Introduction
Coronary heart disease (CHD) is a complex chronic
disease that is caused by an imbalance between blood supply and
demand in myocardium. Various environmental and genetic factors are
known to contribute to onset and development of CHD (1). As of 2010, CHD was the leading cause of
mortality globally, resulting in over 7 million cases of mortality
(2). Therefore, association studies
for CHD biomarkers have been performed worldwide (3–5) for
future forefront diagnostics for the early assessment of cardiac
risks.
The genetic locus at chromosome 9p21 has been
demonstrated to be strongly associated with the risk of CHD
(6,7). Cyclin-dependent kinase inhibitor 2A
(CDKN2A) and cyclin-dependent kinase inhibitor 2B
(CDKN2B) genes both encode putative regulators of
cyclin-dependent kinases on chromosome 9p21. Genome-wide
association studies have identified that some CDKN2A or
CDKN2B genetic variants are susceptible to CHD (8–10). As
recently reported, many human diseases, including cardiovascular
disease, could be influenced by aberrant DNA methylation
modification (11). Aberrant
methylation of cytosine-phosphate-guanine (CpG) islands in gene
promoters is associated with transcription silencing and activity
(3). However, the exact role of
CDKN2A and CDKN2B methylation in cardiovascular
system has not yet been fully elucidated.
CDKN2A gene is involved in the regulation of
cell proliferation, cell aging and apoptosis (12). However, a bidirectional role of
CDKN2A gene expression has been reported in previous
studies. Knösel et al (13)
reported that increased CDKN2A may be linked to oncogene-induced
senescence, whereas the loss of CDKN2A contributes to malignant
progression. Furthermore, Bayoglu et al (14) reported that increased CDKN2A
gene expression in artery plaques may increase the risk of
atherosclerosis and contribute to the development of carotid artery
stenosis. Although methylation-induced CDKN2A downregulation is
observed in multiple human cancer types (15–17), few
studies have evaluated the epigenetic role of CDKN2A in
CHD.
CDKN2B gene lies adjacent to CDKN2A,
and the protein encoded by this gene is associated with controlling
cell cycle G1 progression (18).
CDKN2B has been previously detected as a candidate gene in
CHD (19–21). Kojima et al (22) demonstrated that loss of CDKN2B
promoted advanced development of atherosclerotic plaques, which
suggests a crucial role for CDKN2B in the initiation and
development of CHD. An inverse correlation between CDKN2B
hypermethylation and low expression has previously been found in
CHD (23). However, the potential
for attenuating CDKN2B expression in CHD patients differs in
different CpG regions (23).
The current study aimed to evaluate whether DNA
methylation of CDKN2A and CDKN2B genes is associated
with the risk of CHD. The results of this study may help to provide
a molecular marker for early detection and individual therapy among
CHD patients.
Materials and methods
Patient samples
A total of 189 CHD cases and 190 non-CHD controls
were selected from Ningbo First Hospital (Ningbo, China) between
June 2013 and December 2015. All the participants had undergone
coronary angiography and were reviewed by at least two independent
cardiologists. Those that had ≥50% diameter stenosis in any of the
main coronary arteries, or a history of prior angioplasty, or
coronary artery bypass surgery were placed in the CHD group. Those
who had <50% diameter stenosis in the major coronary artery, or
no history of atherosclerotic vascular disease were placed in the
non-CHD group (24). Demographic
data (age and gender) were collected by researchers. The mean age
of CHD patients was 62.25±5.55 years, including 96 males and 93
females. The mean age of non-CHD controls was 62.07±5.58 years,
including 96 males and 94 females. Biochemical indices
[triglyceride (TG), total cholesterol (TC), high density
lipoprotein cholesterol (HDL-C) and low density lipoprotein
cholesterol (LDL-C) in blood serum] were enzymatically measured
using a CX7 biochemical analyzer (Beckman Coulter, Inc., Brea, CA,
USA). Ethical approval was provided by the Ethics Committee at
Ningbo First Hospital. All patients provided written informed
consent.
DNA extraction and bisulphite
conversion
DNA extraction and quantification was performed as
described previously (25). DNA
samples were converted using an EZ DNA Methylation-Gold Kit (Zymo
Research, Irvine, CA, USA), according to the manufacturer's
instructions.
Methylation-specific polymerase chain
reaction (MSP)
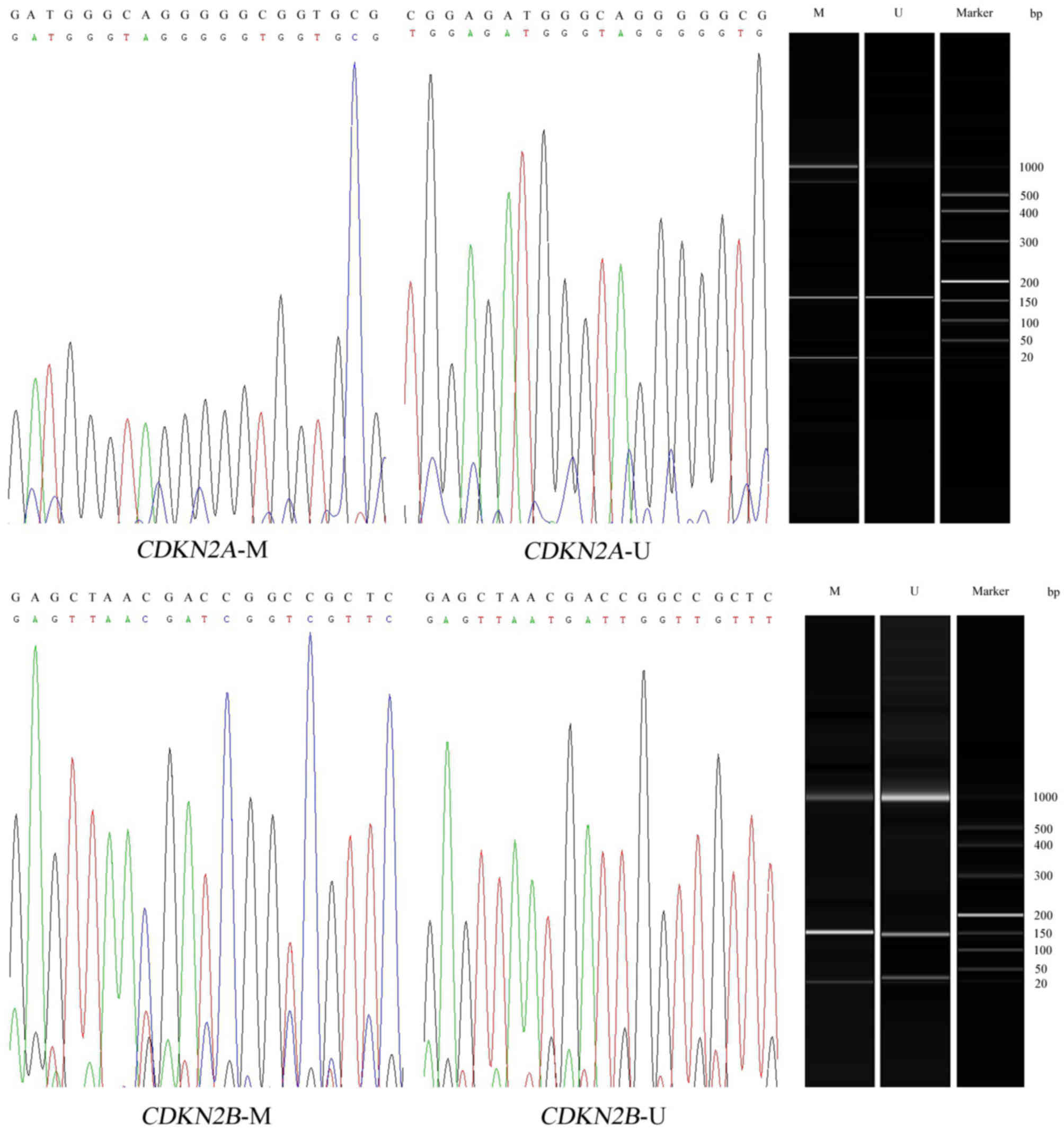
The methylation status of CDKN2A and
CDKN2B was determined by MSP, as described previously
(26). Polymerase chain reaction
(PCR) products were considered as methylated or unmethylated when
clearly visible peaks were produced by a Qsep100 DNA Analyzer
(BiOptic, Inc., Taipei, Taiwan). Further sequencing results
indicated a successful bisulphite conversion and amplification
(Fig. 1). The primer sequences of
methylated and unmethylated primers were as follows: CDKN2A
methylated, forward 5′-GTAGGGTTTAGAGTCGTTTCGA-3′ and reverse
5′-AACTACAAACTAAAACCCACGC-3′; CDKN2A unmethylated, forward
5′-CGTAGGGTTTAGAGTTGTTTTGA-3′ and reverse
5′-AACTACAAACTAAAACCCACACA-3′; CDKN2B methylated, forward
5′-GCGTTCGTATTTTGCGGTT-3′ and reverse 5′-CGTACAATAACCGAACGACCGA-3′;
and CDKN2B unmethylated, forward
5′-TGTGATGTGTTTGTATTTTGTGGTT-3′ and reverse
5′-CCATACATAACCAAACAACCAA-3′. The total amplification involved a
reaction volume of 20 µl, containing 0.5 µl forward and reverse
primers, 1.6 µl bisulphate-converted DNA, 10 µl ZymoTaq™ PreMix
(Zymo Research) and 7.4 µl DNase/RNase-free water. The annealing
temperatures were 55°C for CDKN2A methylation and
unmethylation PCR, 55°C for CDKN2B methylation PCR and 57°C
for CDKN2B unmethylation PCR.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analyses were performed using SPSS 18.0 (SPSS, Inc.,
Chicago, IL, USA) and GraphPad Prism 6.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). The mean subgroup differences for clinical
characteristics were compared using Student's t-test. P-values were
adjusted by age, gender, TG, TC, HDL-C and LDL-C using logistic
regression. The Chi-square test was used to determine the
association between promoter methylation and CHD. Two-sided
P<0.05 was considered to indicate a statistically significant
result.
Results
Patient characteristics
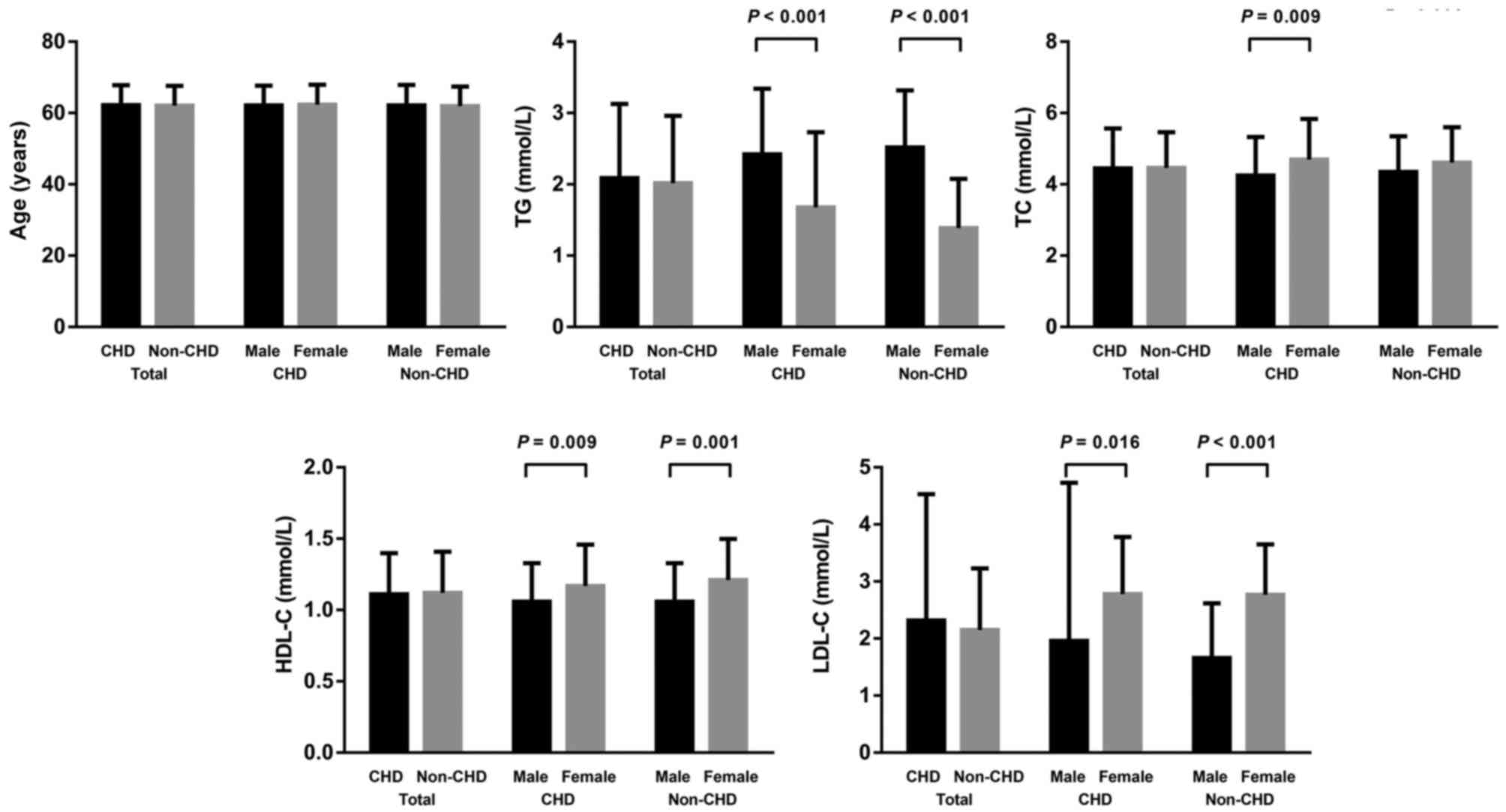
Baseline characteristics of CHD cases and non-CHD
controls are shown in Fig. 2. There
was no significant difference between the age of CHD cases
(62.25±5.55 years) and non-CHD controls (62.07±5.58 years). There
were also no significant differences between levels of TG, TC,
HDL-C or LDL-C between CHD cases and non-CHD controls.
Subsequently, subgroup analysis was performed by gender. The TG
level was significantly higher in males compared with females in
CHD cases (2.42±0.92 vs. 1.68±1.05 mmol/l; P<0.001) and non-CHD
controls (2.52±0.80 vs. 1.39±0.69 mmol/l; P<0.001). The TC level
was significantly lower in CHD males compared with CHD females
(4.25±1.08 vs. 4.70±1.14 mmol/l; P<0.009). The HDL-C level was
significantly lower in males compared with females both in CHD
(1.06±0.27 vs. 1.17±0.29 mmol/l; P=0.009) and non-CHD (1.06±0.27
vs. 1.21±0.29 mmol/l; P=0.001). The LDL-C was significantly lower
in males compared with females both in CHD (1.96±2.77 vs. 2.78±1.00
mmol/l; P=0.016) and non-CHD (1.66±0.96 vs. 2.77±0.88 mmol/l;
P<0.001).
Association analysis between CHD and
methylation of CDKN2A and CDKN2B
In the present study, MSP was used to estimate the
methylation status of CDKN2A and CDKN2B gene
promoters in 189 CHD patients and 190 non-CHD controls. No
associations were found between CDKN2A/CDKN2B gene
promoter methylation and CHD in the total samples or in gender
subgroups (Table I).
 | Table I.Methylation frequencies of
CDKN2A and CDKN2B promoters in CHD cases and non-CHD
controls. |
Table I.
Methylation frequencies of
CDKN2A and CDKN2B promoters in CHD cases and non-CHD
controls.
| Gene | CHD | Non-CHD | P-value | Odds ratio (95%
confidence interval) |
|---|
| Total samples |
|
|
|
|
|
CDKN2A (M/U) | 38/151 | 29/161 | 0.217 | 1.397
(0.821–2.378) |
|
CDKN2B (M/U) | 184/5 | 186/4 | 0.751 | 0.791
(0.209–2.994) |
| Male |
|
|
|
|
|
CDKN2A (M/U) | 12/84 | 11/85 | 0.842 | 1.104
(0.462–2.640) |
|
CDKN2B (M/U) | 92/4 | 92/4 | 1.000 | 1.000
(0.243–4.119) |
| Female |
|
|
|
|
|
CDKN2A (M/U) | 26/67 | 18/76 | 0.156 | 1.638
(0.826–3.250) |
|
CDKN2B (M/U) | 92/1 | 94/0 | 0.497 | 0.495
(0.428–0.572) |
Association analysis between age and
methylation of CDKN2A and CDKN2B
In all participants, the mean age of
CDKN2A-methylated participants was significantly lower
compared with CDKN2A-unmethylated participants (58.73±5.88
vs. 62.62±5.36 years; P<0.001; adjusted P<0.001; Fig. 3). Conversely, the mean age of
CDKN2B-methylated participants was significantly higher
compared with CDKN2B-unmethylated participants (62.26±5.48
vs. 58.33±7.47 years; P=0.036, adjusted P=0.048).
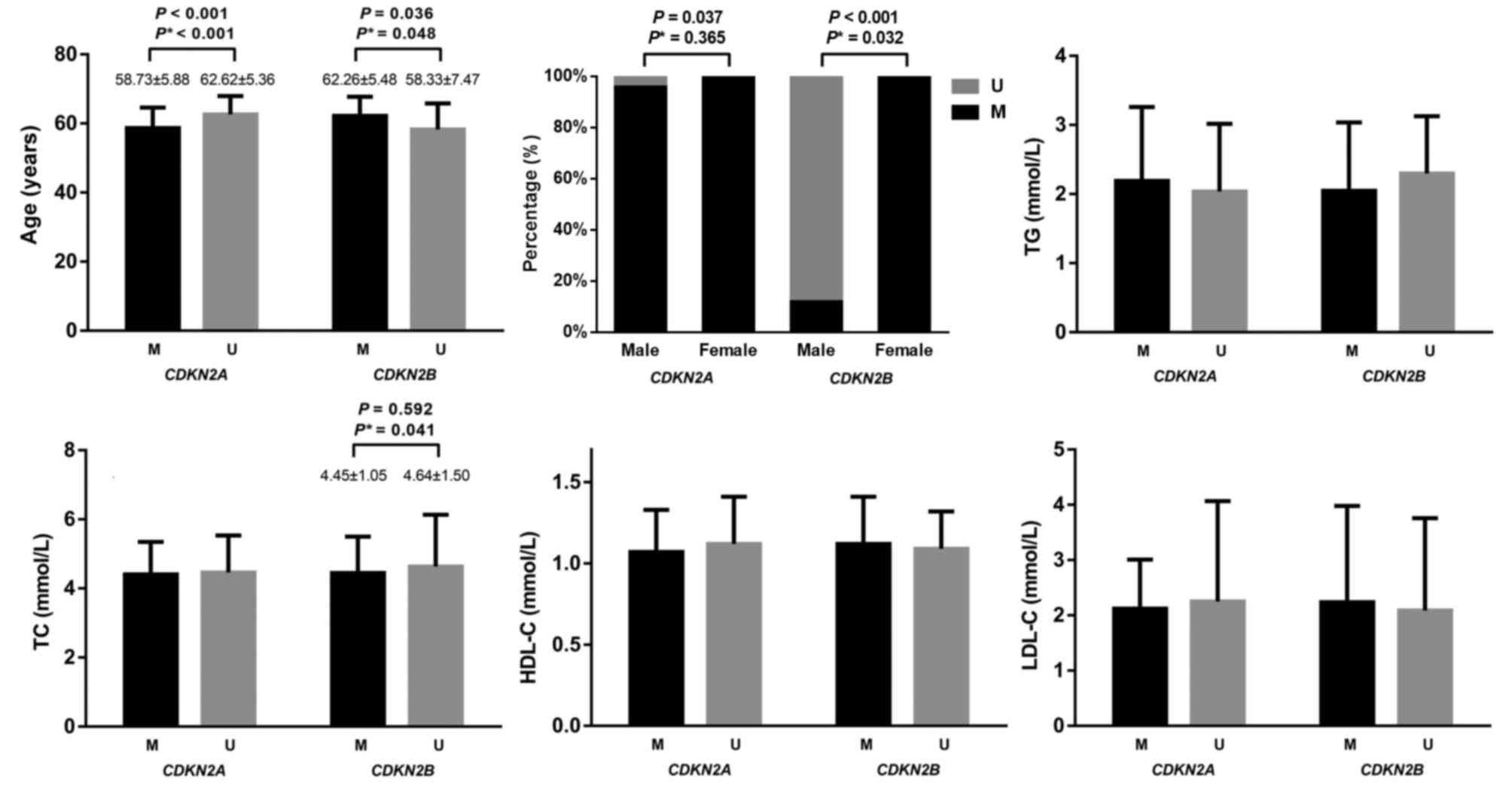 | Figure 3.Association between CDKN2A and
CDKN2B methylation and clinical characteristics in all
subjects. P* is adjusted by age, gender, TG, TC, HDL-C and LDL-C
using logistic regression. CDKN2A, cyclin-dependent kinase
inhibitor 2A; CDKN2B, cyclin-dependent kinase inhibitor 2B;
M, methylation; U, unmethylation; TG, triglyceride; TC, total
cholesterol; HDL-C, high density lipoprotein cholesterol; LDL-C,
low density lipoprotein cholesterol. |
Association analysis between gender
and methylation of CDKN2A and CDKN2B
A significantly larger proportion of female
participants were found to be CDKN2B-methylated compared
with male participants (99.47 vs. 11.98%; P<0.001, adjusted
P=0.032; Fig. 3). Furthermore,
99.47% of female participants were CDKN2A-methylated and
95.83% of male participants were CDKN2A-methylated (P=0.037;
adjusted P=0.365).
Association analysis between blood
cholesterol level and methylation of CDKN2A and CDKN2B
No significant associations were observed between
the plasma levels of TG, HDL-C and LDL-C and CDKN2A/CDKN2B
methylation status (Fig. 3). In
addition, no significant association was observed between TC level
and CDKN2A methylation status. TC level was significantly
lower in methylated CDKN2B compared with unmethylated
CDKN2B (4.45±1.05 vs. 4.64±1.50 mmol/l; adjusted P=0.041;
Fig. 3).
Discussion
The purpose of this study was to investigate the
association between CDKN2A and CDKN2B promoter
methylation and CHD risk. Through a series of statistical analyses,
no notable relationship was found between the methylation status of
CDKN2A or CDKN2B and CHD. However, it was noteworthy
that the methylation of CDKN2A and CDKN2B promoters
was associated with age in all participants. CDKN2A
methylation was associated with younger age, whereas CDKN2B
methylation was associated with older age. Moreover, female
participants were found to be more frequently
CDKN2B-methylated compared with male participants.
DNA methylation is one of the major epigenetic
modifications (3). Accumulating
studies have indicated that DNA methylation changes are associated
with an increased risk of CHD (27–29).
CDKN2A and CDKN2B genes have been previously reported
as hypermethylated tumor suppressor genes in leukemia (30), parathyroid tumor (31) and breast cancer (32), suggesting a potential epigenetic
regulation on cell proliferation and apoptosis. Using
pyrosequencing and MethyLight methods, Zhuang et al
(23) demonstrated that
p15INK4b and p16INK4a
methylation was an important event in CHD. However, the current
data indicated that the methylation of CDKN2A and
CDKN2B genes was not significantly associated with the risk
of CHD, which might be explained by different target fragments and
testing methods.
In the present study, it was demonstrated that age
was associated with gene promoter methylation changes. Alterations
of epigenetic marks such as DNA methylation have been linked to
cancer in older patients (33).
Age-dependent gene methylation may also contribute to the
phenotypic changes associated with skin aging (34). A previous study demonstrated that
age-related DNA methylation affected the essential hypertension
status (25). For the CDKN2A
gene, older patients were more likely to be unmethylated in the
present study, even when assessed independent of blood cholesterol
and gender. An elevated level of CDKN2A in artery plaques may
increase the risk of atherosclerosis (14); it is hypothesized that this may
result from the regulatory effect of demethylation on gene active
expression, or from dysregulation of DNA integrity and function. In
the current study, CDKN2B gene methylation was associated
with older age, which is in accordance with the hypothesis that the
pathogenic role of this cancer suppressor gene in vascular disease
may be associated with its DNA methylation.
Gender is a variable that must be taken into
consideration in studies of chronic diseases, including CHD
(35). The prevalence and incidence
of cardiovascular events are different between males and females
(36). A previous study reported
that women with a low TG/HDL ratio have substantially lower CHD
rates compared with men with a low TG/HDL ratio (37). CDKN2B polymorphism was found
to be independently associated with increased TG/HDL ratio change
(38). In the present study, it was
indicated that methylation of the promoter of CDKN2B was
significantly more likely in females compared with males. No gender
dimorphism was observed for methylation of the CDKN2A
gene.
There were some limitations to the present study.
Firstly, the study involved 189 CHD patients and 190 non-CHD
controls. However, power analysis indicated insufficient powers
(5.0–29.4%) for overall test and gender subgroup analyses. A lack
of power existed in the current study due to small sample size,
thus further replication studies with larger sample sizes are
required. Secondly, only Chinese Han people were recruited,
therefore validations of the findings are required in other ethnic
populations. Furthermore, DNA methylation status was measured using
a qualitative method, and a quantitative method should be explored
in the future.
In conclusion, the present study indicates that
there is an association between age and CDKN2A and
CDKN2B gene promoter methylation status, as well as an
association between gender and CDKN2B methylation. However,
no association was observed between the methylation of these genes
and the risk of CHD. Further investigations are needed to verify
these results and explore the role of DNA methylation in CHD in
more detail.
Acknowledgements
The current study was supported by grants from the
National Natural Science Foundation of China (grant no. 81371469),
the Natural Science Foundation of Zhejiang Province (grant no.
LR13H020003), Ningbo City Medical Science and Technology projects
(grant no. 2014A20) and K. C. Wong Magna Fund in Ningbo
University.
References
|
1
|
Yin YW, Sun QQ, Zhang BB, Hu AM, Liu HL,
Wang Q and Hou ZZ: Association between apolipoprotein E gene
polymorphism and the risk of coronary artery disease in Chinese
population: Evidence from a meta-analysis of 40 studies. PLoS One.
8:e669242013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lozano R, Naghavi M, Foreman K, Lim S,
Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, et
al: Global and regional mortality from 235 causes of death for 20
age groups in 1990 and 2010: A systematic analysis for the Global
Burden of Disease Study 2010. Lancet. 380:2095–2128. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiaoying C, Huadan Y, Qingxiao H, Annan Z,
Linlin T and Shiwei D: The effects of DNA methylation on the
homeostasis in vascular diseases. Yi Chuan. 37:221–232. 2015.(In
Chinese). PubMed/NCBI
|
|
4
|
Krishna SM, Dear A, Craig JM, Norman PE
and Golledge J: The potential role of homocysteine mediated DNA
methylation and associated epigenetic changes in abdominal aortic
aneurysm formation. Atherosclerosis. 228:295–305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim M, Long TI, Arakawa K, Wang R, Yu MC
and Laird PW: DNA methylation as a biomarker for cardiovascular
disease risk. PLoS One. 5:e96922010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schunkert H, Götz A, Braund P, McGinnis R,
Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg
C, Stark K, et al: Repeated replication and a prospective
meta-analysis of the association between chromosome 9p21.3 and
coronary artery disease. Circulation. 117:1675–1684. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chan K, Patel RS, Newcombe P, Nelson CP,
Qasim A, Epstein SE, Burnett S, Vaccarino VL, Zafari AM, Shah SH,
et al: Association between the chromosome 9p21 locus and
angiographic coronary artery disease burden: A collaborative
meta-analysis. J Am Coll Cardiol. 61:957–970. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qi L, Parast L, Cai T, Powers C, Gervino
EV, Hauser TH, Hu FB and Doria A: Genetic susceptibility to
coronary heart disease in type 2 diabetes: 3 independent studies. J
Am Coll Cardiol. 58:2675–2682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeemon P, Pettigrew K, Sainsbury C,
Prabhakaran D and Padmanabhan S: Implications of discoveries from
genome-wide association studies in current cardiovascular practice.
World J Cardiol. 3:230–247. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lettre G, Palmer CD, Young T, Ejebe KG,
Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A,
Dreisbach A, et al: Genome-wide association study of coronary heart
disease and its risk factors in 8,090 African Americans: The NHLBI
CARe Project. PLoS Genet. 7:e10013002011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liyanage VR, Jarmasz JS, Murugeshan N, Del
Bigio MR, Rastegar M and Davie JR: DNA modifications: Function and
applications in normal and disease States. Biology (Basel).
3:670–723. 2014.PubMed/NCBI
|
|
12
|
Kim WY and Sharpless NE: The regulation of
INK4/ARF in cancer and aging. Cell. 127:265–275. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knosel T, Altendorf-Hofmann A, Lindner L,
Issels R, Hermeking H, Schuebbe G, Gibis S, Siemens H, Kampmann E
and Kirchner T: Loss of p16(INK4a) is associated with reduced
patient survival in soft tissue tumours, and indicates a senescence
barrier. J Clin Pathol. 67:592–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bayoglu B, Arslan C, Gode S, Dagistanli
Kaya F, Arapi B, Deser Burc S, Dirican A and Cengiz M: The severity
of internal carotid artery stenosis is associated with the
cyclin-dependent kinase inhibitor 2A gene expression. J Atheroscler
Thromb. 21:659–671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shan M, Yin H, Li J, Li X, Wang D, Su Y,
Niu M, Zhong Z, Wang J, Zhang X, et al: Detection of aberrant
methylation of a six-gene panel in serum DNA for diagnosis of
breast cancer. Oncotarget. 7:18485–18494. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang T, Chen X, Hong Q, Deng Z, Ma H, Xin
Y, Fang Y, Ye H, Wang R, Zhang C, et al: Meta-analyses of gene
methylation and smoking behavior in non-small cell lung cancer
patients. Sci Rep. 5:88972015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang CC, Mao WM and Ling ZQ: DNA
methylation status of RARβ2 and p16 (INK4α) in peripheral blood and
tumor tissue in patients with esophageal squamous cell carcinoma.
Zhonghua Zhong Liu Za Zhi. 34:441–445. 2012.(In Chinese).
PubMed/NCBI
|
|
18
|
Ragione FD and Iolascon A: Inactivation of
cyclin-dependent kinase inhibitor genes and development of human
acute leukemias. Leuk Lymphoma. 25:23–35. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Helgadottir A, Thorleifsson G, Manolescu
A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A,
Sigurdsson A, Baker A, Palsson A, et al: A common variant on
chromosome 9p21 affects the risk of myocardial infarction. Science.
316:1491–1493. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pilbrow AP, Folkersen L, Pearson JF, Brown
CM, McNoe L, Wang NM, Sweet WE, Tang WH, Black MA, Troughton RW, et
al: The chromosome 9p21.3 coronary heart disease risk allele is
associated with altered gene expression in normal heart and
vascular tissues. PLoS One. 7:e395742012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Motterle A, Pu X, Wood H, Xiao Q, Gor S,
Ng FL, Chan K, Cross F, Shohreh B, Poston RN, et al: Functional
analyses of coronary artery disease associated variation on
chromosome 9p21 in vascular smooth muscle cells. Hum Mol Genet.
21:4021–4029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kojima Y, Downing K, Kundu R, Miller C,
Dewey F, Lancero H, Raaz U, Perisic L, Hedin U, Schadt E, et al:
Cyclin-dependent kinase inhibitor 2B regulates efferocytosis and
atherosclerosis. J Clin Invest. 124:1083–1097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhuang J, Peng W, Li H, Wang W, Wei Y, Li
W and Xu Y: Methylation of p15INK4b and expression of ANRIL on
chromosome 9p21 are associated with coronary artery disease. PLoS
One. 7:e471932012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu L, Chen X, Ye H, Hong Q, Xu M and Duan
S: Association of four CpG-SNPs in the vascular-related genes with
coronary heart disease. Biomed Pharmacother. 70:80–83. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fan R, Wang WJ, Zhong QL, Duan SW, Xu XT,
Hao LM, Zhao J and Zhang LN: Aberrant methylation of the GCK gene
body is associated with the risk of essential hypertension. Mol Med
Rep. 12:2390–2394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen X, Hu H, Liu J, Yang Y, Liu G, Ying
X, Chen Y, Li B, Ye C, Wu D and Duan S: FOXF2 promoter methylation
is associated with prognosis in esophageal squamous cell carcinoma.
Tumour Biol. 39:10104283176922302017.PubMed/NCBI
|
|
27
|
Nazarenko MS, Markov AV, Lebedev IN,
Freidin MB, Sleptcov AA, Koroleva IA, Frolov AV, Popov VA,
Barbarash OL and Puzyrev VP: A comparison of genome-wide DNA
methylation patterns between different vascular tissues from
patients with coronary heart disease. PLoS One. 10:e01226012015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng P, Wang L, Yang X, Huang X, Ba Y,
Chen X, Guo J, Lian J and Zhou J: A preliminary study of the
relationship between promoter methylation of the ABCG1, GALNT2 and
HMGCR genes and coronary heart disease. PLoS One. 9:e1022652014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu L, Zheng D, Wang L, Jiang D, Liu H, Xu
L, Liao Q, Zhang L, Liu P, Shi X, et al: GCK gene-body
hypomethylation is associated with the risk of coronary heart
disease. Biomed Res Int. 2014:1517232014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang D, Hong Q, Shen Y, Xu Y, Zhu H, Li
Y, Xu C, Ouyang G and Duan S: The diagnostic value of DNA
methylation in leukemia: A systematic review and meta-analysis.
PLoS One. 9:e968222014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Starker LF, Svedlund J, Udelsman R, Dralle
H, Akerström G, Westin G, Lifton RP, Björklund P and Carling T: The
DNA methylome of benign and malignant parathyroid tumors. Genes
Chromosomes Cancer. 50:735–745. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Murria R, Palanca S, de Juan I, Egoavil C,
Alenda C, García-Casado Z, Juan MJ, Sánchez AB, Santaballa A,
Chirivella I, et al: Methylation of tumor suppressor genes is
related with copy number aberrations in breast cancer. Am J Cancer
Res. 5:375–385. 2014.PubMed/NCBI
|
|
33
|
Christensen BC, Houseman EA, Marsit CJ,
Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF,
Bueno R, et al: Aging and environmental exposures alter
tissue-specific DNA methylation dependent upon CpG island context.
PLoS Genet. 5:e10006022009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grönniger E, Weber B, Heil O, Peters N,
Stäb F, Wenck H, Korn B, Winnefeld M and Lyko F: Aging and chronic
sun exposure cause distinct epigenetic changes in human skin. PLoS
Genet. 6:e10009712010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang Y, Ye H, Hong Q, Xu X, Jiang D, Xu
L, Dai D, Sun J, Gao X and Duan S: Association of CDKN2BAS
polymorphism rs4977574 with coronary heart disease: A case-control
study and a meta-analysis. Int J Mol Sci. 15:17478–17492. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huxley R, Barzi F and Woodward M: Excess
risk of fatal coronary heart disease associated with diabetes in
men and women: Meta-analysis of 37 prospective cohort studies. BMJ.
332:73–78. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Abdel-Maksoud MF, Eckel RH, Hamman RF and
Hokanson JE: Risk of coronary heart disease is associated with
triglycerides and high-density lipoprotein cholesterol in women and
non-high-density lipoprotein cholesterol in men. J Clin Lipidol.
6:374–381. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
An P, Feitosa M, Ketkar S, Adelman A, Lin
S, Borecki I and Province M: Epistatic interactions of
CDKN2B-TCF7L2 for risk of type 2 diabetes and of CDKN2B-JAZF1 for
triglyceride/high-density lipoprotein ratio longitudinal change:
Evidence from the Framingham Heart Study. BMC Proc. 3 Suppl
7:S712009. View Article : Google Scholar : PubMed/NCBI
|