Introduction
Osteosarcoma (OS) is the most common type of primary
bone cancer in children and adolescents (1). Despite medical advances and the
development of novel treatments, the 5-year survival rate (65%) for
patients with OS remains poor (2)
and the underlying mechanisms of bone carcinogenesis have remained
elusive. Therefore, research into these underlying molecular
mechanisms is urgently required.
Long non-coding (lnc)RNAs are evolutionarily
conserved sections of RNA that are >200 nucleotides in length.
Numerous lncRNAs do have known biological functions, such as the
transcriptional and translational regulation of protein coding
sequences 2 (3). Recent studies have
demonstrated that lncRNAs serve a key role within diverse
biological processes, including cell cycle progression,
differentiation, proliferation, apoptosis and tumorigenesis, by
regulating gene expression at the transcriptional and
post-transcriptional stages (4,5). The
urothelial cancer associated 1 lncRNA is highly expressed in
bladder cancer samples and its elevated expression has been
correlated with an increased rate of tumor recurrence (6). Metastasis associated lung
adenocarcinoma transcript 1 (MALAT1) lncRNA exhibits abnormal
expression in numerous types of human carcinoma, including colon,
lung, breast and ovarian cancer (7–10). It
has been reported that MALAT1 could be a novel biomarker for
predicting tumor recurrence after liver transplantation (11). Hox transcript antisense intergenic
RNA (HOTAIR) lncRNA is known to bind directly to the polycomb
repressive complex 2 (PRC2), and cause silencing of the HOXD
antisense growth-associated lncRNA gene (12). Previous studies have reported that
HOTAIR is involved in the regulation of tumor invasion and
metastasis (13,14). In breast cancer, the overexpression
of HOTAIR significantly increases tumor invasion and promotes
cancer metastasis by altering the methylation of histone H3 lysine
27 (15). HOTAIR was also identified
to be significantly upregulated in ovarian cancer, but knockdown of
HOTAIR was revealed to repress cell invasion and the viability of
ovarian cancer cells by controlling the expression of Ras-related
protein Rab-22A (16). Unlike in
other types of cancer, the underlying mechanism by which lncRNA
HOTAIR affects the development of OS remains unknown.
In the present study it was demonstrated that HOTAIR
was overexpressed in OS samples, and that the silencing of HOTAIR
inhibited the growth and invasiveness of MG-63 OS cells. These
results suggest that lncRNA HOTAIR is involved in the progression
of OS and may serve an oncogenic role in promoting the malignancy
of OS cells.
Materials and methods
Patient samples
A total of 60 samples of OS tissues and adjacent
non-tumor tissues were obtained from patients who underwent surgery
at Tianjin Hospital between February 2011 and January 2015. Two
pathologists independently evaluated the histological diagnosis and
differentiation of the tissue samples, according to the World
Health Organization classification system (5). All of the tissue samples collected were
immediately snap-frozen in liquid nitrogen and stored at −80°C
until required. The present study was approved by the Research
Ethics Committee of Tianjin Hospital (Tianjin, China), and informed
consent was obtained from all patients. The clinicopathological
characteristics of the patients are provided in Table I; a low expression of HOTAIR was
considered to be <1.2 times the normal level and a high
expression of HOTAIR was considered to be >1.2 times the normal
level.
 | Table I.LncRNA HOTAIR expression and the
clinicopathological features of the patients with OS. |
Table I.
LncRNA HOTAIR expression and the
clinicopathological features of the patients with OS.
|
|
| HOTAIR expression
(no. of patients) |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of patients
(%) | Low (%) | High (%) | P-value |
|---|
| Age (years) |
|
|
| 0.795 |
|
<20 | 34 (100) | 16 (47) | 18 (53) |
|
| ≥20 | 26 (100) | 14 (54) | 12 (46) |
|
| Gender |
|
|
| 0.599 |
| Male | 38 (100) | 22 (58) | 16 (42) |
|
|
Female | 22 (100) | 11 (50) | 11 (50) |
|
| Tumor size
(cm) |
|
|
| 1.000 |
|
≥10 | 20 (100) | 10 (50) | 10 (50) |
|
|
<10 | 40 (100) | 19 (48) | 21 (52) |
|
| Tumor grade |
|
|
| 0.035 |
| I | 9
(100) | 7
(78) | 2
(22) |
|
| II | 15 (100) | 6
(40) | 9
(60) |
|
|
III | 36 (100) | 11 (31) | 25 (69) |
|
| Distant
metastasis |
|
|
| 0.007 |
|
Absent | 37 (100) | 27 (73) | 10 (27) |
|
|
Present | 23 (100) | 8
(35) | 15 (65) |
|
Cell culture
The OS cell line MG-63 was purchased from the
American Type Culture Collection (Manassas, VA, USA). Cells were
maintained in Dulbecco's modified Eagles medium (DMEM) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), 100 U/ml penicillin and 100 mg/ml
streptomycin in a humidified environment at 37°C with 5%
CO2.
RNA interference and transfection
Specific small interfering RNA (siRNA/siR) targeting
HOTAIR mRNA (siR-HOTAIR1, 3′-GAACGGGAGUACAGAGAGAUU-5′; siR-HOTAIR2,
3′-CCACAUGAACGCCCAGAGAUU-5′) and negative control with scrambled
sequence (siR-NC) were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China), and transfected using Invitrogen
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. A total of
3×105 MG-63 cells were seeded into 6-well plates and
divided into four groups as follows: Mock group, cells without
transfection; siR-NC group, cells transfected with the negative
control siRNA; siR-HOTAIR1 group, cells transfected with
siR-HOTAIR1; and siR-HOTAIR2 group, cells transfected with
siR-HOTAIR2. After 48 h the effects of gene silencing were measured
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis. HOTAIR was silenced in order to explore the
association between the expression of HOTAIR and the behavior of
MG-63 cells.
Cell proliferation
The proliferation of MG-63 cells was measured using
a Cell Counting Kit (CCK)-8 assay (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) according to the manufacturer's protocol,
after siRNA transfection for 0,12, 24 and 48 h. A total of
3×103 MG-63 cells were seeded in 96-well plates and 10
µl CCK-8 was added and the samples were incubated for 4 h at 37°C.
Subsequently, the OD value was measured at 450 nm using a Bio-Rad
3550 microplate reader (Bio-Rad Laboratories, Hercules, CA,
USA).
Cell cycle analysis
MG-63 cells (3×103) were seeded in 6-well
plates. After digestion and centrifugation, the plates were fixed
in 70% ethanol at 4°C overnight and then washed three times in PBS.
A total of 50 µg/ml propidium iodide (PI; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was added and the plates were incubated
for 30 min at room temperature in the dark. Measurement of the cell
cycle was then performed using a BD FACSCalibur™ flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and analysis
was performed using ModFit LT software (Verity Software House,
Topsham, ME, USA).
Cell apoptosis analysis
Cell apoptosis was measured using an Annexin
V-FITC/PI Apoptosis Detection kit (Beyotime Institute of
Biotechnology, Haimen, China) according to the manufacturer's
protocol. Briefly, cells (3×105) were cultured in 6-well
plates, trypsinized 48 h after transfection and then washed with
cold PBS. Cells were then stained by adding the Annexin V reaction
mixture consisting of 10 µl Annexin V and 5 µl PI, and were
incubated at room temperature for 15 min in the dark. The stained
cells were measured using a BD FACSCalibur according to the
manufacturer's protocol and analysis was performed using FlowJo 6
software (FlowJo, LLC, Ashland, OR USA).
RT-qPCR analysis
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) from MG-63 OS cells and tissue
samples. RNA Lysis Buffer (RLA) was provide by Promega Corporation
(Madison, WI, USA). Complementary (c)DNA was synthesized using a
cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China)
in accordance with the manufacturer's protocol. Then, a
SYBR® Premix Ex Taq™ II kit (Takara
Biotechnology Co., Ltd.) was used following the manufacturer's
protocol to amplify the cDNA. The PCR condition s were as follows:
Denaturation at 94°C for 3 min, followed by 30 cycles at 94°C for
30 sec, 55°C for 30 sec and a final elongation at 72°C for 60 sec,
then 72°C for 10 min. The expression of GAPDH was used as a
reference to normalize the amount of lncRNA or mRNA in each sample.
The results were quantified using the 2−ΔΔCq method
(17). The PCR primers used were as
follows: HOTAIR forward, 5′-GGTAGAAAAAGCAACCACGAAGC-3′ and reverse,
5′-ACATAAACCTCTGTCTGTGAGTGCC-3′; G1/S-specific cyclin D1
(cyclin D1) forward, 5′-TCTCCAAAATGCCAGAGGCGAGGAAGTTGTTGGGGCTCCT;
cyclin E forward, 5′-GTGGCTCCGACCTTTCAGTC-3′ and reverse,
5′-CACAGTCTTGTCAATCTTGGCA-3′; cyclin-dependent kinase (CDK)-4
forward, 5′-TGCCAATTGCATCGTTCACCGAG-3′ and reverse,
5′-TGCCCAACTGGTCGGCTTCA-3′; CDK2 forward, 5′-GCCTAATCTCACCCTCTCC-3′
and reverse, 5′-CCCTTTCACCCCTGTATTCC-3′; p21 forward,
5′-GTCCTGGATGTTGACTGCCTTGA-3′ and reverse,
5′-GTCCAGCAAATCCAAGCTGTCTC-3′; p27 forward,
5′-CATTAACCCACCGGAGCTGTTTAC-3′ and reverse,
5′-GGTTAGCGGAGCAGTGTCCA-3′; matrix metalloproteinase (MMP)2
forward, 5′-GTGGATGATGCCTTTGCTC-3′ and reverse,
5′-CAGGAGTCCGTCCTTACC-3′; MMP9 forward,
5′-AATCTCTTCTAGAGACTGGGAAGGAG-3′ and reverse,
5′-AGCTGATTGACTAAAGTAGCTGGA-3′; epithelial (E)-cadherin forward,
5′-CGGGAATGCAGTTGAGGATC-3′ and reverse, 5′-AGGATGGTGTAAGCGATGGC-3′;
neural (N)-cadherin forward, 5′-ACAGTGGCCACCTACAAAGG-3′ and
reverse, 5′-CCGAGATGGGGTTGATAATG-3′; and CD44 forward,
5′-TCCCAGACGAAGACAGTCCCTGGAT-3′ and reverse,
5′-CACTGGGGTGGAATGTGTCTTGGTC-3′. GAPDH was the control for the PCR
assay, with a forward primer sequence of
5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and reverse primer sequence of
5′-CATGTGGGCCATGAGGTCCACCAC-3′.
Western blot analysis
MG-63 cells were transfected with siR-HOTAIR1 or
siR-NC for 48 h, and the proteins were extracted using a
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China). The Bradford assay was used to
determine the protein concentration. A total of 50 µg proteins were
separated by 10% SDS-PAGE (Beyotime Institute of Biotechnology) and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked with 5% non-fat milk at 4°C overnight and then
incubated at 4°C overnight with rabbit or mouse antibodies specific
for the following human proteins: CD44 (1:1,500, sc-65265), CDK4
(1:1,500, sc-23896), MMP2 (1:1,500, sc-13594) and MMP9 (1:1,500,
sc-21733) (all from Santa Cruz Biotechnology, Inc., Dallas, TX,
USA), cyclin D1 (1:1,500, #2926), cyclin E (1:1,500, #2925), p27
(1:1,500, #3686) (all from Cell Signaling Technology, Inc.,
Danvers, MA, USA). Antibodies against phosphoinositide 3-kinase
(PI3K, 1:1,500, #4255), RAC α serine/threonine-specific protein
kinase (AKT, 1:1,500, #4060), mammalian target of rapamycin (mTOR,
1:1,500, #2983), phosphorylated (p)-PI3K (tyr458, 1:1,000, #4228S),
p-AKT (ser473, 1:1,000, #4060) and p-mTOR (ser2448, 1:1,000, #5536)
were purchased from Cell Signaling Technology, Inc. Mouse
anti-human GAPDH antibodies (1:3,000, G8795), which were used as a
control, were purchased from Sigma-Aldrich (Merck KGaA). The next
day the membranes were probed with secondary anti-rabbit IgG
HRP-linked antibody (1:5,000; #7074) or anti-mouse IgG HRP-linked
antibody (1:5,000; #4408) (both from Cell Signaling Technology,
Inc.). After washing three times in TBS-Tween 20, the membranes
were incubated with the corresponding secondary antibodies for 1 h
and then detected by an enhanced Pierce chemiluminescence system
(ECL; Thermo Fisher Scientific, Inc.).
Cell migration and invasion assay
Cell migration and invasion were assayed using a
Transwell invasion assay (invasion Transwell chambers; EMD
Millipore, Billerica, MA, USA) with and without
Matrigel™ (BD Biosciences). The cells were seeded at a
density of 1×105 cells on the upper chamber with 200 µl
serum-free DMEM. Following transfection for 48 h, 600 µl DMEM
supplemented with 20% FBS, which served as a chemoattractant, was
added to the lower chamber. After 48 h incubation at 37°C, the
upper side of the membrane was wiped with cotton wool to remove
noninvasive cells; the membranes were then fixed with 4% methanol
at 4°C and stained with 0.1% crystal violet at room temperature.
Five visual fields with a magnification ×200 were randomly selected
from each membrane and the cell numbers were counted using a light
microscope.
Statistical analysis
All data are expressed as the mean ± standard
deviation (n=3). The significance of differences between two groups
was estimated using a Student's t-test. The significance of
differences between multiple groups was determines using one-way
analysis of variance followed by a post hoc Tukey's range test.
SPSS software (version 21.0; IBM Corp., Armonk, NY, USA) was used
to perform the statistical analysis. All tests performed were two
sided. P<0.05 was considered to indicate a statistically
significant difference.
Results
HOTAIR expression is significantly
increased and correlated with lymph node metastasis in patients
with OS
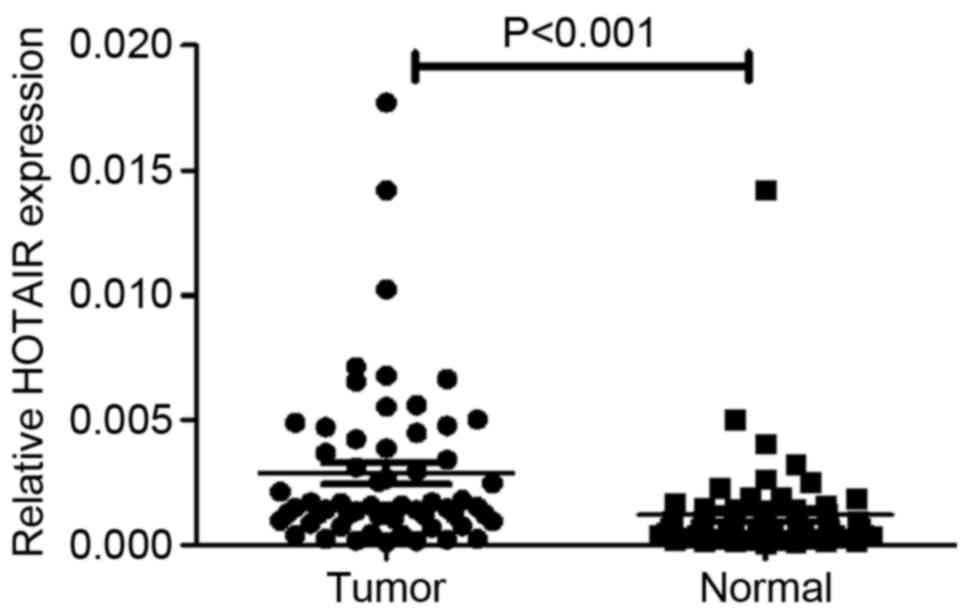
RT-qPCR analysis determined that the expression of
HOTAIR was significantly increased in clinical OS specimens
compared with adjacent normal tissues (P<0.001; Fig. 1). Clinicopathological analysis
demonstrated that when the 60 tissue pairs were tested, the
expression of HOTAIR demonstrated no difference between age, gender
or tumor size; however, it was significantly associated with tumor
grade (P=0.035) and distant metastasis (P=0.007) (Table I). These results indicate that HOTAIR
serves an oncogenic role in OS.
HOTAIR silencing reduces the
proliferation of MG-63 cells
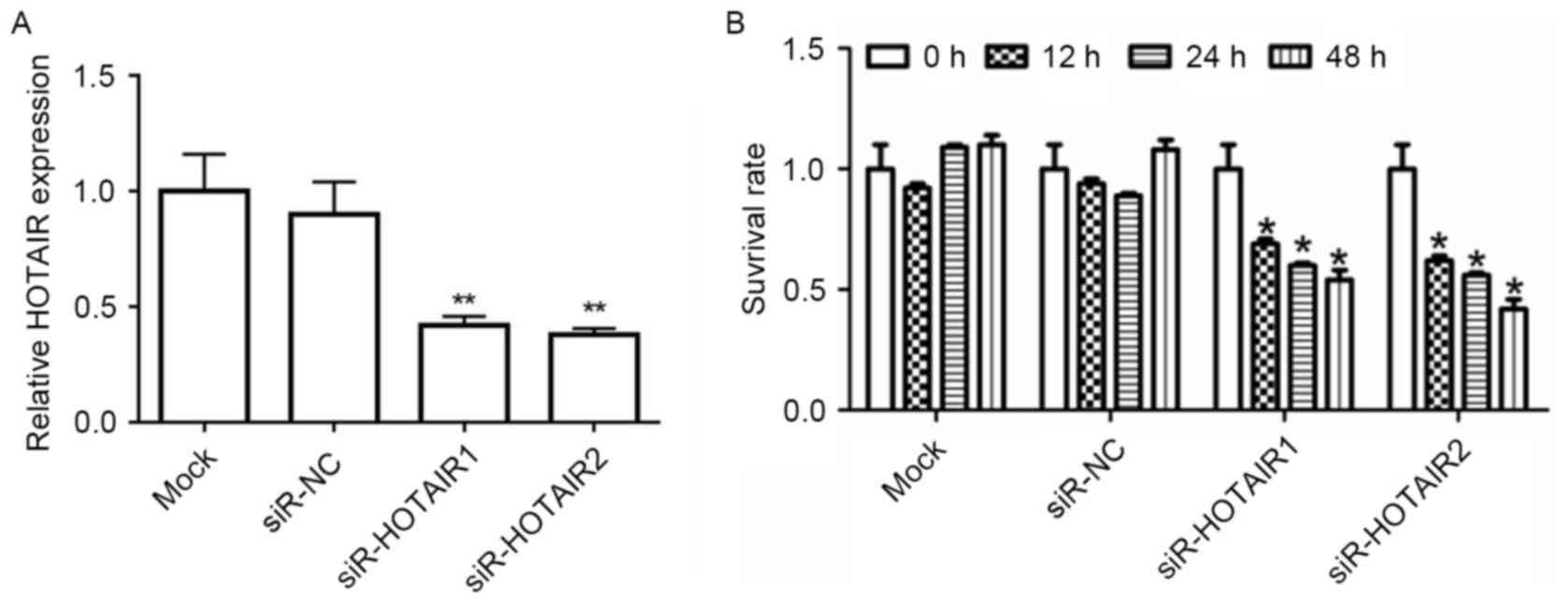
To determine whether HOTAIR affects tumorigenesis,
MG-63 cell lines were transfected with siR-HOTAIR1/2 or siR-NC.
RT-qPCR results revealed that, after being transfected with
siR-HOTAIR1/2, HOTAIR mRNA expression in the MG-63 cells decreased
significantly when compared to the mock group (P<0.01; Fig. 2A). A CCK-8 assay demonstrated that
when HOTAIR was knocked down, the survival rate of MG-63 cells was
significantly inhibited (P<0.05; Fig.
2B). This data supports the hypothesis that HOTAIR serves a
role in OS cell proliferation.
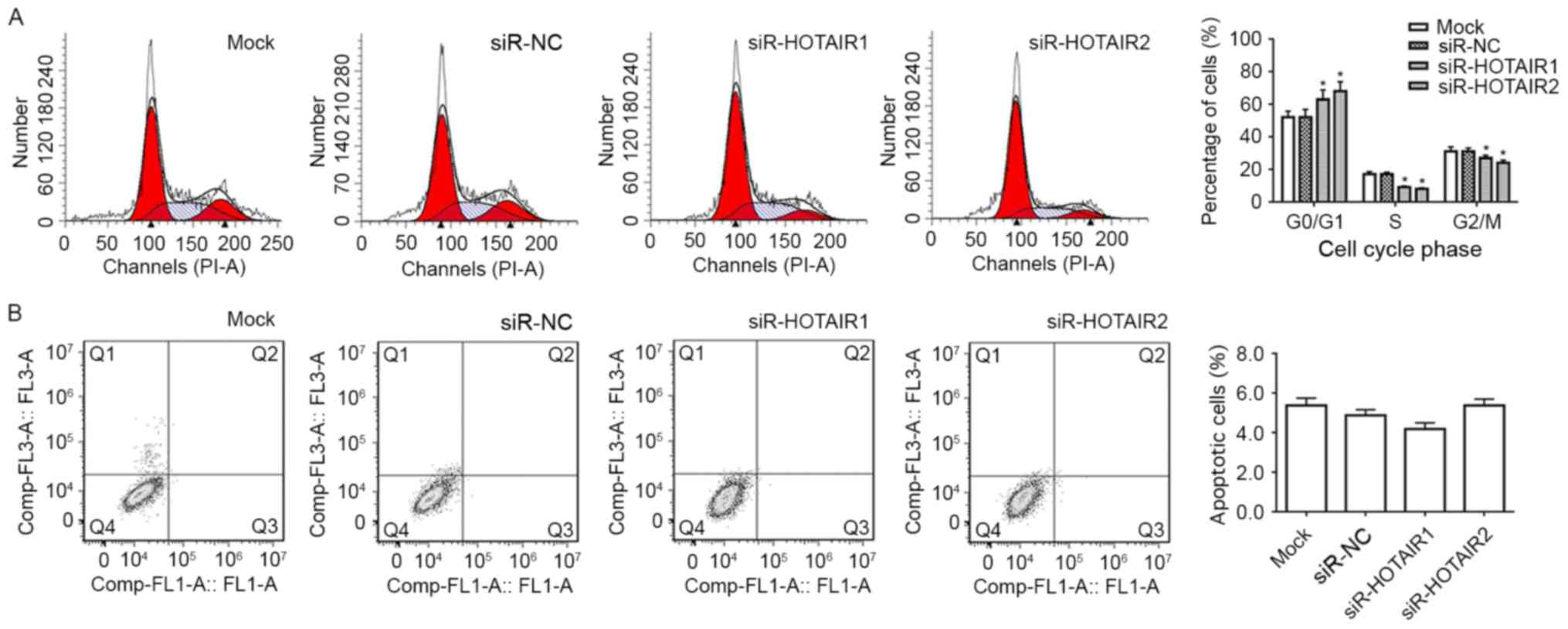
HOTAIR silencing induces the
G1 phase cell cycle arrest of MG-63 cells
Flow cytometry analysis was performed to determine
whether the reduced expression of HOTAIR contributed to cell cycle
arrest or apoptosis. It was revealed that MG-63 cells of
siR-HOTAIR1 group and siR-HOTAIR2 group had a significant increase
in the proportion of cells in the G1 phase compared with
the mock group (P<0.05; Fig. 3A).
This suggests that the cells have been prevented from moving into
the next phase of the cell cycle and have therefore been prevented
from proliferating. It was also revealed that the reduction in
HOTAIR expression did not significantly influence the apoptosis of
MG-63 cells (Fig. 3B). This data
suggests that the inhibitory effect HOTAIR silencing has on the
proliferation of MG-63 cells is through cell cycle arrest and not
apoptosis.
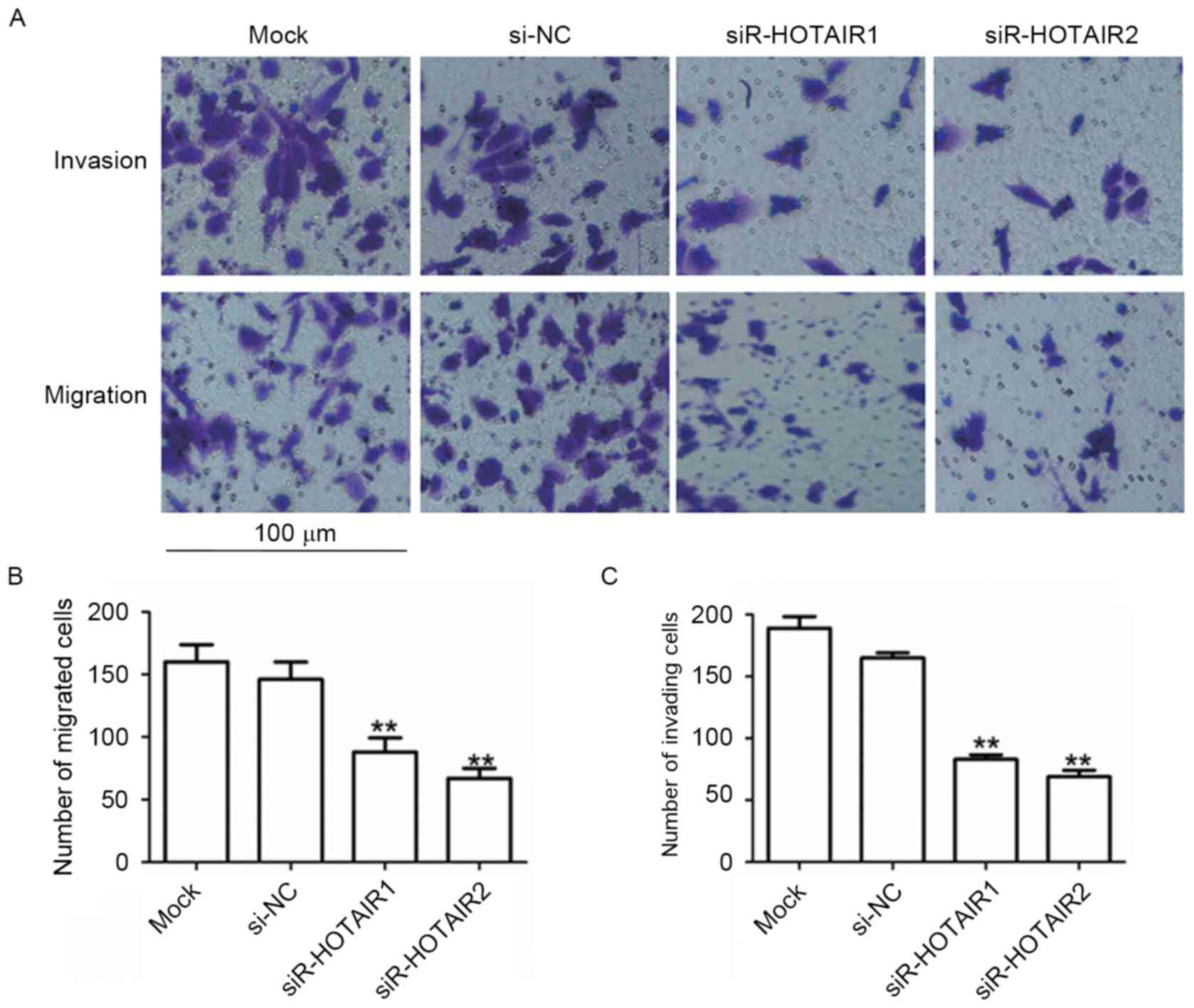
Knockdown of HOTAIR inhibits the
migration and invasion of MG-63 cells
To assess the effects of HOTAIR on cell migration
and invasion, a Transwell migration assay was performed. MG-63
cells transfected with siR-HOTAIR1 or siR-HOTAIR2 exhibited a
significant reduction in migration compared to the mock group
(P<0.01; Fig. 4A and B). A Boyden
chamber coated with Matrigel was used to determine the effect of
siR-HOTAIR on cell invasion after incubation for 48 h. The results
revealed that HOTAIR silencing significantly decreased the invasion
of MG-63 cells (P<0.01; Fig. 4A and
C). The suppressive effects of siR-HOTAIR explain the
association observed between high HOTAIR expression and distant
metastasis in patients with OS. This data indicates that siR-HOTAIR
can inhibit a migratory and invasive phenotype in OS cells.
HOTAIR regulates the expression of
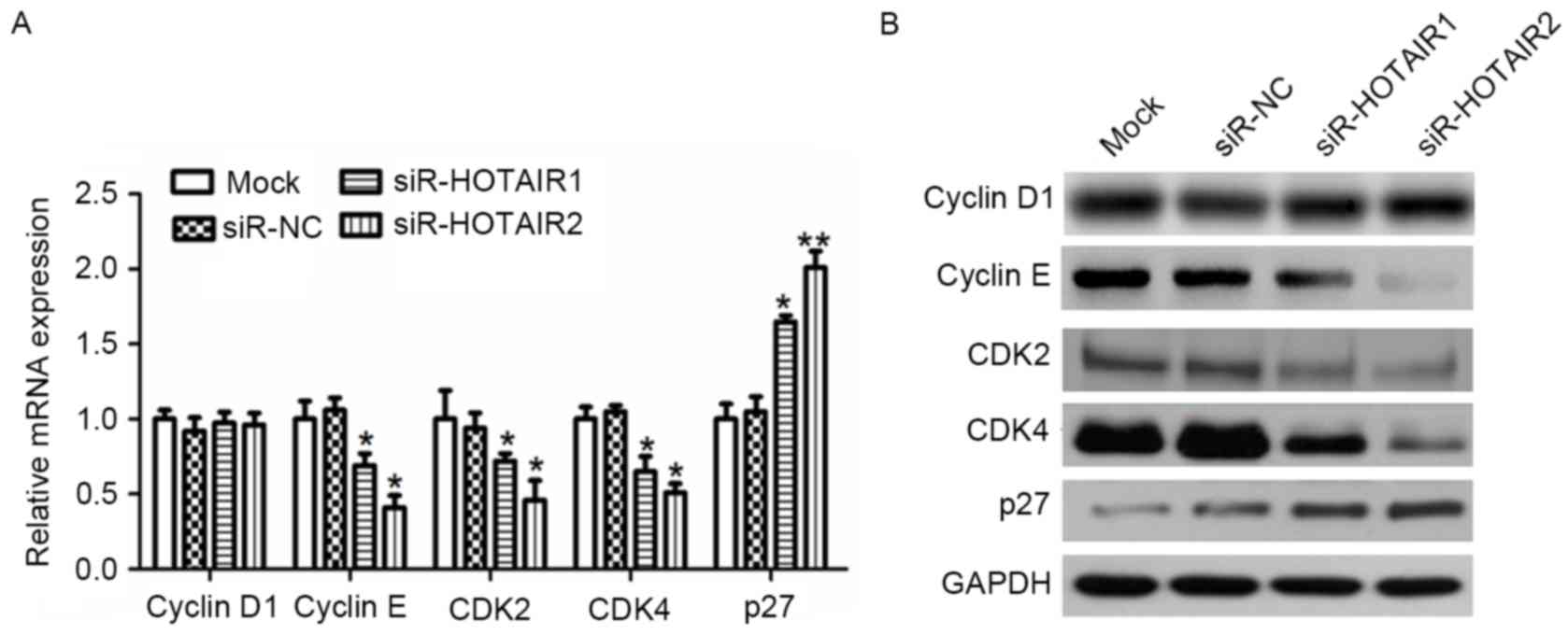
cyclin D1, cyclin E, CDK2, CDK4 and p27 within OS cells
To investigate the evidence that siR-HOTAIR serves a
role in G1/S arrest, RT-qPCR (Fig. 5A) and western blot analysis (Fig. 5B) were used to evaluate the effect of
HOTAIR silencing on the expression of CDK2, CDK4, cyclin D1 and
cyclin E, all of which are associated with progression from the
G1 to S phase in the cell cycle. The CDK inhibitor p27
was also studied. The results demonstrated that HOTAIR silencing
significantly reduced the mRNA expression of CDK2, CDK4 and cyclin
E compared to the controls (P<0.01), while cyclin D1 exhibited
little variation. Conversely, the mRNA expression of p27 was
significantly increased compared to the control (P<0.05).
Similar results were observed for protein expression. These results
suggest that HOTAIR mediates OS cell proliferation by affecting
cyclin E, CDK2, CDK4 and p27 expression.
Expression of MMP2, MMP9, CD44,
E-cadherin and N-cadherin is involved in the HOTAIR-mediated
promotion of OS cell metastasis
To explore the molecular mechanisms underlying the
effect of HOTAIR on the invasion and metastasis of OS cells,
potential targets involved in tumor invasion and metastasis were
investigated. The results revealed a marked decrease in CD44, MMP2,
MMP9 and N-cadherin expression, and a notable increase in
E-cadherin expression at the protein level (Fig. 6A). At the mRNA level a similar trend
was observed, which was significant in the siR-HOTAIR groups
compared to the mock group (P<0.01; Fig. 6B). This data suggests an association
between HOTAIR and MMP2, MMP9, CD44, E-cadherin and N-cadherin
expression, as HOTAIR treatment resulted in increased expression
levels of MMP2, MMP9, CD44 and N-cadherin, but decreased expression
levels of E-cadherin, which indicates that these factors may serve
an important role in the HOTAIR-mediated promotion of OS cell
invasion and metastasis.
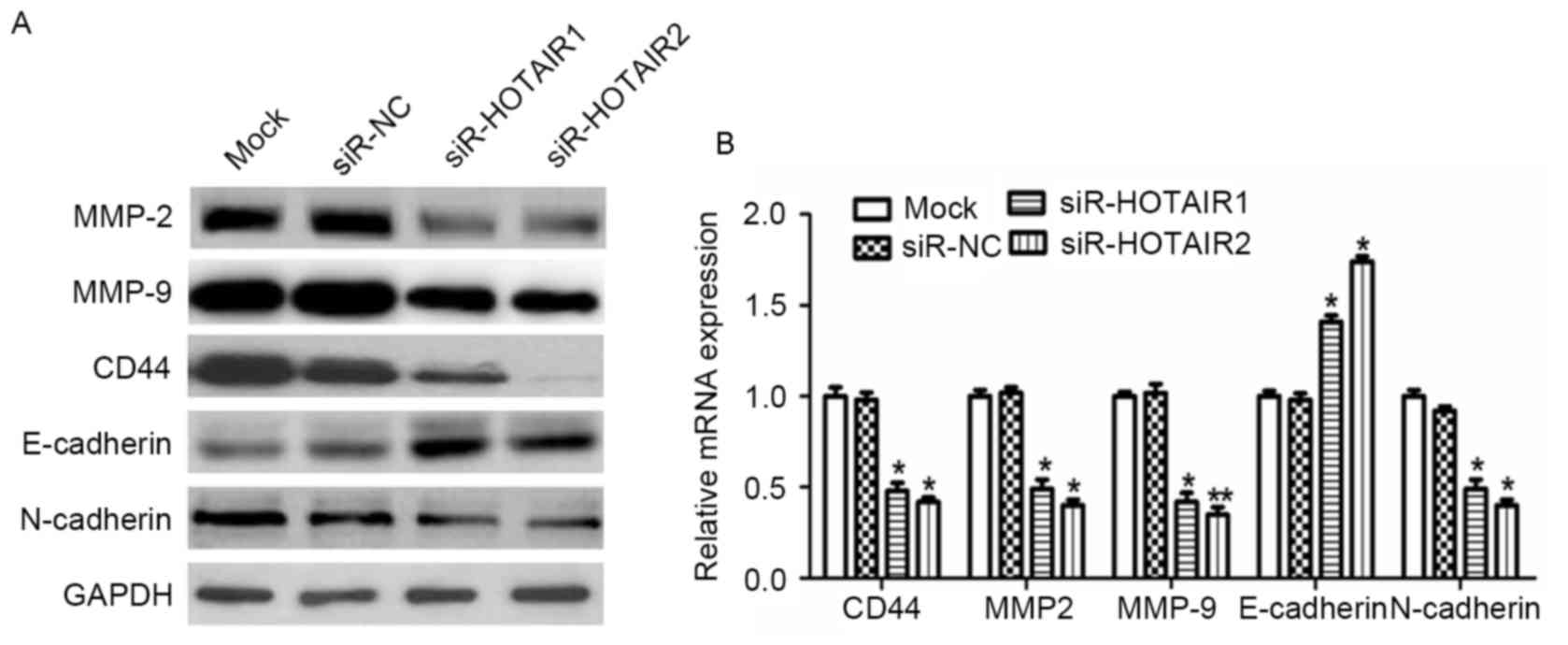 | Figure 6.HOTAIR silencing alters the expression
of proteins associated with tumor invasion and metastasis in MG-63
cells. The protein and mRNA expression of MMP2, MMP9, CD44,
E-cadherin and N-cadherin were detected by (A) western blot
analysis and (B) reverse transcription-quantitative polymerase
chain reaction analysis. *P<0.05, **P<0.01 vs. the mock
group. HOTAIR, Hox transcript antisense intergenic RNA; siR, small
interfering RNA; NC, negative control. MMP, matrix
metalloproteinase; E-cadherin, epithelial-cadherin; CD, cluster of
differentiation. |
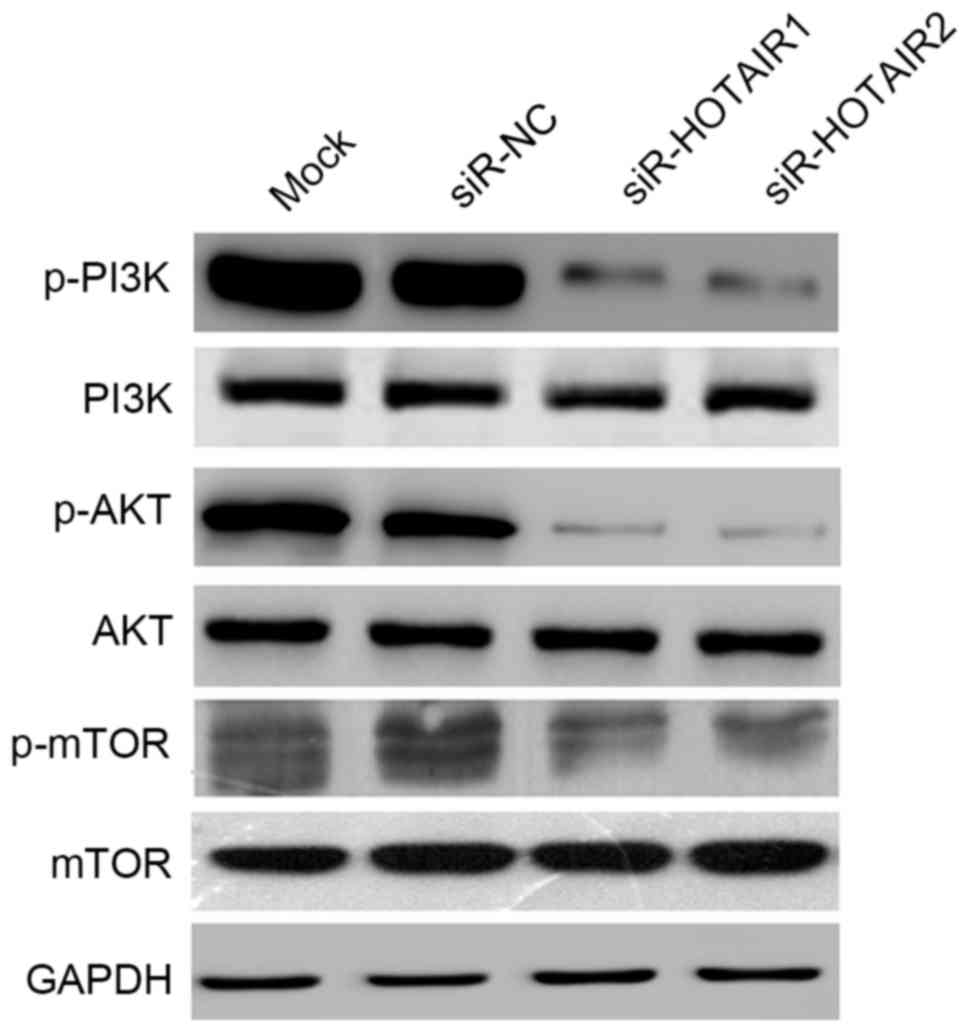
HOTAIR promotes OS cell growth through
activation of the AKT/mTOR signaling pathway
The AKT/mTOR signaling pathway serves a key role in
regulating cell growth, survival and metabolism. To further explore
the molecular mechanisms by which HOTAIR affects OS cell
proliferation and metastasis, the effects of HOTAIR silencing on
the AKT/mTOR signaling pathway were investigated. In the present
study the expression of three key proteins, p-mTOR (ser2448),
p-PI3K (tyr458) and p-AKT (ser473), was detected. HOTAIR silencing
reduced the expression of p-mTOR, p-PI3K and p-AKT, while no
notable changes were observed in the unphosphorylated proteins
(Fig. 7). These results suggest that
HOTAIR promotes OS cell growth through activation of the AKT/mTOR
signaling pathway.
Discussion
Numerous studies have demonstrated that changes in
the expression of certain lncRNAs are closely associated to the
occurrence and development of OS tumors (18,19). It
has also been revealed that lncRNAs serve an important role in the
invasion and metastasis of OS cells (20,21).
These lncRNAs can act as oncogenes or tumor suppressors (3,4).
Therefore, an improved understanding of the aberrant expression of
lncRNAs within OS cells may help to clarify the underlying
mechanisms of OS metastasis and provide novel targets for
therapies.
Recent studies have revealed that the lncRNA HOTAIR
is an oncogene in several different types of cancer, including
breast cancer, bladder cancer, colorectal cancer and OS (22–24). In
the present study, it was revealed that HOTAIR was upregulated in
clinical OS tissue samples, indicating that HOTAIR is a potential
molecular marker for OS. The present study used siRNA technology to
effectively knockdown HOTAIR mRNA expression in human MG-63 OS
cells. It was hypothesized that HOTAIR silencing may suppress OS
cell proliferation, adhesion, migration and invasion ability, and
thus it may serve a tumor suppressor role within OS cells.
Cyclin D1/E and CDK2/4 are considered important
tumor proliferation-associated genes (25,26). To
determine the molecular mechanism by which HOTAIR affects the
growth of OS cells, the effect of HOTAIR silencing on the
regulation of the cell cycle was examined. Cyclin E binds to
G1 phase CDK2 to form a complex, which is required for
the transition from the G1 to S phase of the cell cycle
(27). Cyclin D1 forms a complex
with and functions as a regulatory subunit of CDK4, whose activity
is also required for G1/S transition (25). In the present study it was revealed
that HOTAIR silencing reduced Cyclin E, but not Cyclin D1
expression. p27 binds to and prevents the activation of cyclin
E-CDK2 and cyclin D-CDK4 complexes, and thus controls the cell
cycle progression at the G1 stage (26). HOTAIR silencing resulted in the
upregulation of p27. These results indicate that HOTAIR decreases
the expression of p27, reducing control over the activation of
cyclin E/CDK2 and cyclin D-CDK4 complexes, resulting in
dysregulation of the cell cycle and thus the proliferation of OS
cells.
Tumor metastasis and invasion are closely associated
to the degradation of a variety of metastasis-associated proteins
and the destruction of the extracellular matrix (ECM). MMP-2/9,
CD44, E-cadherin and N-cadherin are all important tumor
metastasis-associated proteins. MMPs are considered the most
important set of proteases that degrade the ECM. MMP2 and MMP9 are
the most thoroughly studied MMPs (28). CD44 is a cell-surface glycoprotein
involved in cell-cell interactions, cell adhesion and migration.
CD44 is a receptor for hyaluronic acid and can also interact with
other ligands, including osteopontin, collagens and MMPs (29). E-cadherin, whose function is critical
for cell-cell adhesion and developmental morphogenesis, serves a
key role in cellular invasion. The loss of E-cadherin contributes
to the movement of tumor cells from their primary location. Often
tumors that have an increased degree of malignancy also have an
increased degree of invasion. E-cadherin can reduce the invasion
ability of a number of types of tumor cells (30). N-cadherin is a transmembrane protein
expressed in multiple tissues, which can provide a mechanism for
the transendothelial migration of cancer cells. If the
proto-oncogene tyrosine-protein kinase Src (Src) signaling pathway
is upregulated, Src phosphorylates β-catenins attached to
N-cadherin and E-cadherin, which can lead to the failure of the
intercellular connection, allowing the cancer cells to metastasize
(31).
The AKT/mTOR signaling pathway is a tumor-associated
signaling pathway. Activated AKT is known to regulate downstream
signaling proteins involved in cell survival, cell growth, cell
cycle progression (32). and
apoptosis (26). Overexpression of
p-AKT is associated with advanced human prostate cancer (33). mTOR, a protein downstream of AKT,
also serves a vital role in regulating cell proliferation, growth,
differentiation and survival. The present study revealed that the
silencing of HOTAIR expression notably reduced the phosphorylation
of mTOR and its upstream kinase AKT, suggesting that HOTAIR
silencing suppresses tumorigenesis by inhibiting the AKT/mTOR
signaling pathway and its multiple downstream signaling components
in OS cells.
In conclusion, the results of the present study
provide evidence that the expression of the lncRNA HOTAIR is
increased in OS and that it is associated with the progression of
OS. HOTAIR silencing inhibited the growth, adhesion, migration and
invasion of MG-63 OS cells. Therefore, HOTAIR silencing may
potentially form the basis of novel cancer treatments.
References
|
1
|
Cortini M, Avnet S and Baldini N:
Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett.
405:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu W, Tang L, Lin F, Yao Y, Shen Z and
Zhou X: The role of chemotherapy for metastatic, relapsed and
refractory osteosarcoma. Surg Oncol. 1:9–15. 2015. View Article : Google Scholar
|
|
3
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lavorgna G, Vago R, Sarmini M, Montorsi F,
Salonia A and Bellone M: Long non-coding RNAs as novel therapeutic
targets in cancer. Pharmacol Res. 110:131–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang CF and Zhu XZ: The fourth edition of
WHO classification of tumours of bone: An introduction. Zhonghua
Bing Li Xue Za Zhi. 10:652–654. 2013.(In Chinese).
|
|
6
|
Xue M, Li X, Wu W, Zhang S, Wu S, Li Z and
Chen W: Upregulation of long non-coding RNA urothelial carcinoma
associated 1 by CCAAT/enhancer binding protein α contributes to
bladder cancer cell growth and reduced apoptosis. Oncol Rep.
5:1993–2000. 2014. View Article : Google Scholar
|
|
7
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li J, Wang J, Chen Y, Li S, Jin M, Wang H,
Chen Z and Yu W: LncRNA MALAT1 exerts oncogenic functions in lung
adenocarcinoma by targeting miR-204. Am J Cancer Res. 6:1099–1107.
2016.PubMed/NCBI
|
|
9
|
Miao Y, Fan R, Chen L and Qian H: Clinical
significance of long Non-coding RNA MALAT1 expression in tissue and
serum of breast cancer. Ann Clin Lab Sci. 46:418–424.
2016.PubMed/NCBI
|
|
10
|
Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li
J and Sheng X: The long noncoding RNA MALAT-1 is highly expressed
in ovarian cancer and induces cell growth and migration. PLoS One.
11:e01552502016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
64:379–391. 2014. View Article : Google Scholar
|
|
13
|
Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren
X, Wei F, Yu W, Liu T, Wang X, et al: Long non-coding RNA HOTAIR
promotes tumor cell invasion and metastasis by recruiting EZH2 and
repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol.
46:2586–2594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee NK, Lee JH, Park CH, Yu D, Lee YC,
Cheong JH, Noh SH and Lee SK: Long non-coding RNA HOTAIR promotes
carcinogenesis and invasion of gastric adenocarcinoma. Biochem
Biophys Res Commun. 45:171–178. 2014. View Article : Google Scholar
|
|
15
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Z, Cheng J, Wu Y, Qiu J, Sun Y and
Tong X: LncRNA HOTAIR controls the expression of Rab22a by sponging
miR-373 in ovarian cancer. Mol Med Rep. 14:2465–2472. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Cui T, Sun L, Peng N and Yang C:
Long noncoding RNA LeXis promotes osteosarcoma growth through
upregulation of CTNNB1 expression. Am J Cancer Res. 7:1577–1587.
2017.PubMed/NCBI
|
|
19
|
Li Z, Dou P, Liu T and He S: Application
of long noncoding RNAs in osteosarcoma: Biomarkers and therapeutic
targets. Cell Physiol Biochem. 42:1407–1419. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Zhao L and Wang Q: Overexpression of
long non-coding RNA HOTTIP increases chemoresistance of
osteosarcoma cell by activating the Wnt/β-catenin pathway. Am J
Transl Res. 8:2385–2393. 2016.PubMed/NCBI
|
|
21
|
Li W, Xie P and Ruan WH: Overexpression of
lncRNA UCA1 promotes osteosarcoma progression and correlates with
poor prognosis. J Bone Oncol. 5:80–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun X, Du P, Yuan W, Du Z, Yu M, Yu X and
Hu T: Long non-coding RNA HOTAIR regulates cyclin J via inhibition
of microRNA-205 expression in bladder cancer. Cell Death Dis.
6:e19072015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H,
Tong N, Chen J, Zhang Z and Wang M: Genetic variants in lncRNA
HOTAIR are associated with risk of colorectal cancer. Mutagenesis.
30:303–310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang
K, Wang H, Zhang R and Liu Y: Overexpression of long Non-coding RNA
HOTAIR promotes tumor growth and metastasis in human osteosarcoma.
Mol Cells. 38:432–440. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Takahashi-Yanaga F and Sasaguri T:
GSK-3beta regulates cyclin D1 expression: A new target for
chemotherapy. Cell Signal. 20:581–589. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sheaff RJ, Groudine M, Gordon M, Roberts
JM and Clurman BE: Cyclin E-CDK2 is a regulator of p27Kip1. Genes
Dev. 11:1464–1178. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yue J, Zhang K and Chen J: Role of
integrins in regulating proteases to mediate extracellular matrix
remodeling. Cancer Microenviron. 5:275–283. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu Q and Stamenkovic I: Localization of
matrix metalloproteinase 9 to the cell surface provides a mechanism
for CD44-mediated tumor invasion. Genes Dev. 13:35–48. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Canel M, Serrels A, Frame MC and Brunton
VG: E-cadherin-integrin crosstalk in cancer invasion and
metastasis. J Cell Sci. 126:393–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu
C, Li G and Zhu Y: Silencing of cZNF292 circular RNA suppresses
human glioma tube formation via the Wnt/β-catenin signaling
pathway. Oncotarget. 7:63449–63455. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sokolowski KM, Koprowski S, Kunnimalaiyaan
S, Balamurugan M, Gamblin TC and Kunnimalaiyaan M: Potential
molecular targeted therapeutics: Role of PI3-K/AKT/mTOR inhibition
in cancer. Anticancer Agents Med Chem. 16:29–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tan J, Jiang X, Yin G, He L, Liu J, Long
Z, Jiang Z and Yao K: Anacardic acid induces cell apoptosis of
prostatic cancer through autophagy by ER stress/DAPK3/Akt signaling
pathway. Oncol Rep. 38:1373–1382. 2017.PubMed/NCBI
|