Introduction
Osteoporosis (OP) is a metabolic bone disorder
characterized by low bone mineral density (BMD) and decreased bone
strength, which leads to an increased risk of fractures with a
consequent increase in morbidity and mortality (1). In the Mexican population, OP is a major
public health issue, mainly due to the increase in the life
expectancy of the Mexican population (2). In OP, the balance of bone formation and
resorption is impaired.
Osteoclasts are multinucleated cells that are formed
from the fusion of progenitors of osteoclasts (3). In the process of osteoclast formation,
there is an absolute requirement for cells of hematopoietic origin
of the monocyte/macrophage linage and bone marrow stromal cells to
commit the hematopoietic cell towards osteoclast development
(4).
Circulating monocytes (CMCs) are an appropriate
model for bone-related studies and are important cells that
participate as osteoclast precursors (5,6); they
also secrete osteoclastogenesis-associated factors, including
interleukin-1 (IL-1), IL-6 and tumor necrosis factor-α (TNF-α)
(5,7). In addition, previous studies have
revealed associations of gene and protein expression in CMCs and OP
(8–10). These studies demonstrate that CMCs
are a valuable cell linage with functional relevance in the
pathogenesis of OP and have been well substantiated and accepted as
a suitable model for studies aiming to dissect etiological
mechanisms in the bone field (6).
MicroRNAs (miRNAs or miRs) are a class of non-coding
single-stranded RNAs (~22 nucleotides) that confer a crucial level
of gene expression regulation, primarily by promoting mRNA
degradation or inhibiting translation (11). Altered miRNA expression is associated
with bone-related diseases, including OP, and serves a key role in
bone formation and resorption (12,13).
Substantial evidence has demonstrated that circulating miRNAs may
be used as potential non-invasive biomarkers for various human
diseases (14,15). Recent reports have illustrated the
importance of miRNAs in osteoclastogenesis and OP. From these
studies, miR-21, miR-133a, miR-148, miR-422a and miR-194-5p have
been associated with postmenopausal OP and fractures (16,17).
The present study aimed to identify potentially
useful miRNAs as biomarkers for postmenopausal OP, in CMCs isolated
from postmenopausal Mexican-Mestizo women. A significant increase
was identified in the miR-1270 expression in women with OP compared
with normal ones using combined miRNAs and transcriptomics
microarray technology and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) validation, and a further
bioinformatics analysis of miR-1270 revealed that interferon
regulatory factor 8 (IRF8) is the most important target gene
associated with osteoclastogenesis.
Materials and methods
Subjects and study design
The Ethics Committee of the Mexican Institute of
Social Security and the National Institute of Genomic Medicine
approved the study protocol and informed consent forms. All
participants enrolled in the present study were informed of the
project and provided written informed consent. The women were of
Mexican-Mestizo origin and were recruited from The Mexican Health
Worker Cohort Study, which is a long-term study of workers from a
large health care institution in Cuernavaca, Mexico, that focuses
on lifestyle and chronic illnesses. Between 2011 and 2013 (Wave 2),
a total of 1,026 participants were followed-up, of which 400 were
postmenopausal women (18). The
detailed characteristics of the postmenopausal women are reported
elsewhere (19). The clinical
procedures, data recording and participant follow-up practices were
standardized and validated (20).
A total of 12 unrelated postmenopausal
Mexican-Mestizo women, 6 with normal and 6 with osteoporotic hip
BMD were recruited. The inclusion criteria were the following:
Spine or hip T-score <-2.5 for the OP group (bottom 20% of the
age-, sex- and ethnicity-matched population) and spine or hip
T-score >-1.0, respectively for the normal group (top 20% of the
age, sex and ethnicity-matched population). The BMD
(g/cm2) for the lumbar spine (L2-L4) and total hip were
measured using a Lunar DPX NT dual energy X-ray absorptiometry
instrument (Lunar Radiation Corp., Madison, WI, USA). The standard
calibration of the instrument was performed daily using the
settings intended for the spine and femoral neck phantom, as
provided by the manufacturer. Technicians ensured that the daily
variation coefficient (VC) was within the normal operational
standards and that the in vivo VC was <1.5%. All women
were aged between 63 and 85 years and had been postmenopausal for
≥12 months (their postmenopausal status was defined as the date of
the last menses followed by at least 12 months of no menses).
Exclusion criteria were used to minimize potential effects of any
known non-genetic factors on bone metabolism and BMD determination.
The present study excluded women who presented serious residuals
from cerebral vascular disease, diabetes mellitus, chronic renal
disease manifest by serum creatinine >1.9 mg/dl, chronic liver
diseases or alcoholism, corticosteroid therapy, treatment with
anticonvulsant therapy, rheumatoid arthritis or collagen disease,
significant disease of any endocrine organ that may affect bone
mass, hyperthyroidism and any other illness, treatment (including
bisphosphonates) or condition (including hormone replacement
therapy) that may be an apparent non-genetic factor underlying the
variation BMD.
Monocyte isolation
A total of 80 ml whole blood was obtained from each
participant. Peripheral blood mononuclear cells were obtained by
density gradients using Histopaque-1077 (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) according to the manufacturer's protocol. CD14+
monocytes were obtained by density gradient centrifugation at 400 ×
g for 30 min at room temperature and magnetic bead isolation. Naive
monocytes were isolated using the negative isolation kit EasySep
Human Monocyte Enrichment (Stemcell Technologies, Inc., Vancouver,
BC, Canada) following the manufacturer's protocol. The purity and
viability was ≥85% (data not shown) as determined by flow cytometry
with the fluorescent-labeled antibodies CD14-phycoerythrin (anti
CD14-PE) and CD45-fluorescein isothiocyanate (anti CD45-FITC; cat.
no. 555574 and cat. no. 555748; BD Biosciences, Franklin Lakes, NJ,
USA). Briefly, 1×106 enriched monocytes were suspended
in 100 µl PBS with 2% fetal bovine serum, stained with 20 µl of
anti CD14-PE and anti CD45-FITC for 45 min at room temperature in
the dark. The analysis was performed on a FACSAria I cytometer
using FACSDiva software version 6.1.3 (both BD Biosciences; San
Jose, CA USA).
miRNA profiling
Total RNA was isolated from monocytes using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. The RNA integrity
and quantification was assessed using a Nanodrop (Thermo Fisher
Scientific, Inc.) and 2100 Bioanalyzer (Agilent Technologies, Inc.,
Santa Clara, CA, USA). Each RNA sample exhibited high quality with
a high integrity number (>8.0). In total, 250 ng total RNA was
labeled using a FlashTag Biotin HSR RNA labeling kit (Affymetrix;
Santa Clara, CA USA.) and was subsequently hybridized on an
GeneChip miRNA 4.0 Array (cat. no. 902411; Affimetrix), which
contains 30,434 probes of mature miRNAs, of which 2,578 of the
probes are human miRNAs, according to the Sanger miRBase v.21
(http://www.mirbase.org/). The array was washed
two times with 1× PBS, 0.02% Tween-20 and stained with FlashTag
Biotin HSR labeled RNA sample on a Fluidics Station 450 (cat. no.
901910, Affymetrix) followed by digitization in a GeneChip Scanner
3000 7G (Affimetrix), according to the manufacturer's protocol.
Gene expression microarray
An Affymetrix GeneChip Human U133 Plus 2.0 Array
(Affymetrix) was used to evaluate the genome-wide gene expression
levels. A total of 500 ng total RNA were used for the cDNA and
biotinylated cRNA synthesis using the GeneChip expression 3′
amplification reagents kits by Affymetrix (Thermo Fisher
Scientific, Inc). Fragmented cDNA was applied to the hybridization,
and the scanning of the array was performed using the Affymetrix
GeneChip 3000 7G scanner (Affymetrix). The expression intensity of
each gene was logarithmically transformed to base 2 and was
normalized using quantile normalization using R (programming
language) through the Bioconductor (v3.3.3; https://www.r-project.org/).
Microarray data analysis
The raw data from microarray platforms, microRNA 4.0
and Human U133 Plus 2.0 gene expression were pre-processed using
Robust Multiarray Average (21) for
background correction and the samples were normalized with quantile
normalization (22) methods
available in the Bioconductor oligo (23) affy packages in Bioconductor v.3.3.3
(24).
The differential expression was determined through
linear models, using the limma Bioconductor package v.3.5 (25), and the miRNAs were classified as
differentially expressed according to a fold-change <-0.5 or
>0.5 and P<0.05.
RT-qPCR analysis
RNA was extracted from monocytes using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Then, 100 ng of total RNA were used
for first strand cDNA synthesis using a TaqMan MicroRNA Reverse
Transcription kit (cat. no. 4366596. Applied Biosystems; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The expression profile for miR-1270 (Assay ID. 002807), miR-548×-3p
(Assay ID. 463079_mat) and miR-8084 (Assay ID. 466802_mat) were
examined using TaqMan microRNA assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.). qPCR was performed on an Applied
Biosystems QuantStudio 7 Flex system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The expression levels were normalized
with respect to RNU44 (Assay ID. 4427975; Applied Biosystems;
Thermo Fisher Scientific, Inc.). For the IRF8 and
GAPDH gene expression analyses, cDNA was synthetized from
100 ng total RNA using the High Capacity cDNA Reverse Transcription
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) following
the manufacturer's protocol. qPCR was performed via TaqMan assay
(Assay ID. Hs01128713_m1; Applied Biosystems; Thermo Fisher
Scientific, Inc.) in the QuantStudio 7 Flex system (Applied
Biosystems; Thermo Fisher Scientific, Inc.).
The relative quantity (RQ) of the miRNAs of each
sample was determined by the 2−ΔΔCq (26) method, where the ΔCq=[the average of
the triplicate Cq of the gene target miRNA-the average of the
triplicate Cq of the endogenous control (GAPDH)], and the ΔΔCq=(the
ΔCq-the mean ΔCq of all samples). The RQ data were used to identify
the miRNAs that were differentially expressed between the two
groups in the study via Student's t-test.
Bioinformatics analysis
To predict the potential target genes of miR-1270,
the algorithms from the following different databases were used:
microRNA.org (http://www.microrna.org/microrna/home.do), miRDB
(http://mirdb.org/miRDB/), miRWalk v2.0
(http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/),
PITA v5.0 (https://omictools.com/pita-tool) and TargetScan Human
v7.0 (http://www.targetscan.org/vert_71/), and the target
genes predicted by ≥3 databases were selected for further analysis.
To validate the in silico results, the list of potential
target genes with the expression data from the Affymetrix GeneChip
Human U133 Plus 2.0 Array of monocytes from the same 12 samples
included in the present study were compared, and a proof of concept
was applied so that if the miRNA is upregulated, then the mRNA of
the target gene must be downregulated. The target genes
accomplished by the proof of concept were used to identify the
binding sites for miR-1270 according to complementary sequences of
the target gene in the 3′-untranslated region (UTR) using the miRDB
database (www.miRDB.org) (27,28) and
TargetScan (www.targetscan.org) (29). PhastCons conservation score, which
measures the evolutionary conservation of sequence blocks across
multiple vertebrates using a phylogenetic hidden Markov model
(30), was used to filter out less
conserved predicted target sites.
Functional and pathway enrichment
analysis
The online tool STRING version 10.0 (www.string-db.org/) was used to search the functional
enrichment analyses of potential target genes of miR-1270 by
selecting the Kyoto Encyclopedia of Genes and Genomes (KEGG)
database. A false discovery rate (FDR)-value <0.05 was selected
as the cut-off criterion for the KEGG enrichment analysis. The
PubMed database (https://www.ncbi.nlm.nih.gov/pubmed/) was used to
search for literature reported as ‘BMD’, ‘osteoclasts’, ‘monocytes’
and ‘osteoporosis’ associated genes to improve the potential
candidate genes of the present study and to search for the
interaction association of the predicted genes contained in the
significant KEGG pathways. A confidence score >0.7 was
considered as the cut-off criterion for an interaction. The
Ingenuity Pathways Analysis (IPA) was used to construct the
protein-protein interaction networks based on the genes reported in
the literature and the miRNA data. This analytical tool is based on
prior knowledge of the expected effects between genes and miRNAs
stored in the base Ingenuity knowledge (Qiagen, Inc., Valencia, CA,
USA).
Statistical analysis
Student's t-tests were used as indicated in each
case. Data plotted represent the mean ± standard deviation as
indicated. For statistical analysis of microarray data of miRNAs
and genes, the CEL files were imported using freely available R
language. The expression was performed on log2 transformed
fold-change (FC) data. The limma package in Bioconductor v.3.5 was
used to compare normal and OP groups. The Robust Multiarray
Analysis algorithm was applied for generation of relative signal
values and normalization. For expression comparison of different
groups a moderated t-statistics was used followed by calculation of
FDR (31). Results were expressed as
the average of three repeats of FC. All statistical analysis were
performed using GraphPad Prism v. 6.0 for Mac (GraphPad Software,
La Jolla, CA, USA; www.graphpad.com). P<0.05 was considered to
indicate a statistically significant difference.
Results
Characteristics of the study
subjects
The clinical characteristics of the study subjects
are summarized in Table I. No
significant differences were observed in the age of the women
included in the present study; however, significant differences
were identified in the weight and height between normal and OP
groups (P<0.05). The mean age of the normal group was 66.66±2.73
years, whereas it was 73.16±7.44 years for the OP group, and the
hip and spine BMDs were significantly different between the groups
(P<0.001).
 | Table I.Characteristics of the study
subjects. |
Table I.
Characteristics of the study
subjects.
| Traits | Osteoporosis
(n=6) | Normal (n=6) | P-value |
|---|
| Age (years) |
73.16±7.44 |
66.66±2.73 | 0.088 |
| Height (cm) |
146.50±7.28 |
155.83±5.94 | 0.036 |
| Weight (kg) |
58.75±6.05 |
69.26±9.49 | 0.049 |
| BMI |
26.16±6.36 |
28.48±3.08 | 0.448 |
| Children (n) |
2.83±2.13 |
5.33±2.06 | 0.066 |
| Age of menarcher
(years) |
12.50±1.97 |
13.00±0.89 | 0.589 |
| Spine BMD
(g/cm2) |
0.86±0.12 |
1.15±0.09 | <0.001 |
| Spine t-score |
−3.27±0.76 |
−0.17±0.
68 | <0.001 |
| Hip BMD
(g/cm2) |
0.
63±0.05 |
1.13±0.07 | <0.001 |
| Hip t-score |
−2.97±0.45 |
1.03±0.56 | <0.001 |
miRNAs are expressed differentially
between the OP and normal groups
miRNA profiling of the OP and normal groups was
performed on a microarray platform with probes for 2,578 mature
human miRNAs, and several of the miRNAs were undetectable in a
number of the samples. The comparison of the expression levels of
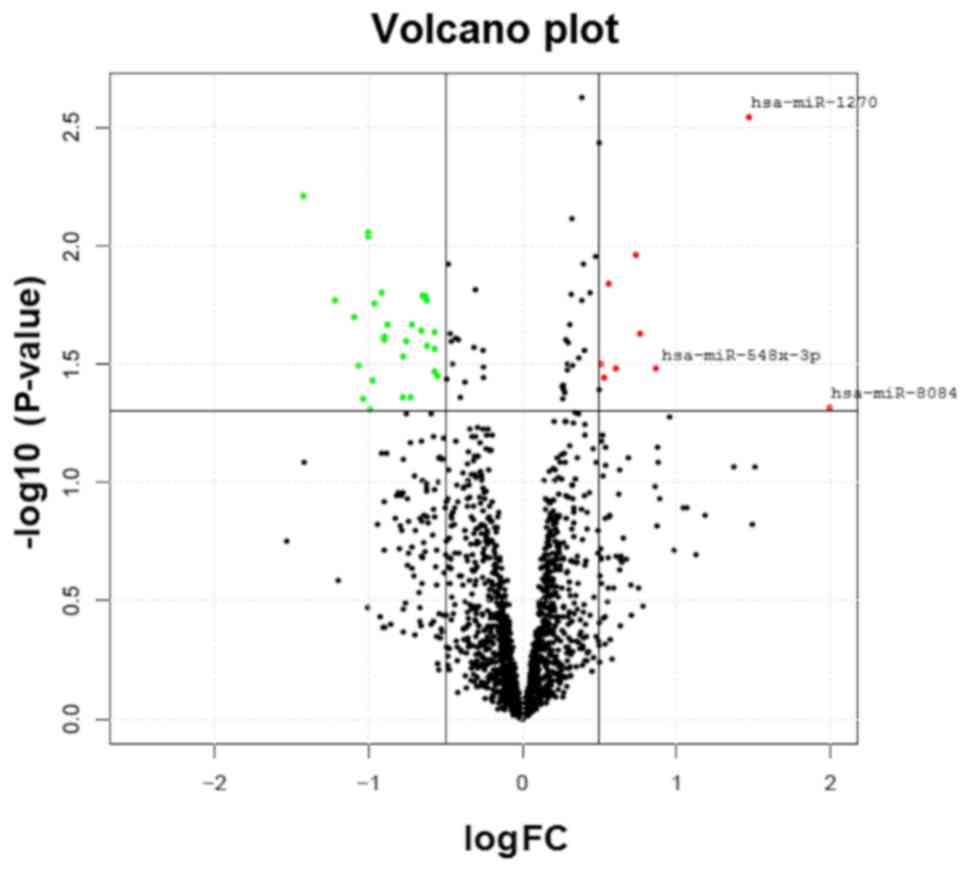
the miRNAs in the OP and normal groups is depicted in Fig. 1. In total, 35 miRNAs revealed a
nominally significant (P<0.05) difference in the microarray
analysis between the two study groups. Table II presents the three miRNAs that
were most markedly upregulated (miR-1270, miR-548×-3p and miR-8084)
and downregulated (miR-6165, miR-6824-5p and miR-6124) in the OP
group. In further analysis, the focus was on the upregulated
miRNAs. The functions of these miRNAs are poorly understood, and
none of them have previously been reported to be associated with
BMD or OP.
 | Table II.miRNAs differentially expressed
between the osteoporotic and normal groups. |
Table II.
miRNAs differentially expressed
between the osteoporotic and normal groups.
| miRNA | Fold-change | P-value |
|---|
| hsa-miR-8084 | 1.999 | 0.047 |
| hsa-miR-1270 | 1.483 | 0.003 |
|
hsa-miR-548×-3p | 0.861 | 0.034 |
| hsa-miR-6165 | −1.098 | 0.022 |
|
hsa-miR-6824-5p | −1.229 | 0.016 |
| hsa-miR-6124 | −1.427 | 0.006 |
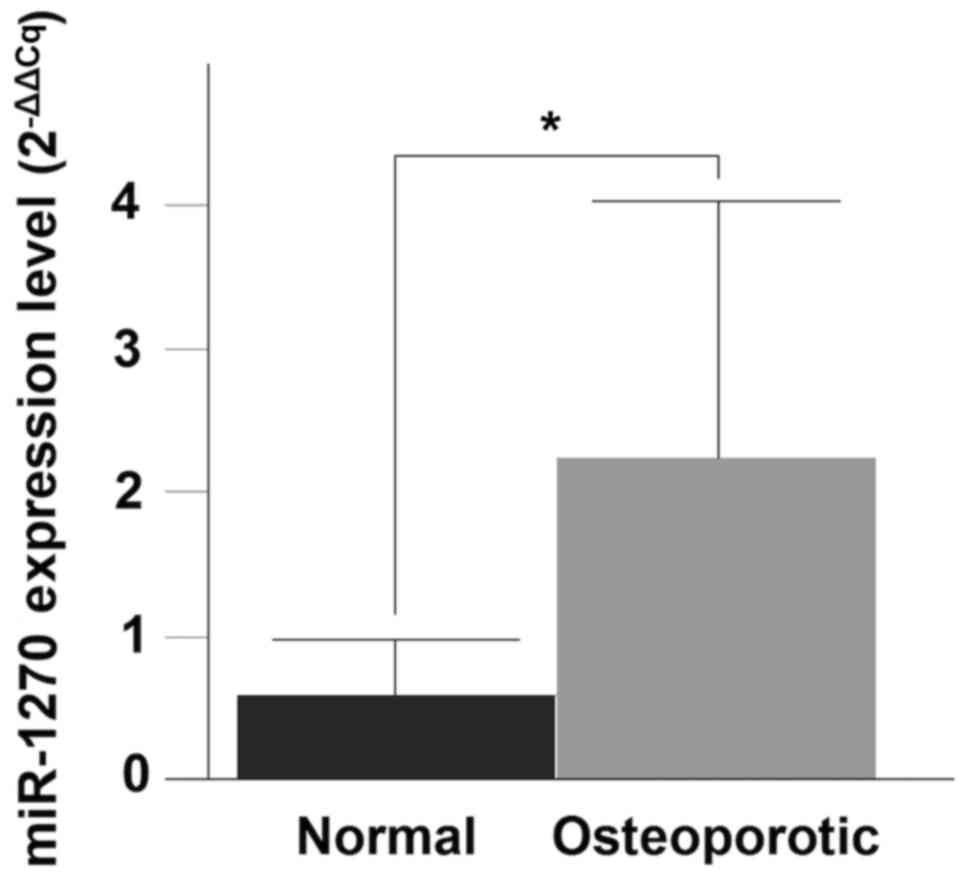
Validation of miRNA expression by
RT-qPCR
RT-qPCR was performed to validate the differential
expression levels of the three-upregulated miRNAs (miR-8084,
miR-1270 and miR-548×-3p). Only miR-1270 demonstrated a significant
upregulation in the OP compared with the normal group (P=0.0043)
(Fig. 2).
Predicted target genes of the
OP-associated miRNA
Target mRNAs were predicted for miR-1270; the
criterion for the target identification was consistent with the
prediction by at least three databases, 3,857 target genes for
miR-1270 were predicted. This list of potential target genes was
linked with data derived from expression changes of the monocytes
of the same samples (data not shown), assuming the association of
the high expression levels of a given miRNA corresponded to the low
level of target gene expression. Downregulation was defined as a
minimum 0.5-FC. Using these criteria 161 putative downregulated
target genes were identified for miR-1270 (data not shown). The
interaction by combining the lists of the target genes generated by
the prediction algorithms of miR-1270 and the expression dataset of
the monocytes is depicted in Fig.
3A.
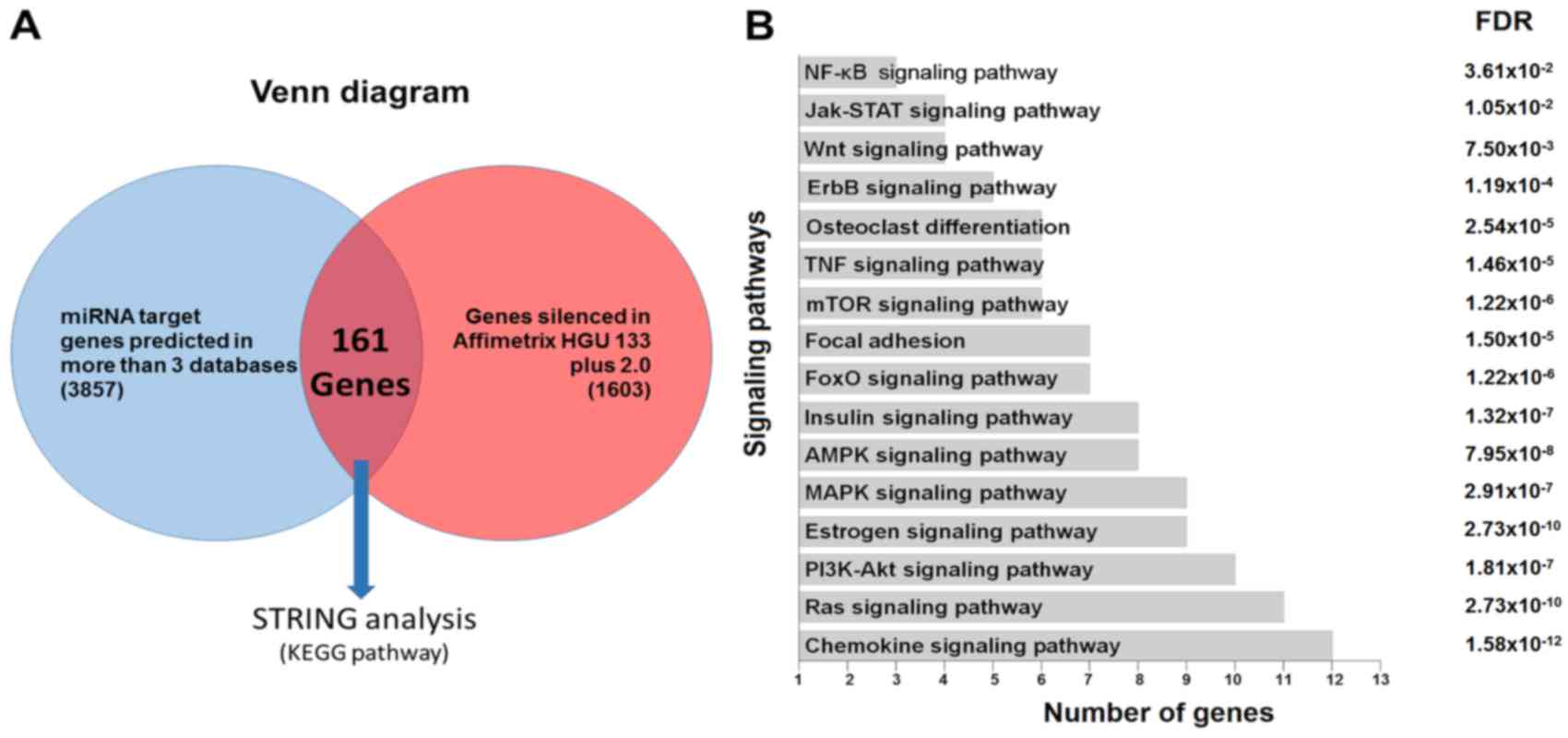 | Figure 3.(A) Venn diagram demonstrating the
basis for selecting the potential target genes of hsa-miR-1270, by
linking the list obtained from the prediction algorithms with those
generated from the expression data of the monocytes of the samples
included in the present study (1,603 genes with fold-change
<-0.5 or >0.5 and P<0.05). (B) KEGG pathway enrichment
analysis of the putative miRNA targets from the previous analysis.
miRNA or miR, microRNA; KEGG, Kyoto Encyclopedia of Genes and
Genomes; NF-κB, nuclear factor-κB; Jak-STAT, Janus kinase/signal
transducers and activators of transcription; TNF, tumor necrosis
factor; mTOR, mechanistic target of rapamycin; FoxO, forkhead box
O; AMPK, adenosine monophosphate-activated protein kinase; MAPK,
mitogen-activated protein kinase; PI3 K,
phosphatidylinositol-3-kinase; FDR, false discovery rate. |
Interaction network of the target
genes and the miRNAs
To explore a potential functional association of the
deregulated miRNAs identified in the present study, the 161 target
gene list was submitted to the online bioinformatics tool STRING
software to identify canonical pathways. The analysis revealed 16
KEGG pathways with overrepresented monocytes, osteoclasts and
OP-related genes. The pathways predicted as most markedly enriched
by the miR-1270 were the chemokine, Ras, estrogen, adenosine
monophosphate-activated protein kinase, insulin,
phosphatidylinositol-3-kinase-protein kinase B, mitogen-activated
protein kinase, forkhead box O, mechanistic target of rapamycin and
TNF signaling pathways, focal adhesion and osteoclast
differentiation. Other pathways were identified including: ErbB,
Wnt, Janus kinase/signal transducers and activators of
transcription and nuclear factor-κB (NF-κB) signaling pathways
(Fig. 3B).
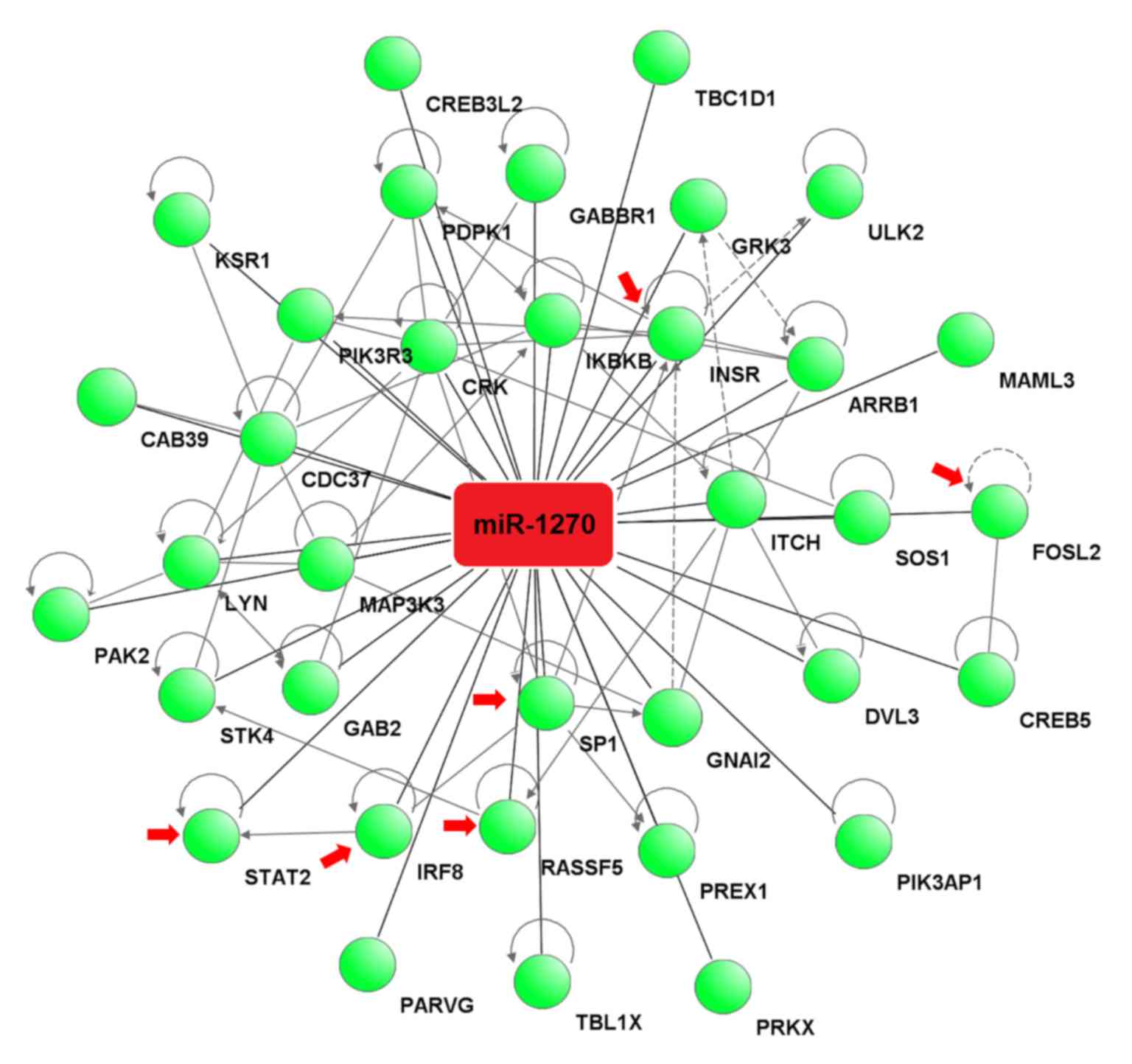
The target genes associated with these pathways were
analyzed by IPA software to construct an interaction network
between miR-1270 and the target genes. The analysis revealed that
miR-1270 binds to the 3′-UTR of 35 potential target genes (Fig. 4).
Target gene prediction and
verification
By performing the bioinformatics analyses, potential
target genes associated with monocytes, osteoclasts and OP were
identified and predicted by at least three databases. By searching
the relevant references for all 35 target genes of miR-1270, it was
revealed that 9 are potential targets genes for miR-1270 and are
associated with osteoclastogenesis, including myocyte enhancer
factor 2C (MEF2C) (32,33), Sp1
transcription factor (SP1) (4), signal transducer and activator of
transcription 2 (STAT2) (34), KRAS (KRAS proto-oncogene,
GTPase) (35) and FOS like 2 (FOSL2)
(36), Inhibitor of κ light
polypeptide gene enhancer in B-cells kinase β (IKBKB)
(37), Ras association domain family
member 5 (RASSF5) (38),
Abelson murine leukemia viral oncogene homolog 1 (ABL1;
non-receptor tyrosine kinase) (39)
and IRF8 (40). The predicted
specific binding sites of miR-1270 to the 3′-UTR of these target
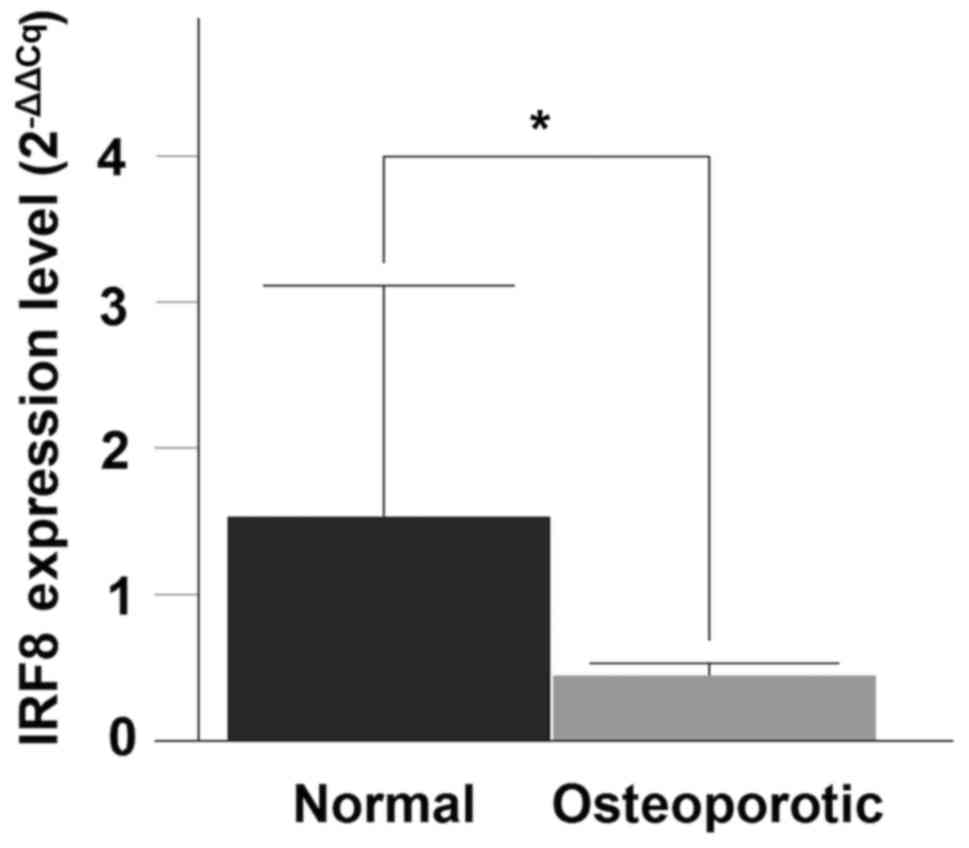
genes are presented in Table III.
The RT-qPCR analysis for the IRF8 gene demonstrated a
significant decrease in mRNA expression between the OP and normal
samples (P=0.0288) (Fig. 5).
 | Table III.Putative binding sites of miR-1270 in
the predicted target genes in humans. |
Table III.
Putative binding sites of miR-1270 in
the predicted target genes in humans.
| Target gene | 3′-UTR
position | Sequence |
| PhastCons
score |
|---|
| KRAS | 2011 |
3′ugUGUCGA-GA-AGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | | | | | | | | | | | | | | | | |
|
|
|
|
|
5′uuACUGCUGCUGUGGAUAUCUCCAu 3′ | KRAS | 0.5726 |
| FOSL2 |
604 |
3′ugUGUCGA-GAAGGUA-UAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | : | | | : | | | | | | | | | | |
|
|
|
|
|
5′ugACGCCUCCCAGUCAUCAUCUCCAg 3′ | FOSL2 | 0.5389 |
| MEF2C | 1948 |
3′ugugUCGAGA-AGGUA-UAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | | | | | | | | | | | | | | | |
|
|
|
|
|
5′gugaAGAUCUGUCGAUUCAUCUCCAa 3′ | MEF2C | 0.6563 |
| IRF8 | 1185 |
3′ugUGUCGAGAAGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| : | | | | | : | | | | | | | |
|
|
|
|
|
5′caAUAG-GCUU-GAAUCUCCAa 3′ | IRF8 | 0.5677 |
| SP1 | 2254 |
3′uguGUCGAGAAGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | : | | | | | | | | | | | |
|
|
|
|
|
5′cucCAUUUGGUCC-UUUCUCCAc 3′ | SP1 | 0.6502 |
| IKBKB |
454 |
3′uguGUCGAGA-AGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | : | | | | | | | | | | | |
|
|
|
|
|
5′agcCUGUCCUCUCCUGCUCUCCAa 3′ | IKBKB | 0.5335 |
| RASSF5 |
597 |
3′ugugUCGA-GAAGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
: | | | | | | | | | | | | | |
|
|
|
|
|
5′cugaGGCUGGCUCAGAGAUCUCCAg 3′ | RASSF5 | 0.4772 |
| ABL1 |
396 |
3′ugugucGAGAAGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
: | | | | | | | | | | | | | |
|
|
|
|
|
5′gccuccUUCUUCCACUUCUCCAa 3′ | ABL1 | 0.4373 |
| STAT2 |
704 |
3′ugugucgaGAAGGUAUAGAGGUc 5′ | hsa-miR-1270 |
|
|
|
|
| | | | : | | | | | | |
|
|
|
|
|
5′aguuuaugCUACCUAGUCUCCAc 3′ | STAT2 | 0.5894 |
Discussion
In the present study, a microarray-based approach
was performed followed by RT-qPCR validation and pathway analysis
to identify important circulating miRNA for postmenopausal OP. The
main observation of the present study is the evidence for miR-1270
as a potential novel biomarker for postmenopausal OP in a
Mexican-Mestizo population, in addition to four previously
recognized miRNAs, miR-21, miR-133a, miR-422a and miR-194-5p in
Asian and Caucasian populations (16,17).
A miRNA microarray platform combined with the
transcript profile of the same samples determined by the Human
Genome U133 plus 2.0 microarray for the high-throughput detection
of hundreds of miRNAs were used. The strength of this method
resides in identifying potential miRNA biomarkers associated with
bone metabolism. However, a major pitfall of this approach is a
high false positive rate (41,42).
Therefore, RT-qPCR is generally performed to validate significant
miRNAs identified by microarray. This experimental approach aims to
reveal novel potential associations between miRNA expression and
the BMD status, assuming an inverse correlation between the levels
of a given miRNA and the expression levels of its targets (43). The results of the present study
identified an upregulation of miR-1270 in the OP group. However,
only miR-1270 expression was validated by RT-qPCR. This may be due
to the lower sensitivity of the qPCR method compared to the
microarray, or an incorrect microarray hybridization, which leads
to erroneous signals.
Using a bioinformatics analysis, target genes for
miR-1270 were identified and classified by a KEGG pathway analysis.
In total 16 KEGG pathways were identified that were significantly
associated with categories associated with bone metabolism. As CMCs
are osteoclast precursors, the focus was on miR-1270 as it was the
most promising signal and its expression was validated by RT-qPCR.
The bioinformatics sequence analyses and literature searches of the
present study identified several potential target genes of miR-1270
associated with monocytes, osteoclastogenesis and OP: KRAS,
FOSL2, MEF2C, IRF8, SP1, IKBKB, RASSF5, ABL1 and
STAT2.
Of these genes, IRF8 serves an important role
in bone osteoclast differentiation (44). It is a gene that encodes a
transcription factor that is expressed in immune cells, including
monocytes (45). Mice deficient in
IRF8 are susceptible to severe OP, accompanied by higher
numbers of osteoclasts (44).
Furthermore, IRF8 has been revealed to suppress
osteoclastogenesis and in vivo bone remodeling, in part, by
inhibiting the function of nuclear factor of activated T cells c1
(NFATc1) (40). A recent study
provided evidence and further emphasized the importance of
IRF8 as a negative regulator of osteoclastogenesis (46). The observations of the present study
demonstrate that miR-1270 may serve an important role in monocyte
differentiation, regulating the expression of IRF8. In the
same context, a recent report demonstrated that interferon-α1
(IFN-α1) mRNA is a target gene for miR-1270,
revealing that IFN-α1 antisense (AS) enhances the
stability of IFN-α1 mRNA. The data indicated that
IFN-α1 AS functions as a competing endogenous RNA to
prevent miR-1270 from acting on IFN-α1 mRNA (47).
This complex regulatory mechanism indicates a key
role for the innate immune system in maintaining specific
physiological type I IFN levels via post-transcriptional
regulatory mechanisms, including miRNAs and implies the involvement
of the interferon pathway in bone metabolism. Previously, it was
reported that IFN-α and IL-1 induce the activation of
signaling molecules, including NF-κB and p38, which are key
molecules in the process of the differentiation of
monocytes-osteoclasts (48). It
highlights the role of type I IFN in osteoclastogenesis,
revealing that an uncontrolled activation of IFN signaling
in Usp18-knockout mice increases osteoclast differentiation
(49). However, additional studies
are required to understand the role of IFN-α1 in CMCs
as osteoclast precursors.
BMD is the single best predictor and is currently
the gold standard, for the diagnosis of OP (50). Several studies indicate that miR-21,
miR-133a, miR-422a and miR-194-5p identified in CMCs could be
considered as biomarkers for postmenopausal OP (16,17,51–53),
however, the present study did not replicate those results.
Furthermore, the discrepancies in the observed results may be
associated with the study design, for example, the astringents
methods used to select the miRNAs, the differences in the study
populations, the different experimental methods, the arrays and
number of participants.
Finally, there were several limitations in the
present study, including the sample size being relatively small
(n=12), although this was similar to previously published studies
(6,43,51–53).
Furthermore, raw, rather than adjusted, P-values were used for the
multiple comparisons which was in agreement with other studies and
was mainly for avoiding further loss of power. In this context, the
significant RT-qPCR P-value confirmed the differential expression
of miR-1270 in the microarray analysis.
In conclusion, the results of the present study in
combination with the functional role of IRF8 in CMCs
indicate that miRNA-1270 may be a viable biomarker for
postmenopausal OP in a Mexican population. Nevertheless, further
studies are required to validate these observations.
Acknowledgements
Mr. Rogelio Frank Jimenez-Ortega is a doctoral
student from the Programa de Doctorado en Ciencias Biomédicas,
Universidad Nacional Autónoma de México (UNAM) and received
fellowship 557758 from CONACYT. The authors are grateful to Dr
Rossana González Sosa for her support in the recruitment and
collection of the patient samples. Furthermore, the authors would
like to thank Miss Nelly Patiño (National Institute of Genomic
Medicine, INMEGEN) for the technical assistance in flow cytometric
analysis. The present study was supported by grants from the
Consejo Nacional de Ciencia y Tecnología (grant no.
CB-2013-1-221628) and partially supported by INMEGEN (grant nos.
132-13/2013/I and 217-14/2015/I) and Fundación Miguel Alemán,
A.C.
References
|
1
|
Kanis JA: Diagnosis of osteoporosis and
assessment of fracture risk. Lancet. 359:1929–1936. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clark P, Carlos F, Luis J and Martínez V:
Epidemiology, costs and burden of osteoporosis in Mexico. Arch
Osteoporos. 5:9–17. 2010. View Article : Google Scholar
|
|
3
|
Kylmäoja E, Nakamura M and Tuukkanen J:
Osteoclasts and remodeling based bone formation. Curr Stem Cell Res
Ther. 11:626–633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yamashita T, Takahashi N and Udagawa N:
New roles of osteoblasts involved in osteoclast differentiation.
World J Orthop. 3:175–181. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hemingway F, Cheng X, Knowles HJ, Estrada
FM, Gordon S and Athanasou NA: In vitro generation of mature human
osteoclasts. Calcif Tissue Int. 89:389–395. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou Y, Deng HW and Shen H: Circulating
monocytes: An appropriate model for bone-related study. Osteoporos
Int. 26:2561–2572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kwan Tat S, Padrines M, Théoleyre S,
Heymann D and Fortun Y: IL-6, RANKL, TNF-alpha/IL-1: Interrelations
in bone resorption pathophysiology. Cytokine Growth Factor Rev.
15:49–60. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng FY, Lei SF, Zhang Y, Zhang YL, Zheng
YP, Zhang LS, Pan R, Wang L, Tian Q, Shen H, et al: Peripheral
blood monocyte-expressed ANXA2 gene is involved in pathogenesis of
osteoporosis in humans. Mol Cell Proteomics. 10:M111.0117002011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Deng FY, Liu YZ, Li LM, Jiang C, Wu S,
Chen Y, Jiang H, Yang F, Xiong JX, Xiao P, et al: Proteomic
analysis of circulating monocytes in Chinese premenopausal females
with extremely discordant bone mineral density. Proteomics.
8:4259–4272. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng Y, Zhang L, Zhu W, Xu C, He H, Zhou
Y, Liu YZ, Tian Q, Zhang JG, Deng FY, et al: Quantitative
proteomics and integrative network analysis identified novel genes
and pathways related to osteoporosis. J Proteomics. 142:45–52.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lian F, Cui Y, Zhou C, Gao K and Wu L:
Identification of a plasma four-microRNA panel as potential
noninvasive biomarker for osteosarcoma. PLoS One. 10:e01214992015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao N, Han D, Liu Y, Li Y, Zeng L, Wang Y
and Feng H: DLX3 negatively regulates osteoclastic differentiation
through microRNA-124. Exp Cell Res. 341:166–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA. 105:pp.
10513–10518. 2008, View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hackl M, Heilmeier U, Weilner S and
Grillari J: Circulating microRNAs as novel biomarkers for bone
diseases - Complex signatures for multifactorial diseases? Mol Cell
Endocrinol. 432:83–95. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meng J, Zhang D, Pan N, Sun N, Wang Q, Fan
J, Zhou P, Zhu W and Jiang L: Identification of miR-194-5p as a
potential biomarker for postmenopausal osteoporosis. PeerJ.
3:e9712015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji X, Chen X and Yu X: MicroRNAs in
osteoclastogenesis and function: Potential therapeutic targets for
osteoporosis: Int J Mol Sci. 17:3492016.
|
|
18
|
Denova-Gutiérrez E, Flores YN,
Gallegos-Carrillo K, Ramírez-Palacios P, Rivera-Paredez B,
Muñoz-Aguirre P, Velázquez-Cruz R, Torres-Ibarra L, Meneses-León J,
Méndez-Hernández P, et al: Health workers cohort study: methods and
study design. Salud Publica Mex. 58:708–716. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Velázquez-Cruz R, García-Ortiz H,
Castillejos-López M, Quiterio M, Valdés-Flores M, Orozco L,
Villarreal-Molina T and Salmerón J: WNT3A gene polymorphisms are
associated with bone mineral density variation in postmenopausal
Mestizo women of an urban Mexican population: Findings of a
pathway-based high-density single nucleotide screening. Age
(Dordr). 36:96352014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Denova-Gutiérrez E, Castañón S, Talavera
JO, Flores M, Macías N, Rodríguez-Ramírez S, Flores YN and Salmerón
J: Dietary patterns are associated with different indexes of
adiposity and obesity in an urban Mexican population. J Nutr.
141:921–927. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bolstad BM: Low level analysis of
high-density oligonucleotide array data: Background, normalization
and sumarizationPhD dissertation. University of California;
Berkeley: 2004
|
|
23
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wettenhall JM and Smyth GK: limmaGUI: A
graphical user interface for linear modeling of microarray data.
Bioinformatics. 20:3705–3706. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang X and El Naqa IM: Prediction of both
conserved and nonconserved microRNA targets in animals.
Bioinformatics. 24:325–332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
30
|
Siepel A, Bejerano G, Pedersen JS,
Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW,
Richards S, et al: Evolutionarily conserved elements in vertebrate,
insect, worm and yeast genomes. Genome Res. 15:1034–1050. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc. 57:289–300. 1995.
|
|
32
|
Feng X: RANKing intracellular signaling in
osteoclasts. IUBMB Life. 57:389–395. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnson ME, Deliard S, Zhu F, Xia Q, Wells
AD, Hankenson KD and Grant SFA: A ChIP-seq-defined genome-wide map
of MEF2C binding reveals inflammatory pathways associated with its
role in bone density determination. Calcif Tissue Int. 94:396–402.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng HT, Cheng T, Steer JH, Joyce DA,
Pavlos NJ, Leong C, Kular J, Liu J, Feng X, Zheng MH and Xu J:
Myocyte enhancer factor 2 and microphthalmia-associated
transcription factor cooperate with NFATc1 to transactivate the
V-ATPase d2 promoter during RANKL-induced osteoclastogenesis. J
Biol Chem. 284:14667–14675. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Westra WH, Sturm P, Drillenburg P, Choti
MA, Klimstra DS, Albores-Saavedra J, Montag A, Offerhaus GJ and
Hruban RH: K-ras oncogene mutations in osteoclast-like giant cell
tumors of the pancreas and liver: Genetic evidence to support
origin from the duct epithelium. Am J Surg Pathol. 22:1247–1254.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bozec A, Bakiri L, Hoebertz A, Eferl R,
Schilling AF, Komnenovic V, Scheuch H, Priemel M, Stewart CL,
Amling M and Wagner EF: Osteoclast size is controlled by Fra-2
through LIF/LIF-receptor signalling and hypoxia. Nature.
454:221–225. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chaisson ML, Branstetter DG, Derry JM,
Armstrong AP, Tometsko ME, Takeda K, Akira S and Dougall WC:
Osteoclast differentiation is impaired in the absence of inhibitor
of kappa B kinase alpha. J Biol Chem. 279:54841–54848. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song H, Kim H, Lee K, Lee DH, Kim TS, Song
JY, Lee D, Choi D, Ko CY, Kim HS, et al: Ablation of Rassf2 induces
bone defects and subsequent haematopoietic anomalies in mice. EMBO
J. 31:1147–1159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X and Li B: Genetic studies of bone
diseases: Evidence for involvement of DNA damage response proteins
in bone remodeling. Int J Biomed Sci. 3:217–228. 2007.PubMed/NCBI
|
|
40
|
Zhao B, Takami M, Yamada A, Wang X, Koga
T, Hu X, Tamura T, Ozato K, Choi Y, Ivashkiv LB, et al: Interferon
regulatory factor-8 regulates bone metabolism by suppressing
osteoclastogenesis. Nat Med. 15:1066–1071. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu YZ, Dvornyk V, Lu Y, Shen H, Lappe JM,
Recker RR and Deng HW: A novel pathophysiological mechanism for
osteoporosis suggested by an in vivo gene expression study of
circulating monocytes. J Biol Chem. 280:29011–29016. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao P, Chen Y, Jiang H, Liu YZ, Pan F,
Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR and Deng HW: In
vivo genome-wide expression study on human circulating B cells
suggests a novel ESR1 and MAPK3 network for postmenopausal
osteoporosis. J Bone Miner Res. 23:644–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
de la Rica L, García-Gómez A, Comet NR,
Rodríguez-Ubreva J, Ciudad L, Vento-Tormo R, Company C,
Álvarez-Errico D, García M, Gómez-Vaquero C and Ballestar E:
NF-κB-direct activation of microRNAs with repressive effects on
monocyte-specific genes is critical for osteoclast differentiation.
Genome Biol. 16:22015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Saito E, Suzuki D, Kurotaki D, Mochizuki
A, Manome Y, Suzawa T, Toyoshima Y, Ichikawa T, Funatsu T, Inoue T,
et al: Down-regulation of Irf8 by Lyz2-cre/loxP accelerates
osteoclast differentiation in vitro. Cytotechnology. 69:443–450.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yáñez A and Goodridge HS: Interferon
regulatory factor 8 and the regulation of neutrophil, monocyte, and
dendritic cell production. Curr Opin Hematol. 23:11–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Boyce BF, Xiu Y, Li J, Xing L and Yao Z:
NF-κB-mediated regulation of osteoclastogenesis. Endocrinol Metab
(Seoul). 30:35–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kimura T, Jiang S, Yoshida N, Sakamoto R
and Nishizawa M: Interferon-alpha competing endogenous RNA network
antagonizes microRNA-1270. Cell Mol Life Sci. 72:2749–2761. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Coelho LF, de Freitas Almeida Magno G,
Mennechet FJ, Blangy A and Uzé G: Interferon-alpha and -beta
differentially regulate osteoclastogenesis: Role of differential
induction of chemokine CXCL11 expression. Proc Natl Acad Sci USA.
102:pp. 11917–11922. 2005, View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yim HY, Park C, Lee YD, Arimoto K, Jeon R,
Baek SH, Zhang DE, Kim HH and Kim KI: Elevated response to type I
IFN enhances RANKL-mediated osteoclastogenesis in usp18-knockout
mice. J Immunol. 196:3887–3895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Johnell O, Kanis JA, Oden A, Johansson H,
De Laet C, Delmas P, Eisman JA, Fujiwara S, Kroger H, Mellstrom D,
et al: Predictive value of BMD for hip and other fractures. J Bone
Miner Res. 20:1185–1194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cao Z, Moore BT, Wang Y, Peng XH, Lappe
JM, Recker RR and Xiao P: MiR-422a as a potential cellular microRNA
biomarker for postmenopausal osteoporosis. PLoS One. 9:e970982014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen C, Cheng P, Xie H, Zhou HD, Wu XP,
Liao EY and Luo XH: MiR-503 regulates osteoclastogenesis via
targeting RANK. J Bone Miner Res. 29:338–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang Y, Li L, Moore BT, Peng XH, Fang X,
Lappe JM, Recker RR and Xiao P: MiR-133a in human circulating
monocytes: A potential biomarker associated with postmenopausal
osteoporosis. PLoS One. 7:e346412012. View Article : Google Scholar : PubMed/NCBI
|