Introduction
Sjögren syndrome (SS) is a chronic systemic
inflammatory disease affecting the exocrine glands, particularly
the salivary and lacrimal glands, resulting in keratoconjunctivitis
sicca and xerostomia (1). The
disease primarily affects middle-aged females. However,
establishing the diagnosis of SS is often challenging, since the
symptoms are nonspecific, while no single laboratory test allows
for a definitive diagnosis and there are no disease-specific
diagnostic criteria (2). Although
the exact pathogenic mechanisms are not yet fully elucidated,
immune system abnormalities are considered to be important in SS
pathogenesis, including hypergammaglobulinemia, increased levels of
autoantibodies against Ro/SS-associated antigen A (Ro/SSA) and
La/SS-associated antigen B (La/SSB) (3), and exocrine gland histology
characterized by focal lymphocyte infiltrates surrounding the
glandular ducts (4,5). However, it is unlikely that the
pathogenesis of SS is solely immunologically mediated, and the
contribution of nonimmune mechanisms should be considered.
Therefore, the investigation of SS is appealing, particularly with
respect to the role of numerous microRNAs (miRNAs or miR) involved
in systemic autoimmune diseases, including their potential as
diagnostic or prognostic biomarkers, as well as their roles in the
pathogenesis of autoimmunity, inflammation and/or organ
dysfunction.
miRNAs are highly conserved, small, noncoding,
single-stranded RNA molecules that are able to regulate gene
expression by binding to complementary sequences within the
3′-untranslated region of targeted mRNAs, leading to translational
repression or degradation (6).
miRNAs participate in various physiological and pathological
processes, including the cellar metabolism, differentiation,
proliferation and apoptosis. These molecules have been demonstrated
to be potential diagnostic/prognostic biomarkers and/or therapeutic
targets in various diseases, including hepatocellular carcinoma,
multiple myeloma and cardiac diseases (7). In addition, miRNAs participate in the
regulation of innate and adaptive immune responses (8). At present, miRNAs have attracted
increasing attention due to their roles in the pathogenesis of
rheumatic diseases. For instance, dysregulation of miRNAs has been
reported to be associated with the disease activity and major organ
involvement in patients with systemic lupus erythematosus and
rheumatoid arthritis (RA) (9).
However, only a limited number of studies have focused on
understanding the miRNA function in SS. It has been reported that
miR-146a, −155 and −181a were overexpressed in peripheral blood
mononuclear cells (PBMCs) collected from SS patients (7,10–13),
while miR-17 to −92 were downregulated in the salivary gland
tissues (12). Although regulated
expression of certain miRNAs have been demonstrated in the salivary
gland tissues of SS patients, a comprehensive description of their
contribution to the pathological grade of SS remains
incomplete.
In the current study, the miRNA profile of labial
salivary glands from SS patients and control subjects were
investigated and the correlation between miRNAs and SS pathological
grade was examined.
Materials and methods
Patients
SS patients (n=28) were recruited from the
Department of Rheumatology of Beijing Shijitan Hospital (Beijing,
China) between August 2008 and August 2013. The inclusion criteria
for SS patients included the following: i) Fulfillment of all
revised criteria for SS diagnosis proposed by the American-European
Consensus (AEC) Group (14); ii)
absence of any other rheumatic or underlying disease; and iii) no
previous treatment with glucocorticoid or immunosuppressive agents.
Furthermore, 18 age- and sex-matched non-SS sicca syndrome patients
recruited from Beijing Shijitan Hospital were used as controls.
Non-SS sicca syndrome was defined as the presence of xerostomia and
xerophthalmia in patients with negative non-organ-specific
autoantibodies and a negative minor salivary gland biopsy according
to the Chisolm and Mason scoring system (15). Detailed information on the SS and
control patients is provided in Table
I. The current study was approved by the Ethics Committee of
the Capital Medical University (Beijing, China). Informed written
consent was obtained from all patients participating in the present
study.
 | Table I.Demographic data and clinical
characteristics of patients with SS and controls. |
Table I.
Demographic data and clinical
characteristics of patients with SS and controls.
| Variable | SS (n=28) | Controls (n=18) |
|---|
| Demographics |
|
|
|
Male/female | 1/27 | 1/17 |
| Age,
years | 50.14±2.65 | 54.23±3.21 |
| Clinical
manifestations |
|
|
| Dry eye,
no. +/− | 22/6 | 11/7 |
| Dry
mouth, no. +/− | 22/6 | 14/4 |
| Organ
involvement, no. +/− | 12/16 | – |
| Laboratory tests |
|
|
| ANA
positivity, no. +/− | 18/10 | – |
|
Anti-Ro(SSA), no. +/− | 14/14 | – |
|
Anti-La(SSB), no. +/− | 8/20 | – |
| RF, no.
+/− | 12/16 |
|
| Elevated
ESR, no. +/− | 18/10 | – |
Salivary gland tissue specimens
As part of the diagnostic workup, all SS and control
patients underwent salivary gland biopsy, according to the AEC
Group guidelines. Briefly, a local anesthetic was injected into the
lower lip, followed by performing a small incision to the right or
left of the midline of the lip. A total of 5 or 6 minor salivary
gland lobules were harvested, and all specimens had a glandular
area of ≥4 mm2. Following harvest, the majority of
specimens were stored at −80°C for miRNA expression profiling,
while the remaining samples were delivered directly to the
pathology department for histopathological evaluation. Evaluation
of biopsy samples from SS and control patients was performed
independently by two oral pathologists. Specimens from SS patients
demonstrated characteristic features of SS, including a salivary
gland pathological focus (SGPF) score of ≥1 and presence of
lymphoepithelial lesions (16),
whereas specimens obtained from the control subjects did not
present such features. Subsequently, SS patients were divided into
two groups based on their SGPF scores, as follows: Scores of <3
(n=16) and scores of ≥3 (n=12) (17).
RNA isolation and microarray
analysis
Total RNA was harvested from the labial salivary
gland tissues from all SS and control subjects using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
miRNeasy mini kit (Qiagen, Hilden, Germany) according to the
manufacturer's protocol, in order to efficiently recover all RNA
species, including miRNAs. The total RNA concentration was then
measured using a NanoDrop 1000 device (Thermo Fisher Scientific,
Inc.) at an optical density of 260/280 nm, and then samples were
labeled using a miRCURY™ Hy3™/Hy5™
miRNA Power Labeling kit (Exiqon; Qiagen) and hybridized on a
miRCURY™ LNA miRNA Array (version 18.0; Exiqon; Qiagen).
Subsequent to washing, arrays were scanned using an Axon GenePix
4000B microarray scanner (Molecular Devices, LLC, Sunnyvale, CA,
USA).
Microarray expression profiling of miRNAs in the
labial salivary gland tissues was performed on 3 randomly selected
female patients with SS, and 3 randomly selected age- and
sex-matched patients with non-SS sicca syndrome. This was performed
using a seventh generation, single-color miR CURY LNA™
miRNA Array that contained 3,100 capture probes, covering all the
human, mouse and rat miRNAs annotated in the miRBase database
(version 18.0; mirbase.org), as well as all the viral
miRNAs associated with these species. In addition, this array
contained capture probes for 25 proprietary miRPlus™
human miRNAs (Exiqon; Qiagen) that were not found in the miRBase
database. Gene expression was normalized using the median
normalization method, as follows: Normalized
data=(foreground-background)/median expression, where median
expression is the 50% quantile of miRNA intensity >50 in all
samples subsequent to background correction. The threshold value
used to define the significant upregulation or downregulation of
miRNAs in patients with SS compared with the controls was a
fold-change in the expression level of >1.5 and a P-value of
<0.05 according to Student's t-test.
The Ro/SSA and La/SSB intracellular
ribonucleoproteins are the major targets of autoimmune humoral
responses in SS (18). A literature
review found that there were ~30 miRNAs, including miR-181a,
miR-16, target Ro/SSA (Ro52/E3 ubiquitin-protein ligase TRIM21 or
Ro60/60 kDa SS-A/Ro ribonucleoprotein) and La/SSB mRNAs (19). miRNA target prediction programs,
TargetScan (targetscan.org, version 7.1) and
MicroRNA (microrna.org, 2010 release), were used
to double-check these miRNAs target genes and validate their
functions. Subsequently, the differential expression of miR-181a
and miR-16 in the tissues of patients with SS were confirmed by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis.
miRNA validation by RT-qPCR
analysis
Subsequent to integrating all database results,
miR-181a and −16 were observed to be associated with La/SSA and
Ro/SSB during SS pathogenesis. Therefore, these two miRNAs were
verified in the labial salivary gland samples of all 28 patients
with SS and 18 control patients. Total RNA was reverse transcribed
into cDNA using a Taqman miRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). qPCR was then
conducted using TaqMan™ Fast Advanced Master Mix (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Briefly, a 20 µl PCR reaction included 2 µl RT product, 1 µl
TaqMan™ Gene Expression Assay (20X), 10 µl
TaqMan™ Fast Advanced Master Mix (10X), 7 µl
nuclease-free water. The primers were synthesized by the Sangon
Biotech Co., Ltd. (Shanghai, China) and were as follows: miR-181a,
5′-GGGAACATTCAACGCTGTCG-3′ (forward) and 5′-GTGCGTGTCGTGGAGTCG-3′
(reverse); miR-16, 5′-GGGTAGCAGCACGTAAATA-3′ (forward) and
5′-CAGTGCGTGTCGTGGAGT-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse). The reactions were carried
out at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. Relative gene expression was calculated by the
2−∆∆Cq method (20) and
snRNA U6 was used as an internal control.
Statistical analysis
The results of the microarray analysis of miRNA
expression between the SS and control patients were compared using
an unpaired Student's t-test, where a P<0.05 and fold-change of
>2.0 were considered statistically significant. Statistical
analysis was performed using SPSS version 19.0 software (IBM Corp.,
Armonk, NY, USA). A Mann-Whitney test was used to assess the
differences between controls and patients with SS, and a P<0.05
was considered as an indicator of statistically significant
differences.
Results
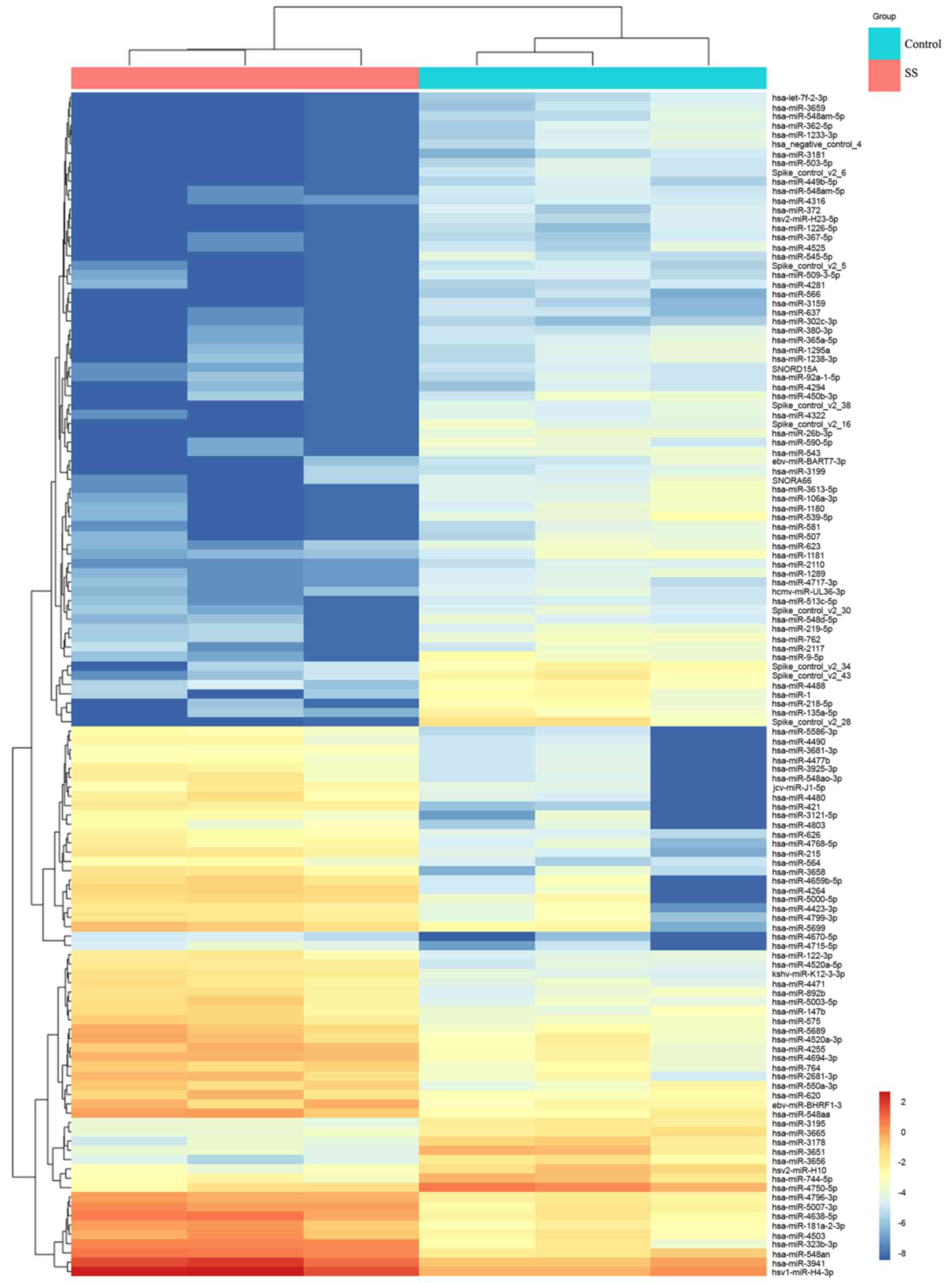
Differentially expressed miRNAs in
labial salivary gland tissues
Since altered expression levels of several miRNAs
have been reported in the PBMCs of patients with SS (21). The present study further investigated
the dysregulation of miRNAs in the labial salivary gland tissue. In
order to identify miRNAs that may correlate with SS, microarray
analysis was used to investigate the miRNA expression profiles in
labial salivary gland tissues from 3 patients with SS and 3
patients with non-SS sicca syndrome. The results revealed a total
of 127 differentially expressed miRNAs in SS patients when compared
with their levels in the control patients, including 76 upregulated
and 51 downregulated miRNAs (Fig.
1).
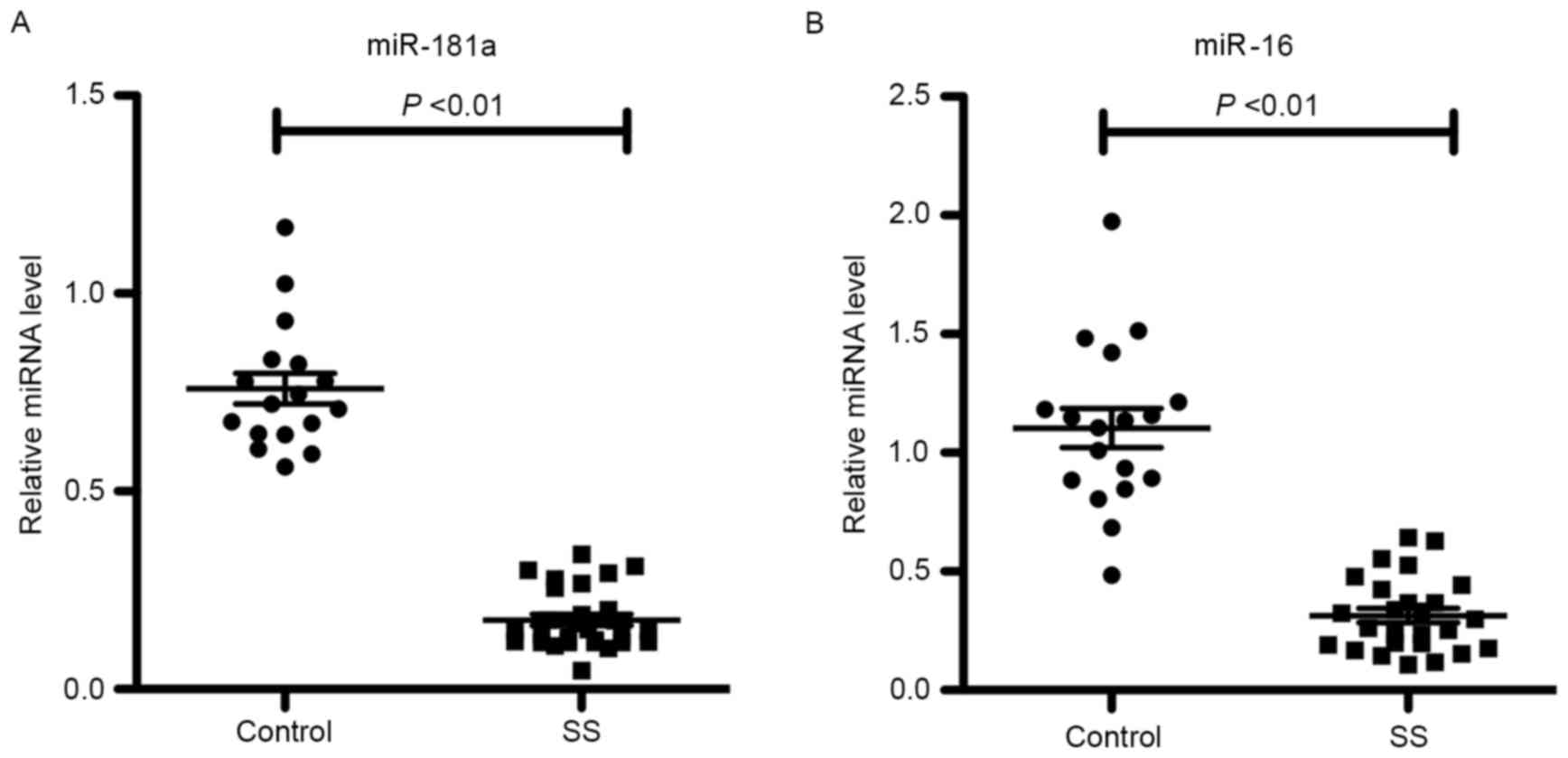
miR-181a and −16 expression levels in
labial salivary gland tissues
In order to identify miRNAs potentially associated
with SS, a literature search and miRNA target prediction was
performed using TargetScan and MicroRNA.org.
Through this analysis two miRNAs, miR-181a and −16, were identified
to be associated with Ro/SSA and La/SSB in patients with SS.
miR-181a and −16 expression levels in 28 SS and 18 control labial
salivary gland tissues were detected by RT-qPCR. As shown in
Fig. 2, the miR-181a and −16
expression levels were significantly downregulated in patients with
SS compared with the controls (P<0.01), suggesting that miR-181a
and −16 may serve a role in the pathogenesis of SS.
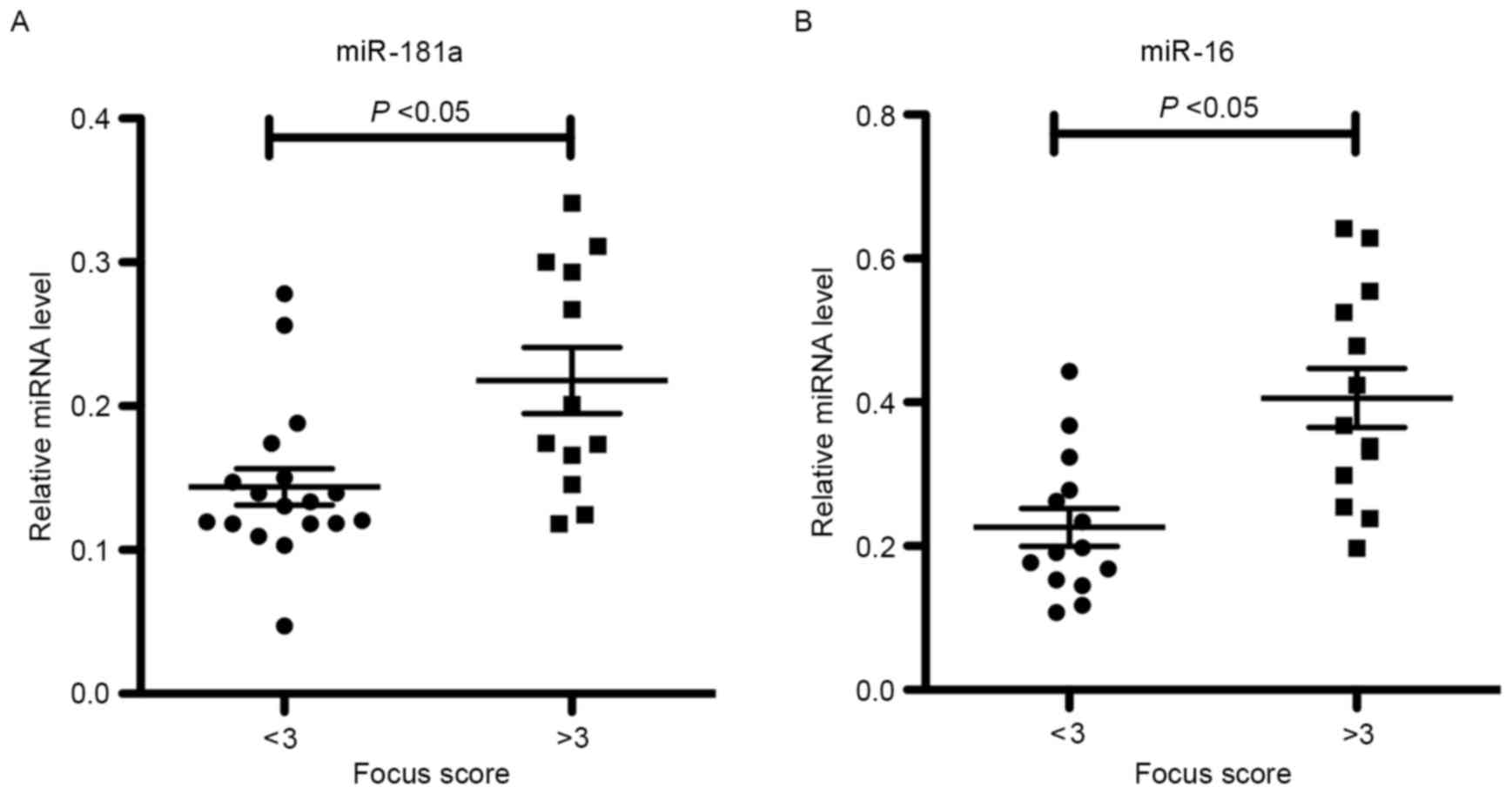
Correlation of miR-181a and −16
expression levels with SGPF scores
To further characterize the relevance of miR-181a
and −16 expression levels in SS labial salivary glands, statistical
analyses were performed on SS patients grouped according to their
SGPF score. It was observed that the levels of miR-181a and −16
were increased in patients with SS who possessed high SGPF scores
of ≥3 (high-grade inflammation) when compared with those in
patients exhibiting low SGPF scores of <3 (low-grade
inflammation; Fig. 3). This
indicates that miR-181a and −16 may be involved in the pathogenesis
of SS.
Discussion
SS is a chronic, inflammatory, autoimmune disease
characterized by lymphocytic infiltration of the exocrine glands,
particularly of the salivary and lacrimal glands, leading to
destruction of their functional components (1). However, SS diagnostic criteria,
including the AECG serological or histopathological criteria in
conjunction with oral and ocular dryness, are frequently
inconsistently applied and cannot optimally differentiate patients
with SS from patients with non-SS sicca or address extraglandular
manifestations. As a result, the diagnosis of SS may be delayed by
6–10 years after the disease onset, while recent attempts to
address these issues by novel diagnostic criteria for early SS that
were reported in 2015 require further large-scale validation
(22). According to the mentioned SS
criteria, labial salivary gland biopsy occupies a very important
position in the diagnosis of SS (14,23),
although this is an invasive procedure. Recently, an increasing
number of studies revealed that miRNAs in PBMCs, sera and saliva
contribute to the pathogenesis of SS and may represent a novel
class of diagnostic biomarkers for this disease (8,10,11,21).
However, studies investigating the miRNA profiles in the salivary
gland tissue of SS patients are currently lacking. In the current
study, microarray analysis demonstrated the upregulation of 76
miRNAs and downregulation of 51 miRNAs in the labial salivary gland
tissues of SS patients relative to the controls. miR-181a and −16
were observed to be associated with and to regulate Ro/SSA and
La/SSB during SS pathogenesis. Therefore, their expression was
further verified in labial salivary gland tissues from 28 patients
with SS and 18 controls included in the present study. The results
revealed that the expression levels of miR-181a and −16 were
decreased in SS patients compared with those in control subjects.
Furthermore, miR-181a and −16 expression levels were associated
with the labial SGPF score.
miR-181a has been reported to be involved in
hematological malignancies (24).
For instance, miR-181a was overexpressed in T-/B-cell
leukemia/lymphoma, while miR-181a-deficient mice demonstrated
severe defects in T-cell development (24–26).
This indicates that miR-181a may be involved in the pathogenesis of
lymphocytic diseases. In the present study, miR-181a expression was
significantly downregulated in the labial salivary gland tissues of
patients with SS in comparison with the controls. In contrast to
the present observations, Peng et al (27) reported that miR-181a expression was
elevated in PBMCs of Chinese patients with SS, and there was a
positive correlation between miR-181a levels and antinuclear
antibody titer. The overexpression of miR-181a contributed to the
B-cell maturation rather than that of T-cells in patients with SS.
Although this observation is inconsistent with the results of the
present study, it should be noted that the miRNA profiles in
different tissues were examined in the two studies. Since the
function and effect of miRNA is dependent on the tissue in numerous
cases, identifying the cell types that present miRNA dysregulation
is imperative to the study of their role in disease (28). Furthermore, miR-181a expression has
been demonstrated to increase the number of B-lymphoid cells, with
a concurrent decrease in T-lymphoid cells (28). Peng et al (27) reported that individual measurement of
the miR-181a level in B- and T-cells isolated from PBMCs indicated
that the B-cell population contributed to the elevation in miR-181a
levels, which may be involved in the increased miR-181a levels in
PBMCs and decreased levels in labial salivary gland tissues.
Another study by Li et al (29) reported that increasing the miR-181a
expression in mature T-cells augments their sensitivity to peptide
antigens, while inhibiting the miR-181a expression in immature
T-cells reduces their sensitivity and impairs the positive and
negative selection. T-cell sensitivity to autoantigens is
quantitatively regulated by managing the expression of miR-181a in
order to enable mature T-cells to recognize antagonists (inhibitory
peptide antigens) as agonists. Li et al (29) also revealed a correlation between
higher miR-181a levels and greater sensitivity in immature T-cells,
which may explain the elevated miR-181a levels in the salivary
glands of SS patients with high-grade inflammation.
Numerous studies have indicated that miR-16 family
members regulate cell cycle-associated genes, as well as promote
tumorigenesis and metastasis (30,31).
Hence, miR-16 family members have been considered as potential
biomarkers in cancer diagnosis (32). Certain studies have recently reported
altered miR-16 expression in rheumatoid disease. For instance,
Pauley et al (33) identified
miR-16 overexpression in the PBMCs of RA patients with active
disease as compared with RA patients with low disease activity or
healthy control patients. Another study demonstrated that higher
levels of miR-16 in the sera of treatment naïve patients with
early-stage RA correlated with attenuation of the disease (34). Thus, monitoring miR-16 levels may be
a useful tool for predicting the disease outcomes in patients with
early-stage RA. In the current study, reduced miR-16 expression was
observed in the labial salivary glands of patients with SS compared
with that in patients with non-SS sicca syndrome (controls),
however, the patients with SS who exhibited high SGPF scores in
salivary glands had elevated level of miR-16.
To the best of our knowledge, the present study is
the first to report decreased expression levels of miR-181a and −16
in the labial salivary gland of SS patients; however, this study
has several limitations. Firstly, the current study only verified
the expression levels of miR-181a and −16 in labial salivary gland
tissues. In the future, we intend to investigate the expression of
miRNAs in PBMCs and labial salivary gland tissues simultaneously in
a large cohort of patients. In addition, the precise mechanism
through which miR-181a and −16 affect the focal lymphocytic
infiltration should be further clarified.
In conclusion, the present investigation of miRNA
profiles in the labial salivary gland of SS and control patients
demonstrated reduced expression levels of miR-181a and −16 in
conjunction with SS. Furthermore, these two miRNAs were increased
in patients with SS who possessed high SGPF scores, suggesting that
miR-181a and −16 may serve a role in the pathogenesis of SS.
Acknowledgements
The present study was supported by the Beijing
National Science Foundation (grant no. 7123220). The authors would
like to thank all the patients enrolled into the present study, and
Dr. Xiaoyang Jin (Institute of Biophysics, Chinese Academy of
Science, Beijing, China) for his special contribution to the
investigation.
References
|
1
|
Tincani A, Andreoli L, Cavazzana I, Doria
A, Favero M, Fenini MG, Franceschini F, Lojacono A, Nascimbeni G,
Santoro A, et al: Novel aspects of Sjögren's syndrome in 2012. BMC
Med. 11:932013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cotrim AP and Alevizos I: Human and viral
microRNA expression in Sjögren syndrome. J Rheumatol. 41:2102–2103.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mackay F, Groom JR and Tangye SG: An
important role for B-cell activation factor and B cells in the
pathogenesis of Sjögren's syndrome. Curr Opin Rheumatol.
19:406–413. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greenspan JS, Daniels TE, Talal N and
Sylvester RA: The histopathology of Sjögren's syndrome in labial
salivary gland biopsies. Oral Surg Oral Med Oral Pathol.
37:217–229. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fox PC: Autoimmune diseases and Sjogren's
syndrome: An autoimmune exocrinopathy. Ann N Y Acad Sci.
1098:15–21. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chua JH, Armugam A and Jeyaseelan K:
MicroRNAs: Biogenesis, function and applications. Curr Opin Mol
Ther. 11:189–199. 2009.PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi H, Zheng LY, Zhang P and Yu CQ:
miR-146a and miR-155 expression in PBMCs from patients with
Sjögren's syndrome. J Oral Pathol Med. 43:792–797. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carlsen AL, Schetter AJ, Nielsen CT, Lood
C, Knudsen S, Voss A, Harris CC, Hellmark T, Segelmark M, Jacobsen
S, et al: Circulating microRNA expression profiles associated with
systemic lupus erythematosus. Arthritis Rheum. 65:1324–1334. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tandon M, Gallo A, Jang SI, Illei GG and
Alevizos I: Deep sequencing of short RNAs reveals novel microRNAs
in minor salivary glands of patients with Sjögren's syndrome. Oral
Dis. 18:127–131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gallo A, Tandon M, Illei G and Alevizos I:
Discovery and validation of novel microRNAs in Sjögren's syndrome
salivary glands. Clin Exp Rheumatol. 32:761–762. 2014.PubMed/NCBI
|
|
12
|
Alevizos I, Alexander S, Turner RJ and
Illei GG: MicroRNA expression profiles as biomarkers of minor
salivary gland inflammation and dysfunction in Sjögren's syndrome.
Arthritis Rheum. 63:535–544. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pauley KM, Stewart CM, Gauna AE, Dupre LC,
Kuklani R, Chan AL, Pauley BA, Reeves WH, Chan EK and Cha S:
Altered miR-146a expression in Sjögren's syndrome and its
functional role in innate immunity. Eur J Immunol. 41:2029–2039.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vitali C, Bombardieri S, Jonsson R,
Moutsopoulos HM, Alexander EL, Carsons SE, Daniels TE, Fox PC, Fox
RI, Kassan SS, et al: Classification criteria for Sjögren's
syndrome: A revised version of the European criteria proposed by
the American-European Consensus Group. Ann Rheum Dis. 61:554–558.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chisholm DM and Mason DK: Labial salivary
gland biopsy in Sjögren's disease. J Clin Pathol. 21:656–660. 1968.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baldini C, Giusti L, Ciregia F, Da Valle
Y, Giacomelli C, Donadio E, Sernissi F, Bazzichi L, Giannaccini G,
Bombardieri S and Lucacchini A: Proteomic analysis of saliva: A
unique tool to distinguish primary Sjögren's syndrome from
secondary Sjögren's syndrome and other sicca syndromes. Arthritis
Res Ther. 13:R1942011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Daniels TE, Cox D, Shiboski CH, Schiødt M,
Wu A, Lanfranchi H, Umehara H, Zhao Y, Challacombe S, Lam MY, et
al: Associations between salivary gland histopathologic diagnoses
and phenotypic features of Sjögren's syndrome among 1,726 registry
participants. Arthritis Rheum. 63:2021–2030. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gourzi VC, Kapsogeorgou EK, Kyriakidis NC
and Tzioufas AG: Study of microRNAs (miRNAs) that are predicted to
target the autoantigens Ro/SSA and La/SSB in primary Sjögren's
Syndrome. Clin Exp Immunol. 182:14–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kapsogeorgou EK, Gourzi VC, Manoussakis
MN, Moutsopoulos HM and Tzioufas AG: Cellular microRNAs (miRNAs)
and Sjögren's syndrome: Candidate regulators of autoimmune response
and autoantigen expression. J Autoimmun. 37:129–135. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen JQ, Papp G, Szodoray P and Zeher M:
The role of microRNAs in the pathogenesis of autoimmune diseases.
Autoimmun Rev. 15:1171–1180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brito-Zerón P, Theander E, Baldini C,
Seror R, Retamozo S, Quartuccio L, Bootsma H, Bowman SJ, Dorner T,
Gottenberg JE, et al: Early diagnosis of primary Sjögren's
syndrome: EULAR-SS task force clinical recommendations. Expert Rev
Clin Immunol. 12:137–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shiboski SC, Shiboski CH, Criswell L, Baer
A, Challacombe S, Lanfranchi H, Schiødt M, Umehara H, Vivino F,
Zhao Y, et al: American College of Rheumatology classification
criteria for Sjögren's syndrome: A data-driven, expert consensus
approach in the Sjögren's International Collaborative Clinical
Alliance cohort. Arthritis Care Res (Hoboken). 64:475–487. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bi C and Chng WJ: MicroRNA: Important
player in the pathobiology of multiple myeloma. BioMed Res Int.
2014:5215862014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Henao-Mejia J, Williams A, Goff LA, Staron
M, Licona-Limón P, Kaech SM, Nakayama M, Rinn JL and Flavell RA:
The microRNA miR-181 is a critical cellular metabolic rheostat
essential for NKT cell ontogenesis and lymphocyte development and
homeostasis. Immunity. 38:984–997. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peng L, Ma W, Yi F, Yang YJ, Lin W, Chen
H, Zhang X, Zhang LH, Zhang F and Du Q: MicroRNA profiling in
Chinese patients with primary Sjögren syndrome reveals elevated
miRNA-181a in peripheral blood mononuclear cells. J Rheumatol.
41:2208–2213. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alevizos I and Illei GG: MicroRNAs in
Sjögren's syndrome as a prototypic autoimmune disease. Autoimmun
Rev. 9:618–621. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li QJ, Chau J, Ebert PJ, Sylvester G, Min
H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, et al:
miR-181a is an intrinsic modulator of T cell sensitivity and
selection. Cell. 129:147–161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer: Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tung YT, Huang PW, Chou YC, Lai CW, Wang
HP, Ho HC, Yen CC, Tu CY, Tsai TC, Yeh DC, et al: Lung
tumorigenesis induced by human vascular endothelial growth factor
(hVEGF)-A165 overexpression in transgenic mice and amelioration of
tumor formation by miR-16. Oncotarget. 6:10222–10238. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cui J: MiR-16 family as potential
diagnostic biomarkers for cancer: A systematic review and
meta-analysis. Int J Clin Exp Med. 8:1703–1714. 2015.PubMed/NCBI
|
|
33
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Filková M, Aradi B, Senolt L, Ospelt C,
Vettori S, Mann H, Filer A, Raza K, Buckley CD, Snow M, et al:
Association of circulating miR-223 and miR-16 with disease activity
in patients with early rheumatoid arthritis. Ann Rheum Dis.
73:1898–1904. 2014. View Article : Google Scholar : PubMed/NCBI
|