Introduction
Epilepsy is one of the common severe chronic
psychiatric disorders, currently, there are ~54 million people
suffering from epilepsy disease globally (1). Now the treatment of epilepsy is mainly
through the use of antiepileptic drugs such as phenytoin,
ethoxylamine, but clinical statistics show that (2), there is no specific treatment for
epilepsy drugs, and because of long-term use of antiepileptic
drugs, ~40% of patients with epilepsy are resistant to epilepsy
drugs (3). Epilepsy pathogenesis and
the cause of antiepileptic is not clear, which directly lead to the
increase of epilepsy mortality year by year. As a neuropathic
syndrome (4), tuberous sclerosis
complex (TSC) is considered to be a genetic disease caused by
chromosomal abnormalities, the main mechanism is the abnormalities
of TSC1 and TSC2 protein (5).
Studies have found that TSC1 protein is mainly involved in cell
proliferation, division and cell adhesion, and in recent years with
the in-depth research on TSC1 gene (6), it was observed that TSC1 protein can
interact with TSC2 protein, when TSC1 coding abnormality occurs, it
will lead to inhibition of cell proliferation processing, causing
accelerated cell proliferation (7).
In addition, by detecting TSC1 gene in patients with epilepsy, skin
pigmentation, and vascular fibroma, it was found that the above
patients had different degree of TSC1 gene mutation (e.g., locus
129 and locus 174), leading to abnormal protein function (8). We investigated the epilepsy patients in
Rizhao hospital, explored the genetic polymorphisms of TSC1 in
epilepsy and healthy people, in order to improve the diagnostic
accuracy of epilepsy disease from the genetic level, and to provide
a certain theoretical and experimental basis for early diagnosis
and early treatment of epilepsy.
Materials and methods
The study subjects were the 38 epilepsy patients
treated in People's Hospital of Rizhao from May 2015 to June 2016,
marked as observation group, including 20 males and 18 females, the
average age 52.3±8.9 years; 38 healthy people at the same period
were selected as the control group, including 20 males and 18
females, with an average age of 51.2±9.5 years. The study was
approved by the Ethics Committee of People's Hospital of Rizhao and
informed consents were signed by the patients and/or guardians.
Inclusion criteria, in the study, the epilepsy
patients selected were based on the criteria of diagnosis of
epilepsy disease in epilepsy and epilepsy syndrome (9).
Exclusion criteria: i) patients with other
neurological disorders; ii) patients younger than 10 years of
age.
Main reagents: Molecular reagents, Pfu high-fidelity
DNA polymerase (Thermo Fisher Scientific, Shanghai, China); dNTP,
6X buffer, RNA extraction kit, reverse transcription kit, PCR
product purification kit and fluorescence quantitative PCR kit
(Takara, Dalian, China); genomic and protein extraction kits
(Axygen, Union City, CA, USA); EcoR72I restriction endonucleases,
TSC1 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
antibodies (NEB Corp., Beijing, China); agarose, GodView (Solarbio
Science and Technology Co., Ltd., Suzhou, China); the rest of the
chemical reagents were purchased from Sangon Biotechnology Co.,
Ltd. (Shanghai, China).
Main instruments: Fluorescence quantitative PCR
(ABI, Foster City, CA, USA), protein electrophoresis (Beijing Liuyi
Biotechnology Co., Ltd., Beijing, China), multifunctional
microplate reader (Bio-Rad, Hercules, CA, USA).
Methods
Genome extraction
Five milliliters of the elbow vein blood was taken
from the observation group and the control group, centrifuged at
1,000 × g for 5 min, the blood cells were collected. The genome of
healthy people and epilepsy patients was extracted according to the
instructions of the Axygen genome extraction kit (10).
Polymerase chain reaction-restriction fragment
length polymorphism (RFLP). The TSC1 gene sequence was
searched from NCBI website, and primers were designed based on the
sequence. The TSC1 gene sequence of healthy people and epilepsy
patients was amplified. The primer sequences are shown in Table I.
 | Table I.TSC1 primer sequence. |
Table I.
TSC1 primer sequence.
| Name of primer | Length | Sequence |
|---|
| TSC1-F | 21 bp |
ATGAGTCGTAGCTAGTCGAAG |
| TSC1-R | 24 bp |
TACGTCGGAGCTGATCGATGCTAC |
The obtained PCR product was purified by PCR product
purification kit, then ligated with pMD19T simple vector, and the
recombinant vector was introduced into Escherichia coli, and
then the positive clones were screened in the resistant plate (LB +
AMP) and sequenced.
TSC1 gene polymorphism digestion test
TSC1 polymorphism test, the TSC1 gene was amplified
by PCR method using the extracted genome of healthy people and
epilepsy patients as template, and then the PCR product was
digested by EcorR72I restriction enzyme according to the
restriction site predicted by primer (ABI) (11).
Fluorescence quantitative PCR
RNA extraction. In this study, the blood of the
observation group and the control group was taken. RNA was
extracted and the extraction quality was determined.
In order to study the difference of TSC1 mRNA
expression in different treatment on tissues, fluorescence
quantitative PCR was conducted using the cDNA obtained by RNA
reverse transcription as template. The primer sequences are shown
in Table II.
 | Table II.Fluorescence PCR primers. |
Table II.
Fluorescence PCR primers.
| Name of primer | Sequence |
|---|
| qTSC1-F |
TGCTAGCTGAGTCGATCGTACG |
| qTSC1-R |
CGTAGCTGATGCTAGTCGAC |
| GAPDH-F |
CGTAGGGATCGTAGCTAGC |
| GAPDH-R |
CGTAGTCGATGCTAGCTGCG |
Enzyme-linked immunoreaction
Five milliliters of the elbow vein blood was taken
from the observation group and the control group, centrifuged at
1,000 × g for 5 min, the blood cells were collected, the
intracellular total protein was extracted by Axygen kit. After the
total protein was quantified by Coomassie Brilliant Blue, the TSC1
protein in different samples was quantified by enzyme-linked
immunosorbent assay (ELISA) kit.
Western blotting
Five milliliters of the elbow vein blood was taken
from the observation group and the control group, centrifuged at
1,000 × g for 5 min, the blood cells were collected, the
intracellular total protein was extracted by Axygen kit, then
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) was conducted (0.2 mg total protein). After the protein
was transferred, 5% skim milk powder was added for closing at room
temperature for 2 h, then 1:5,000 diluted TSC1 primary antibody was
added (Thermo Fisher Scientific), and incubated at room temperature
for 2 h, then washed by TBST (3 times, 5 min each), then
HRP-labeled secondary antibody was added, incubated at room
temperature for 2 h, and then washed by TBST (3 times, each 5 min)
(internal reference was processed with reference to TSC1 protein
detection method), and color development solution was added for
observation (12).
Statistical analysis
In this study, we analyzed the data by SPSS 19.0
software (SPSS Inc., Chicago, IL, USA). The data were expressed as
(mean ± SD), and the difference between the control group and the
observation group was analyzed by t-test. P<0.05 for the
difference was considered as statistically significant.
Results
TSC1 gene amplification and
sequencing
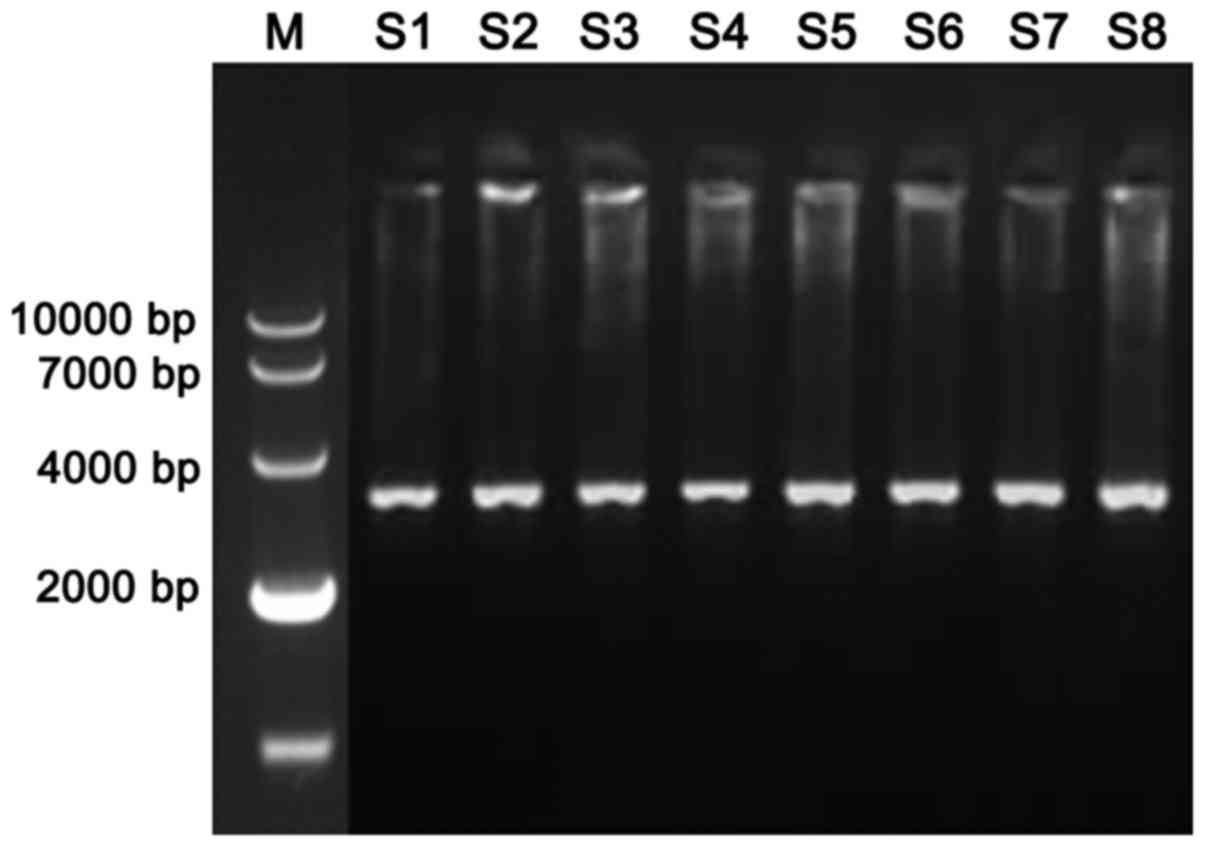
The TSC1 gene of the epilepsy patients and the
healthy population (Fig. 1) was
obtained by using the genome of the control group and the
observation group as template. By sequencing TSC1 of epilepsy
patients and healthy population (Table
III), we found that there were three genotypes at locus 142 in
healthy population, CC (79.3%), CA (13.9%) and AA (6.8%), there
were also three genotypes at locus 142 in epilepsy patients, CC
(21.3%), CA (26.4%) and AA (52.3%). There was significant
difference between the epilepsy patients and healthy population in
terms of the genotype CC and AA (P<0.05).
 | Table III.Results of TSC1 gene sequencing in
epilepsy patients and healthy population. |
Table III.
Results of TSC1 gene sequencing in
epilepsy patients and healthy population.
|
| Genotype |
|---|
|
|
|
|---|
| Group | CC | CA | AA |
|---|
| Control | 79.3% | 13.9% | 6.8% |
| Observation | 21.3% | 26.4% | 52.3% |
| t-value | 4.31 | 2.14 | 4.86 |
| P-value | <0.05 | >0.05 | <0.05 |
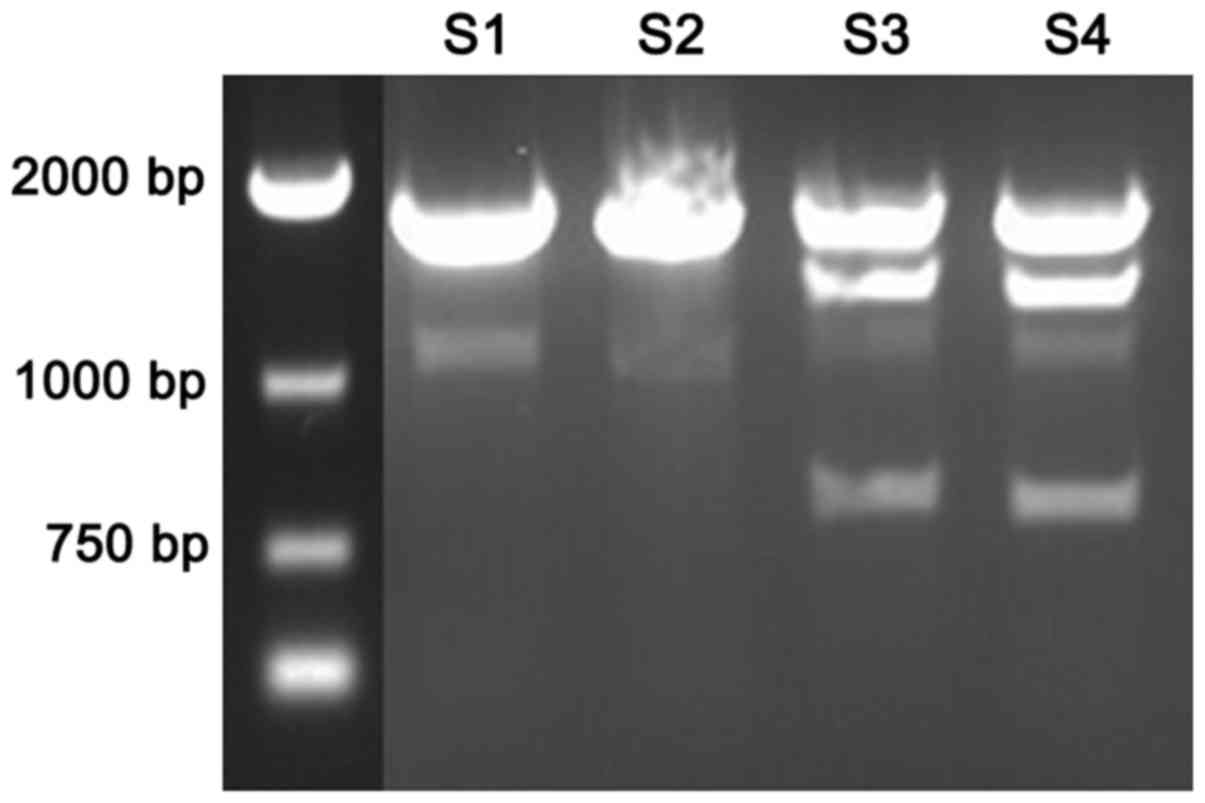
TSC1 polymorphism digestion
By analyzing the sequence of TSC1 gene, it was found
that locus 142 was the recognition site for EcoR72I (Fig. 2). There were 2 bands in epilepsy
patients (S1-S2) TSC1 after digested by EcoR72I; there were 3 bands
in healthy population (S3-S4) TSC1 gene after digested by
EcoR72I.
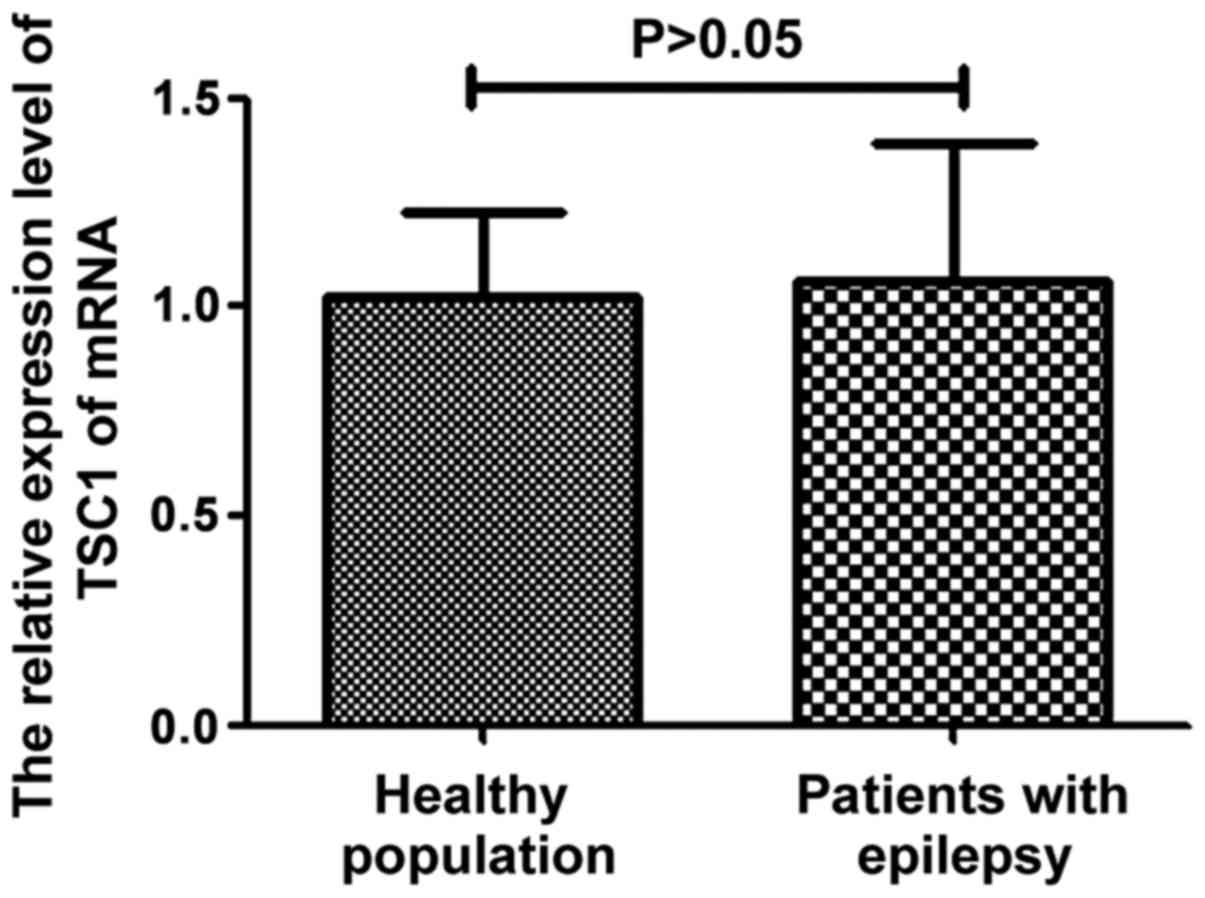
TSC1 mRNA expression in the control
and observation groups
The total RNA was extracted from patients with
epilepsy and healthy people. The difference of TSC1 mRNA expression
in different subjects was determined by fluorescence quantitative
PCR. The results are shown in Fig.
3. It can be seen from the figure that there was no significant
difference in TSC1 mRNA expression between epilepsy patients and
healthy people (P>0.05).
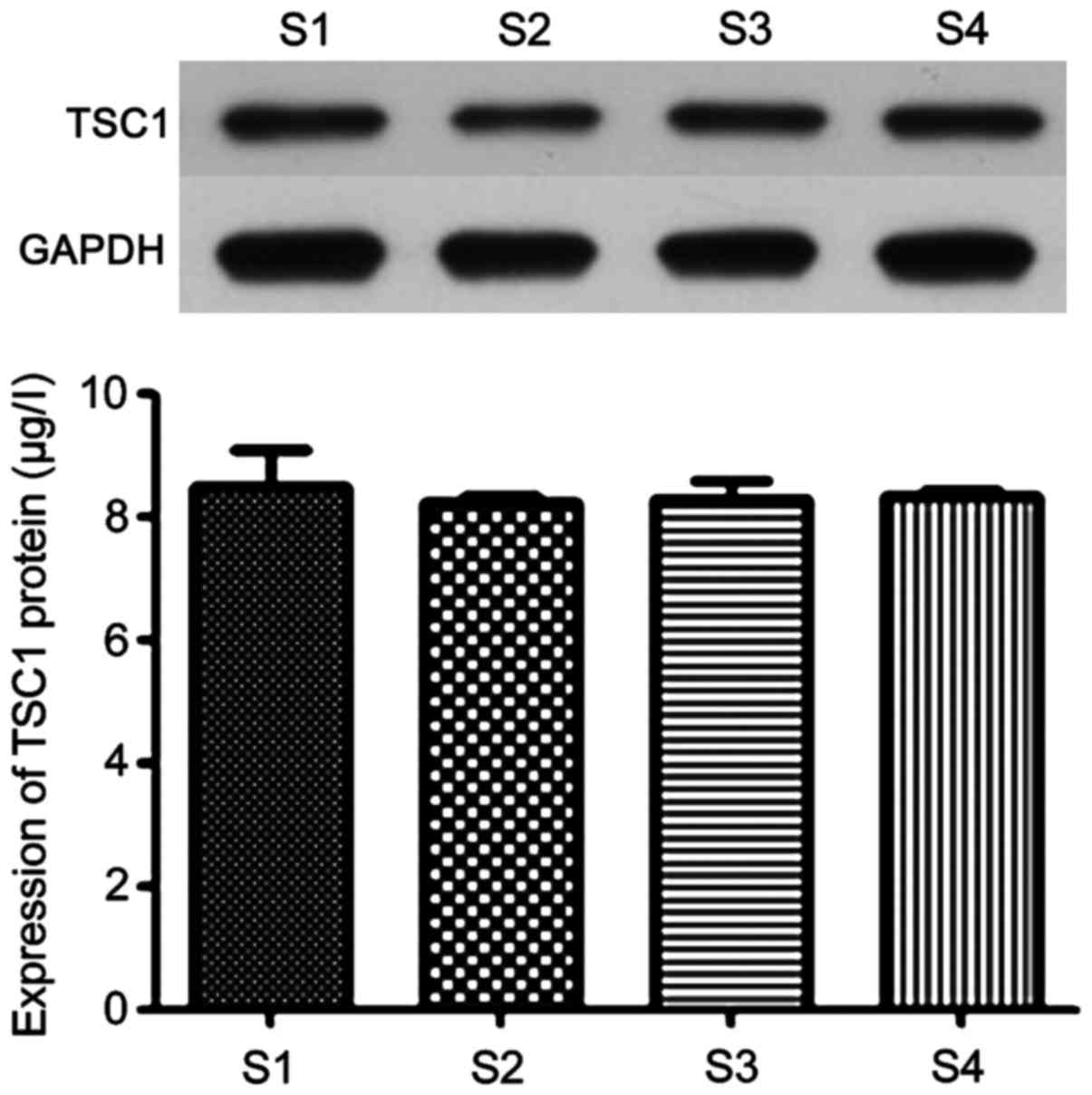
TSC1 protein expression in the control
and observation groups
The expression of TSC1 protein in epilepsy patients
and healthy people was determined by western blot method. The
results are shown in Fig. 4. It can
be seen from the figure that there was no significant difference in
the protein expression of TSC1 protein between epilepsy and healthy
people (P>0.05), which indicated that polymorphism of locus 142
did not affect TSC1 gene expression.
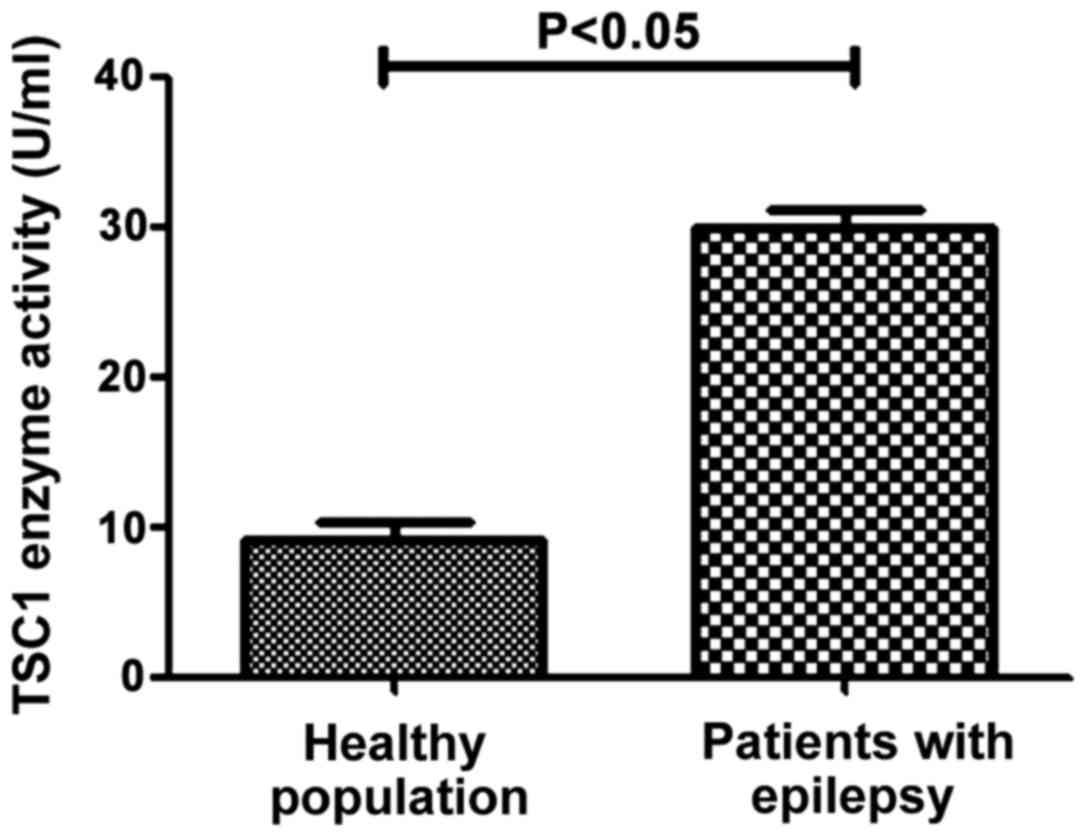
ELISA detection of TSC1 protein
activity in the control and observation groups
The activity of TSC1 protein in different samples
was determined by ELISA method. The results in Fig. 5 shows that the activity of TSC1
protein in healthy population (8.95±2.41 U/ml) was significantly
lower than that in patients with epilepsy (29.27±4.06 U/ml), the
difference was statistically significant (P<0.05). This
suggested that mutations of TSC1 in patients with epilepsy can lead
to elevated TSC1 enzyme activity.
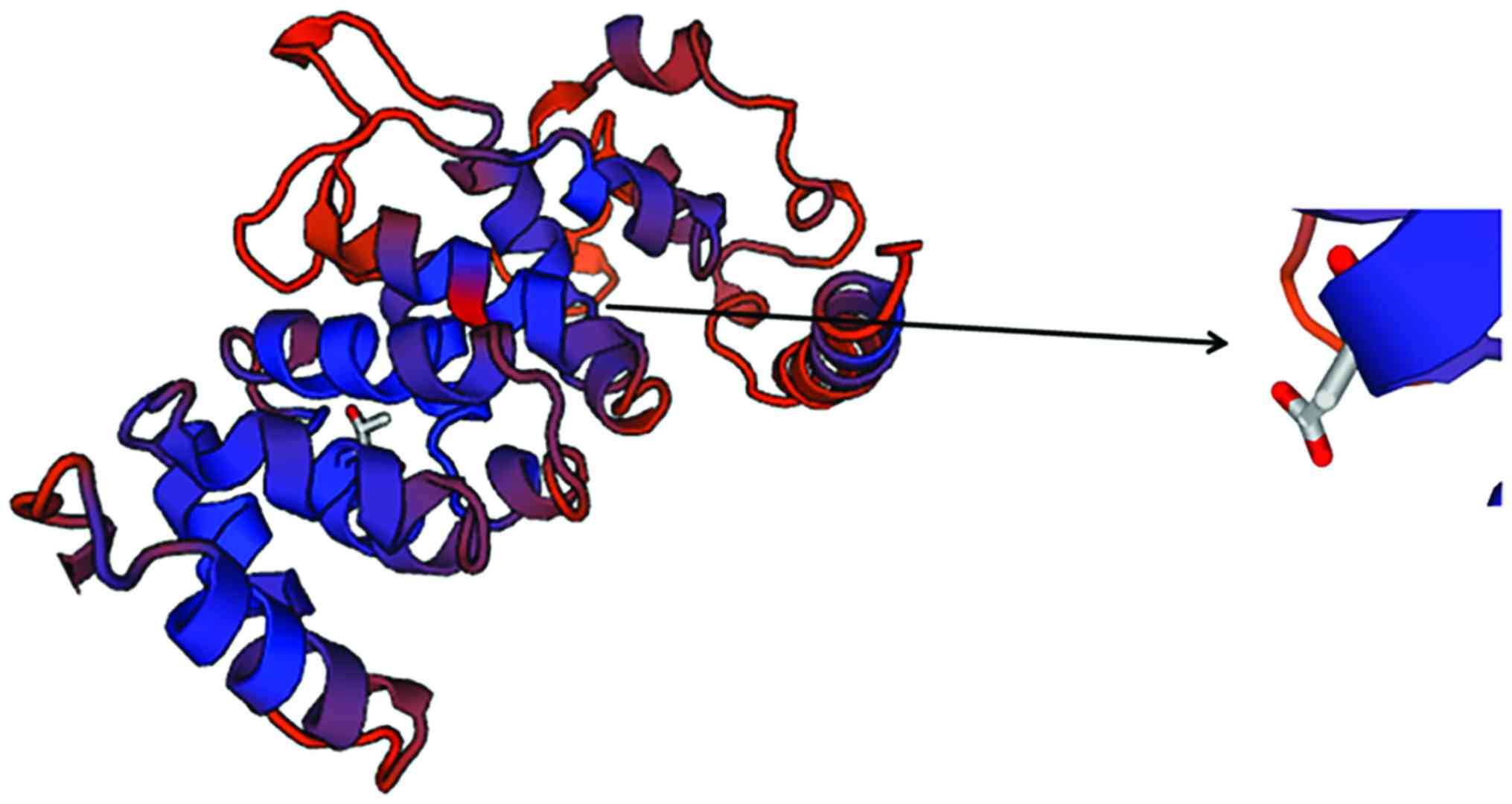
SWISS-MODEL homology modeling
analysis
In this study, we found that TSC1 activity in
patients with epilepsy was significantly higher than that in
healthy people. The three-dimensional structure of TSC1 protein was
predicted by SWISS-MODEL software (Swiss Institute of
Bioinformatics, Basel, Switzerland), the results are shown in
Fig. 6. It was found that locus 142
was located at the TSC1 protein active center, which revealed the
reason of increased activity of TSC1 after locus 142 mutation in
epilepsy patients.
Discussion
Clinical data show that (13), the sensitivity of epilepsy drugs
against epilepsy is gradually reducing. With the deepening of
epilepsy research in recent years, it was found that the main cause
of antiepileptic symptoms was the mutation of antiepileptic drug
target site in the human body or the change of the
three-dimensional structure of the protein, making the original
antiepileptic drug identification site wrapped within the
three-dimensional structure of the protein (14–16), so
that the antiepileptic drug cannot bind with the target site and
eventually lost effect. For example, the study found that (17), GABAAR, as the current recognized
antiepileptic drug recognition site, the main structure is ion
channel composed of five subunits (including α1-6, β1-4 and λ1-3),
studies found that in patients with epilepsy disease, the
expression of β1-4 protein decreased, and with the duration of
disease gradually extending, the protein expression showed a
gradual decreasing trend (18,19).
This suggests that changes in antiepileptic drug interaction target
site and mutations can both lead to occurrence and progression of
epilepsy in patients (20). In this
study, we found that TSC1 gene in the epileptic patients and
healthy human showed a certain polymorphism, that is, there were
significant differences in the base at locus 142, locus 142 in
healthy population was mainly CC (79.3%), while in the epilepsy
patients was predominantly CA (26.4%) and AA (52.3%), there was
significant difference between the two groups (P<0.05). However,
there was no significant difference between the two groups in terms
of the TSC1 gene mRNA and protein expression between the healthy
population and epilepsy patients (P>0.05). The ELISA results
showed that the activity of TSC1 protein in patients with epilepsy
(29.27±4.06 U/ml) was significantly higher than that in healthy
population (8.95±2.41 U/ml), which indicated that locus 142
polymorphisms could promote the occurrence of epilepsy, that is,
the increased CC content was more likely to cause epilepsy. By
SWISS-MODEL, it was found that the locus 142 was mainly located in
the TSC1 protein activity center, and the change of the base in the
locus could significantly affect the protein activity.
References
|
1
|
Faure JB, Marques-Carneiro JE, Akimana G,
Cosquer B, Ferrandon A, Herbeaux K, Koning E, Bar-belivien A,
Nehlig A and Cassel JC: Attention and executive functions in a rat
model of chronic epilepsy. Epilepsia. 55:644–653. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fernández-Gajardo R, Matamala JM, Carrasco
R, Gutiérrez R, Melo R and Rodrigo R: Novel therapeutic strategies
for traumatic brain injury: Acute antioxidant reinforcement. CNS
Drugs. 28:229–248. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rowley S and Patel M: Mitochondrial
involvement and oxidative stress in temporal lobe epilepsy. Free
Radic Biol Med. 62:121–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korczyn AD, Schachter SC, Brodie MJ, Dalal
SS, Engel J Jr, Guekht A, Hecimovic H, Jerbi K, Kanner AM,
Johannessen Landmark C, et al: Epilepsy, cognition, and
neuropsychiatry (Epilepsy, Brain, and Mind, part 2). Epilepsy
Behav. 28:283–302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Figueiredo SM, Filho SA,
Nogueira-Machado JA and Caligiorne RB: The anti-oxidant properties
of isothiocyanates: A review. Recent Pat Endocr Metab Immune Drug
Discov. 7:213–225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kan MC, Wang WP, Yao GD, Li JT, Xie T,
Wang W and Ma WQ: Anticonvulsant effect of dexmedetomidine in a rat
model of self-sustaining status epilepticus with prolonged amygdala
stimulation. Neurosci Lett. 543:17–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Howard BA and Lu P: Stromal regulation of
embryonic and postnatal mammary epithelial development and
differentiation. Semin Cell Dev Biol. 25–26:43–51. 2014. View Article : Google Scholar
|
|
8
|
Liu X, Ory V, Chapman S, Yuan H, Albanese
C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al:
ROCK inhibitor and feeder cells induce the conditional
reprogramming of epithelial cells. Am J Pathol. 180:599–607. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Düvel K, Yecies JL, Menon S, Raman P,
Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S,
et al: Activation of a metabolic gene regulatory network downstream
of mTOR complex 1. Mol Cell. 39:171–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bissler JJ, Kingswood JC, Radzikowska E,
Zonnenberg BA, Frost M, Belousova E, Sauter M, Nonomura N,
Brakemeier S, de Vries PJ, et al: Everolimus for angiomyolipoma
associated with tuberous sclerosis complex or sporadic
lymphangioleiomyomatosis (EXIST-2): A multicentre, randomised,
double-blind, placebo-controlled trial. Lancet. 381:817–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu NK and Xu XM: Neuroprotection and its
molecular mechanism following spinal cord injury. Neural Regen Res.
7:2051–2062. 2012.PubMed/NCBI
|
|
12
|
Colín-González AL, Orozco-Ibarra M,
Chánez-Cárdenas ME, Rangel-López E, Santamaría A, Pedraza-Chaverri
J, Barrera-Oviedo D and Maldonado PD: Heme oxygenase-1 (HO-1)
upregulation delays morphological and oxidative damage induced in
an excitotoxic/pro-oxidant model in the rat striatum. Neuroscience.
231:91–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morroni F, Tarozzi A, Sita G, Bolondi C,
Moraga Zolezzi JM, Cantelli-Forti G and Hrelia P: Neu-roprotective
effect of sulforaphane in 6-hydroxydopamine-lesioned mouse model of
Parkinson's disease. Neurotoxicology. 36:63–71. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou H, Wang N, Xu L, Huang HL and Yu CY:
Clinical study on anti-epileptic drug with B vitamins for the
treatment of epilepsy after stroke. Eur Rev Med Pharmacol Sci.
21:3327–3331. 2017.PubMed/NCBI
|
|
15
|
Sanchez RM, Ribak CE and Shapiro LA:
Synaptic connections of hilar basal dendrites of dentate granule
cells in a neonatal hypoxia model of epilepsy. Epilepsia. 53 Suppl
1:98–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vittos O, Toana B, Vittos A and Moldoveanu
E: Lipoprotein-associated phospholipase A2 (Lp-PLA2): A review of
its role and significance as a cardiovascular biomarker.
Biomarkers. 17:289–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao T, Li Y, Dai X, Wang J, Qi Y, Wang J
and Xu K: Effects of retrograde gene transfer of brain-derived
neurotrophic factor in the rostral spinal cord of a compression
model in rat. Mol Biol Rep. 39:8045–8051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakano N, Nakai Y, Seo TB, Yamada Y, Ohno
T, Yamanaka A, Nagai Y, Fukushima M, Suzuki Y, Nakatani T, et al:
Characterization of conditioned medium of cultured bone marrow
stromal cells. Neurosci Lett. 483:57–61. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uriarte G, Paternain L, Milagro FI,
Martínez JA and Campion J: Shifting to a control diet after a
high-fat, high-sucrose diet intake induces epigenetic changes in
retroperitoneal adipocytes of Wistar rats. J Physiol Biochem.
69:601–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lovinsky-Desir S and Miller RL:
Epigenetics, asthma, and allergic diseases: A review of the latest
advancements. Curr Allergy Asthma Rep. 12:211–220. 2012. View Article : Google Scholar : PubMed/NCBI
|