′UTR of Tgif2 mRNA. Therefore it
was speculated that miR-34a suppressed the proliferation and
migration and promoted the apoptosis of HUVECs by downregulating
Tgif2 expression and aggravating vascular endothelial injury.
In summary, miR-34a was upregulated in the
peripheral blood of patients with hypertension, and overexpression
of miR-34a may have promoted vascular endothelial injury through
targeting of Tgif2. Therefore, miR-34a may be a potential marker
for the clinical diagnosis and treatment of primary hypertension
and vascular injury.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372473) and the
China Postdoctoral Science Foundation (grant no. 2014M550766).
References
|
1
|
Bönner G: Use of behavioral therapies in
hypertention. MMW Fortschr Med. 157:63–64. 2015. View Article : Google Scholar
|
|
2
|
Iaitskiĭ NA, Bedrov AIa, Maslevtsov DV,
Tsvetkova EA and Moiseev AA: Ischemic heart disease and arterial
hypertention as risk factors of surgical treatment of patients with
infrarenal segment of aortic aneurysm. Vestn Khir Im I I Grek.
172:11–15. 2013.
|
|
3
|
Leopold JA: Catheter-based therapies for
patients with medication-refractory pulmonary arterial
hypertension. Circ Cardiovasc Interv. 8:e0033322015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu H, Zheng H, Huang J, Shen Y and Luo M:
T-cell subsets are associated with serum homocysteine concentration
in patients with essential hypertension. Clin Exp Hypertens.
39:377–381. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang S, Zeng W, Qin J and Jiang M:
Diagnosis and treatment of portal hypertension caused by superior
mesenteric arteriovenous fistula. Zhonghua Gan Zang Bing Za Zhi.
23:638–640. 2015.(In Chinese). PubMed/NCBI
|
|
6
|
Balduino Mendes AB, Giollo-Junior LT, de
Andrade DO, Gregório ML, Yugar-Toledo JC and Vilela-Martin JF: How
to investigate the vascular changes in resistant hypertension. Curr
Hypertens Rev. 12:139–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jenkins D: Pulmonary endarterectomy: The
potentially curative treatment for patients with chronic
thromboembolic pulmonary hypertension. Eur Respir Rev. 24:263–271.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaillancourt M, Ruffenach G, Meloche J and
Bonnet S: Adaptation and remodelling of the pulmonary circulation
in pulmonary hypertension. Can J Cardiol. 31:407–415. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Caillon A, Mian MOR, Fraulob-Aquino JC,
Huo KG, Barhoumi T, Ouerd S, Sinnaeve PR, Paradis P and Schiffrin
EL: γδ T cells mediate angiotensin II-induced hypertension and
vascular injury. Circulation. 135:2155–2162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X, Yang Y, Yang D, Tong G, Lv S, Lin
X, Chen C and Dong W: Tetrandrine prevents monocrotaline-induced
pulmonary arterial hypertension in rats through regulation of the
protein expression of inducible nitric oxide synthase and cyclic
guanosine monophosphate-dependent protein kinase type 1. J Vasc
Surg. 64:1468–1477. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao Y, Jiang Z, Zeng Z, Liu Y, Gu Y, Ji Y,
Zhao Y and Li Y: Bcl-2 silencing attenuates hypoxia-induced
apoptosis resistance in pulmonary microvascular endothelial cells.
Apoptosis. 21:69–84. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matkovich SJ, Dorn GW II, Grossenheider TC
and Hecker PA: Cardiac disease status dictates functional mRNA
targeting profiles of individual microRNAs. Circ Cardiovasc Genet.
8:774–784. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kriegel AJ, Baker MA, Liu Y, Liu P, Cowley
AW Jr and Liang M: Endogenous microRNAs in human microvascular
endothelial cells regulate mRNAs encoded by hypertension-related
genes. Hypertension. 66:793–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lorenzen JM: Vascular and circulating
microRNAs in renal ischaemia-reperfusion injury. J Physiol.
593:1777–1784. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang MM, Fang MX, Chen LG, Wang HQ, Liu HJ
and Tang HL: Differential expression of microRNA in endothelial
cells incubated with serum of hypertension patients with
blood-stasis syndrome. Chin J Integr Med. 21:817–822. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Hong Z, Wu L, Ding B, Bi Y, Gu Z
and Li W: Dietary intake and cardiometabolic biomarkers in relation
to insulin resistance and hypertension in a middle-aged and elderly
population in beijing, China. Appl Physiol Nutr Metab. 42:869–875.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
DeCicco D, Zhu H, Brureau A, Schwaber JS
and Vadigepalli R: MicroRNA network changes in the brain stem
underlie the development of hypertension. Physiol Genomics.
47:388–399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cengiz M, Yavuzer S, Kılıçkıran Avcı B,
Yürüyen M, Yavuzer H, Dikici SA, Karataş ÖF, Özen M, Uzun H and
Öngen Z: Circulating miR-21 and eNOS in subclinical atherosclerosis
in patients with hypertension. Clin Exp Hypertens. 37:643–649.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qi H, Liu Z, Liu B, Cao H, Sun W, Yan Y
and Zhang L: micro-RNA screening and prediction model construction
for diagnosis of salt-sensitive essential hypertension. Medicine
(Baltimore). 96:e64172017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Z and Kaye DM: Mechanistic insights
into the link between a polymorphism of the 3′UTR of the SLC7A1
gene and hypertension. Hum Mutat. 30:328–333. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Potus F, Graydon C, Provencher S and
Bonnet S: Vascular remodeling process in pulmonary arterial
hypertension, with focus on miR-204 and miR-126 (2013 Grover
Conference series). Pulm Circ. 4:175–184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bockmeyer CL, Maegel L, Janciauskiene S,
Rische J, Lehmann U, Maus UA, Nickel N, Haverich A, Hoeper MM,
Golpon HA, et al: Plexiform vasculopathy of severe pulmonary
arterial hypertension and microRNA expression. J Heart Lung
Transplant. 31:764–772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang L, Wang X, Miao Y, Chen Z, Qiang P,
Cui L, Jing H and Guo Y: Magnetic ferroferric oxide nanoparticles
induce vascular endothelial cell dysfunction and inflammation by
disturbing autophagy. J Hazard Mater. 304:186–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Renga B, Cipriani S, Carino A, Simonetti
M, Zampella A and Fiorucci S: Reversal of endothelial dysfunction
by GPBAR1 agonism in portal hypertension involves a AKT/FOXOA1
dependent regulation of H2S generation and endothelin-1. PLoS One.
10:e01410822015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng L, Blanco FJ, Stevens H, Lu R,
Caudrillier A, McBride M, McClure JD, Grant J, Thomas M, Frid M, et
al: MicroRNA-143 activation regulates smooth muscle and endothelial
cell crosstalk in pulmonary arterial hypertension. Circ Res.
117:870–883. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye Z, Fang J, Dai S, Wang Y, Fu Z, Feng W,
Wei Q and Huang P: MicroRNA-34a induces a senescence-like change
via the down-regulation of SIRT1 and up-regulation of p53 protein
in human esophageal squamous cancer cells with a wild-type p53 gene
background. Cancer Lett. 370:216–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Hu, Qingha Pu, Bin Cui and Jia Lin:
MicroRNA-34a inhibits tumor invasion and metastasis in gastric
cancer by targeting Tgif2. Int J Clin Exp Pathol. 8:8921–8928.
2015.PubMed/NCBI
|
|
29
|
Wang H, Zhao X, Guo C, Ren D, Zhao Y, Xiao
W and Jiao W: Aptamer-dendrimer bioconjugates for targeted delivery
of miR-34a expressing plasmid and antitumor effects in non-small
cell lung cancer cells. PLoS One. 10:e01391362015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li SZ, Hu YY, Zhao J, Zhao YB, Sun JD,
Yang YF, Ji CC, Liu ZB, Cao WD and Qu Y: MicroRNA-34a induces
apoptosis in the human glioma cell line, A172, through enhanced ROS
production and NOX2 expression. Biochem Biophys Res Commun.
444:6–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mizuno S, Bogaard HJ, Gomez-Arroyo J,
Alhussaini A, Kraskauskas D, Cool CD and Voelkel NF:
MicroRNA-199a-5p is associated with hypoxia-inducible factor-1α
expression in lungs from patients with COPD. Chest. 142:663–672.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Willer A, Jakobsen JS, Ohlsson E, Rapin N,
Waage J, Billing M, Bullinger L, Karlsson S and Porse BT: TGIF1 is
a negative regulator of MLL-rearranged acute myeloid leukemia.
Leukemia. 29:1018–1031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Powers SE, Taniguchi K, Yen W, Melhuish
TA, Shen J, Walsh CA, Sutherland AE and Wotton D: Tgif1 and Tgif2
regulate nodal signaling and are required for gastrulation.
Development. 137:249–259. 2010. View Article : Google Scholar : PubMed/NCBI
|