Introduction
Tuberculosis (TB), which is caused by mycobacterium
TB, is a major cause of human mortality worldwide, with two million
deaths and ten million new cases of TB occurring annually (1). Children are more susceptible to the
infection of mycobacterium TB due to their having a relatively
weaker immune system compared with adults (2,3). The
World Health Organization (WHO) reported that almost one million
children were infected with the mycobacterium TB in 2015 (4). India, Indonesia, China, Nigeria,
Pakistan and South Africa account for 60% of newly identified cases
(5). There are more than 30,000 new
children cases of multidrug-resistant TB in 2015 worldwide
(6).
Vaccination with BacilleCalmette-Guerin (BCG) is an
effective form of prevention of TB. The BCG vaccine has 60–80%
protective effect against severe types of TB in children,
especially meningitis (7). The Xpert
Mycobacterium tuberculosis/rifampicin (MTB/RIF) assay can be
used to diagnose TB and yield reliable results. Zar et al
reported that Xpert MTB/RIF was a useful assay for the rapid and
reliable diagnosis of paediatric TB in African children, using
induced sputum and nasopharyngeal as the specimens (8). Gous et al also used the Xpert
MTB/RIF assay to diagnose TB in childhood (9). Fiebig et al used the nucleic
acid amplification tests and culture of gastric aspirates to detect
bacteriological confirmation of TB in German children. Those
authors found that the combined use of molecular assay and culture
method had an improved test accuracy rate (10).
Protein-protein interactions (PPI) play an important
role in all biological processes. The interaction networks can be
used to explore the intricate protein organizations and
cellprocesses (11,12). Safaei et al carried out a PPI
network study on cirrhosis liver disease. Authors of that study
found that the regulation of cell survival and lipid metabolism
were pivotal biological processes in cirrhosis disease (13). In ovarian cancer, 12-gene network
modules have been identified using the differential co-expression
PPI network. The gene expression data and PPI networks can be used
to develop effective biomarkers for understanding disease
mechanisms (14). Ramadan et
al combined the PPI and gene co-expression network (GCN) to
analyze breast cancer (15).
In the present study, the PPI network and GCN were
employed to analyze the latent and active period of TB in children.
Thirteen seed genes were found in the differential gene
co-expression networks (DCNs), and eight multiple differential
modules (M-DMs) were identified based on the DCNs (16). The identified M-DMs provided new
insights into the development of TB in children.
Materials and methods
Gene expression data
The Array Express Archive of Functional Genomics
Data is a functional genomics database at the European
Bioinformatics Institute. The microarray data of E-GEOD-39940 were
downloaded from the Array Express database. The data contained the
gene expression profilings of patients who were HIV-negative,
suffered from latent period of TB (n=54) and active period of TB
(n=70).
In order to eliminate the influence of non-specific
hybridization, the robust multichip average method was used to
correct background. The quantile-based algorithm was carried out to
normalize the data. The probes were discarded when they did not
match any genes. In total, 13,997 genes were obtained after the
mapping between gene IDs and probe IDs.
PPI data
Human related PPI data were obtained from the The
Search Tool for the Retrieval of Interacting database, containing
787,896 pairs and 16,730 genes. The genes that were included in
gene expressions and PPIs were selected to construct DCN. After
processing, 501,736 PPI pairs and 12,310 genes were obtained.
Construction of DCNs
The absolute value of the Pearson's correlation
coefficient of PPI pairs of the active TB samples were calculated.
The PPIs were selected if the corresponding absolute value was
>0.8. Finally, 3,820 edges (PPIs) and 1,359 nodes (genes) were
obtained to construct the DCNs.
wi,j={(logpi+logpj)1/2(2*maxɩ∈V|logpɩ)1/2,ifcor(i,j)≥δ,0,ifcor(i,j)<δ,
The one-tailed t-test was used to calculate the
P-value of differentially expressed genes in the latent and active
TB. The weight value of each interaction was calculated based on
the P-values of genes according to EdgeR (17) as follows:
Where pi and pj are the P-values of the differential
expression of gene i and gene j, respectively. V is the node set of
the co-expression network. In addition, cor(i,j) indicates the
absolute value of Pearson's correlation between gene i and j.
g(i)=∑j∈N(i)Aij′g(j)
Construction of M-DMs
The construction of M-DMs consists of three steps:
i) Seed genes prioritization, ii) module search based on each gene,
and iii) the refinement of candidate modules. i) The importance of
each gene in the networks was calculated as:
where g(i), the importance of vertex i in the
network; N(i), the adjacent set of gene i; A', the degree
normalized weighted adjacent set, which is calculated as A' =
D−1/2AD1/2, where D is the diagonal set of
A.
The g (i) = z-score, and the genes were then ranked
by the z-scores. The genes with the highest 1% z-scores were
selected as the seed genes. ii) For each seed gene v ϵ V, it was
selected as one differential module C. Then the gene u, which was
adjacent to the gene v in the network was incorporated into this
module, designated as module C'. The entropy change of the two
modules was assessed as: ΔH(C',C)=H(C')-H(C).
ΔH(C',C)>0 exhibited that the connectivity of
module C was increased by the joining of gene u. This was then
joined to the adjacent gene u, which potentially increased the ΔH
in module C until the ΔH was no longer able to increase. iii) The
candidate module was removed if it contained <5 nodes. If the
overlapping degree between two modules was ≥0.5, the two modules
were merged into one module.
The statistical significant test of
candidate M-DMs
In total, 3,820 edges were selected randomly from
501,736 edges and formed the random network. The module searching
was carried out following the above mentioned steps. The random
networks were constructed 100 times, and 2,318 modules were
constructed. The empirical P-value of the candidate module was
calculated as the probability of the module, which has the observed
score or smaller score by chance. The Benjamini-Hochberg algorithm
was used to correct the P-value (16). The modules that had the P-value of
≤0.05 were selected as the differential modules.
Results
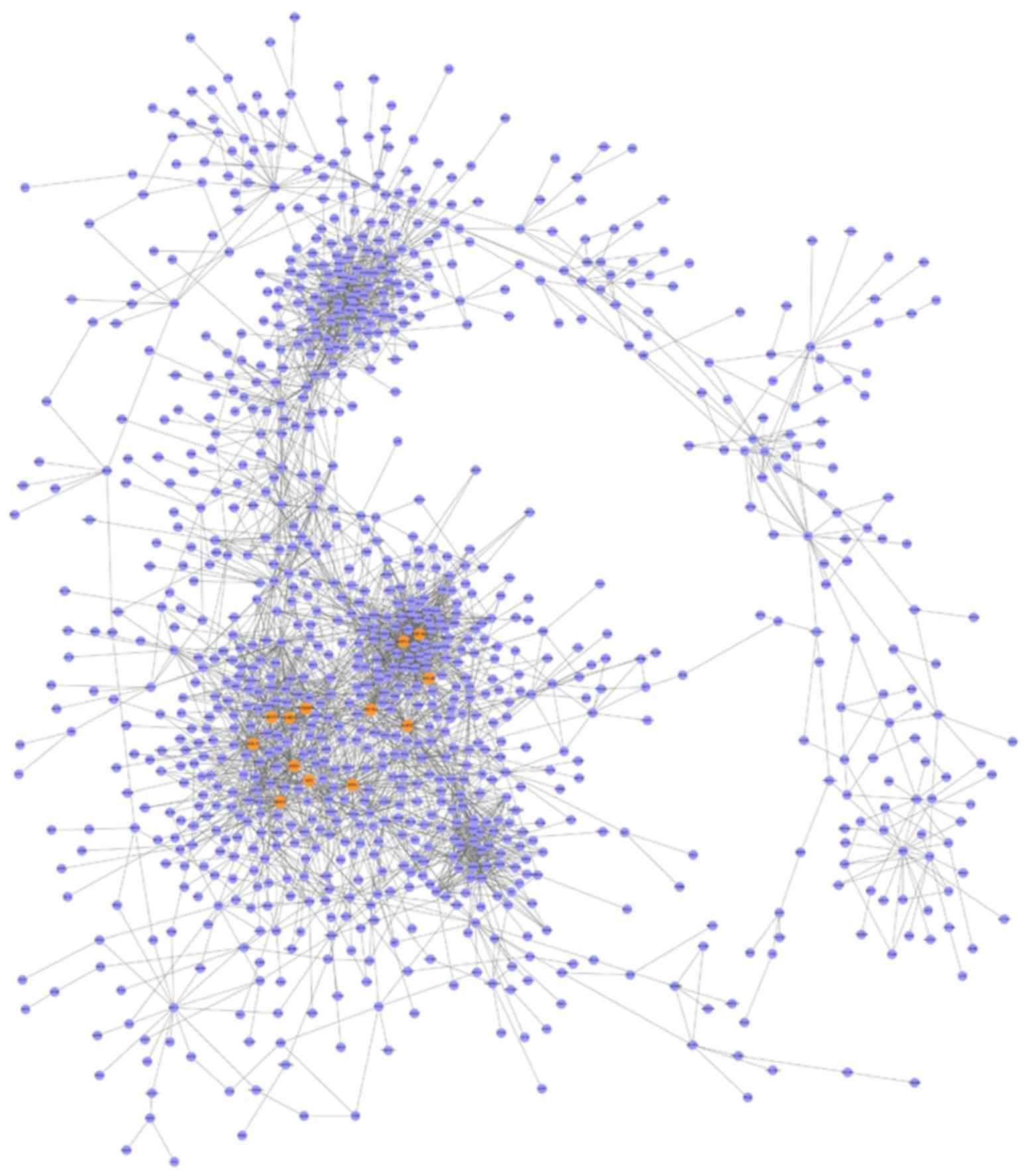
Construction of DCNs
The human-related PPI and gene expression data were
downloaded to construct the DCNs. Based on the criteria of absolute
value of Pearson's correlation coefficient >0.8, 3,820 edges
(PPIs) and 1,359 nodes (genes) were obtained (Fig. 1). The DCNs consisted of these edges
and nodes.
Identification of candidate M-DMs
The genes which had the highest 1% z-scores in DCNs
were selected as the seed genes. On aggregate, 13 seed genes were
obtained (Table I). The z-scores
ranged from 284.5787 to 473.111. The seed genes contained
SS18L2, NOL11, ADSL, ILF2,
DDX18, DDX1, CLNS1A, ENOPH1,
MTERF3, MRPL32, NUP37, RPL35 and
EEF1B2. After the modules were investigated and refined, 11
modules were obtained.
 | Table I.Genes with highest 1% z-scores in
DCNs were selected as the seed genesa. |
Table I.
Genes with highest 1% z-scores in
DCNs were selected as the seed genesa.
| Gene name | z-score |
|---|
| SS18L2 | 473.111 |
| NOL11 | 457.7947 |
| ADSL | 438.8713 |
| ILF2 | 365.7652 |
| DDX18 | 345.6201 |
| DDX1 | 330.0789 |
| CLNS1A | 306.1616 |
| ENOPH1 | 300.3362 |
| MTERF3 | 300.3337 |
| MRPL32 | 294.2793 |
| NUP37 | 287.2869 |
| RPL35 | 285.7214 |
| EEF1B2 | 284.5787 |
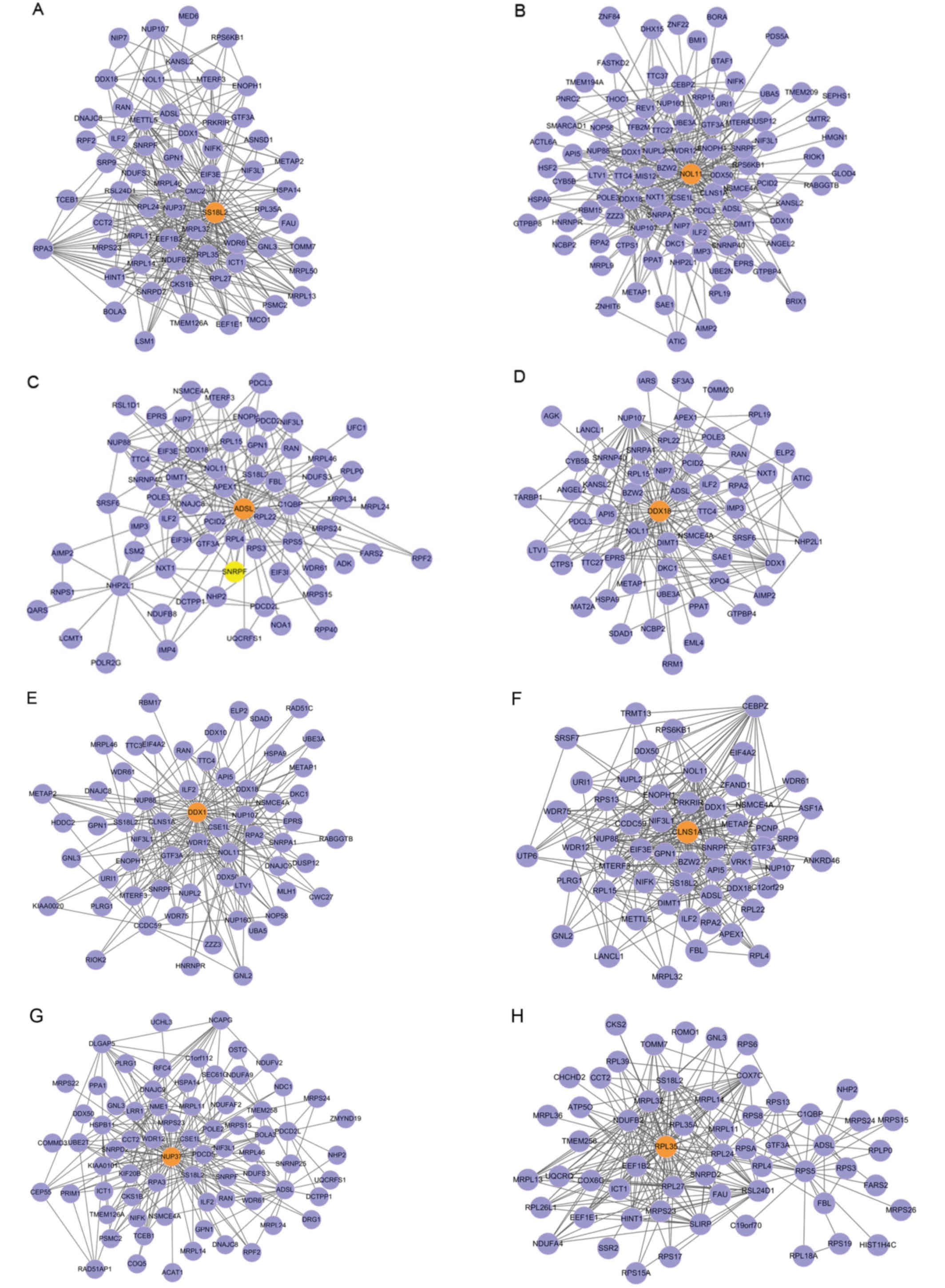
Identification of candidate M-DMs
The P-value of the 11 candidate M-DMs were
calculated and corrected using the Benjamini-Hochberg algorithm.
The modules with P≤0.05 were regarded as the objective modules.
Finally, 8 modules were selected as significant differential
modules (Table II and Fig. 2). The module entropy ranged from
0.687 to 0.851.
 | Table II.The P-value of 11 candidate M-DMs was
calculated using the Benjamini-Hochberg algorithma. |
Table II.
The P-value of 11 candidate M-DMs was
calculated using the Benjamini-Hochberg algorithma.
| Modules | P-values | Entropy |
|---|
| 1 | 0 | 0.847 |
| 2 | 0 | 0.687 |
| 3 | 0 | 0.739 |
| 5 | 0 | 0.721 |
| 6 | 0 | 0.775 |
| 7 | 0 | 0.851 |
| 11 | 0 | 0.798 |
| 12 | 0 | 0.716 |
Discussion
From a systematic biology point of view, diseases
are caused by the fluctuations to the gene expression network. Such
fluctuations change significantly during the disease progressions
(18). Schwarz et al combined
the PPI works and expression genes to examine the biological
processes and genes related with schizophrenia (19). The PPI and gene-gene functional
interaction networks were constructed to identify potential
biomarkers of pediatric adreno cortical carcinoma (20).
In the present study, we introduced a new method
based on M-DMs to identify new biomarkers to better understand the
molecular mechanisms and search for potential biomarkers of TB. We
identified 8 modules associated with TB.
Humans possess two SS18 homologous genes,
SS18L1 and SS18L2. The SS18L2 gene has three
exons and is mapped to chromosome 3, with band p21 (21). de Bruijn reported that SS18
encoded nuclear proteins and functioned as a transcriptional
co-activator. The fusion of either SSX genes or SS18
is a hallmark of human synovial sarcoma (22).
Nuclear protein 11 (NOL11) is a
metazoan-specific protein and is involved in ribosome biogenesis.
NOL11 also plays an important role in the maturation of 18S
RNA and pathogenesis of North American Indian childhood cirrhosis
(23).
Human adenylosuccinatelyase (ADSL) is a
bifunctional enzyme acting in two pathways of purine nucleotide
metabolism including de novo purine synthesis and purine nucleotide
recycling (24). The human liver
ADSL gene was cloned and mapped to chromosome 22 (25,26).
The antisense oligonucleotides (ASOs) combine with
RNA to form heteroduplexes, which can be specifically recognized by
the interleukin enhancer-binding factor 2 and 3 complex
(ILF2/3). The combination of ASO and ILF2/3 modulates
gene expression by alternative splicing (27). ILF2 mRNA accumulates in the
pachytene spermatocytes. ILF2 is also expressed in the adult
ovary and different embryo tissues (28).
DEAD-Box Helicase 1 (DDX1) was found in a
high-molecular complex containing a series of Drosha-associated
polypeptides (29). Low DDX1
levels are associated with poor clinical outcome in serious ovarian
cancer by the cancer genome atlas and DDX1 plays an
important role in the modulation of miRNA maturation (30).
Nevertheless, there are some drawbacks to the
present study. The study included 124 samples, which is not a
sufficient amount of samples to support the conclusions and future
studies are to be conducted to confirm the findings. In addition,
the results were not verified by clinical experiments.
In conclusion, in the present study, we identified 8
significant different modules using the new bioinformatic methods.
We believe that the present study will benefit the understanding of
TB in children and provide new therapeutic methods to combat the
disease.
Glossary
Abbreviations
Abbreviations:
|
WHO
|
World Health Organization
|
|
BCG
|
Bacille Calmette-Guerin
|
|
NAAT
|
nucleic acid amplification test
|
|
PPI
|
protein-protein interaction
|
|
GCN
|
gene co-expression network
|
|
DCN
|
differential gene co-expression
network
|
|
M-DM
|
multiple differential module
|
|
RMA
|
robust multichip average
|
|
GGI
|
gene-gene functional interaction
|
|
NAIC
|
North American Indian childhood
cirrhosis
|
|
ASO
|
antisense oligonucleotide
|
|
TB
|
tuberculosis
|
References
|
1
|
Rawat J, Sindhwani G and Juyal R:
Clinico-radiological profile of new smear positive pulmonary
tuberculosis cases among young adult and elderly people in a
tertiary care hospital at Deheradun (Uttarakhand). Indian J Tuberc.
55:84–90. 2008.PubMed/NCBI
|
|
2
|
Starke JR: Resurgence of tuberculosis in
children. Pediatr Pulmonol Suppl. 11:16–17. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith S, Jacobs RF and Wilson CB:
Immunobiology of childhood tuberculosis: A window on the ontogeny
of cellular immunity. J Pediatr. 131:16–26. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yoo AS, Staahl BT, Chen L and Crabtree GR:
MicroRNA-mediated switching of chromatin-remodelling complexes in
neural development. Nature. 460:642–646. 2009.PubMed/NCBI
|
|
5
|
World Health Organisation (WHO), . Global
health observatory data. 2017, http://www.who.int/gho/hiv/en/
|
|
6
|
Hamilton CD, Swaminathan S, Christopher
DJ, Ellner J, Gupta A, Sterling TR, Rolla V, Srinivasan S, Karyana
M, Siddiqui S, et al: RePORT International: Advancing tuberculosis
biomarker research through global collaboration. Clin Infect Dis.
61:155–159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trunz BB, Fine P and Dye C: Effect of BCG
vaccination on childhood tuberculous meningitis and miliary
tuberculosis worldwide: A meta-analysis and assessment of
cost-effectiveness. Lancet. 367:1173–1180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zar HJ, Workman L, Isaacs W, Dheda K,
Zemanay W and Nicol MP: Rapid diagnosis of pulmonary tuberculosis
in African children in a primary care setting by use of Xpert
MTB/RIF on respiratory specimens: A prospective study. Lancet Glob
Health. 1:97–104. 2013. View Article : Google Scholar
|
|
9
|
Gous N, Scott LE, Khan S, Reubenson G,
Coovadia A and Stevens W: Diagnosing childhood pulmonary
tuberculosis using a single sputum specimen on Xpert MTB/RIF at
point of care. S Afr Med J. 105:1044–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fiebig L, Hauer B, Brodhun B, Balabanova Y
and Haas W: Bacteriological confirmation of pulmonary tuberculosis
in children with gastric aspirates in Germany, 2002–2010. Int J
Tuberc Lung Dis. 18:925–930. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stelzl U, Worm U, Lalowski M, Haenig C,
Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A,
Koeppen S, et al: A human protein-protein interaction network: A
resource for annotating the proteome. Cell. 122:957–968. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ferrari R, Forabosco P, Vandrovcova J,
Botía JA, Guelfi S, Warren JD, Momeni P, Weale ME, Ryten M and
Hardy J: UK Brain Expression Consortium (UKBEC): Frontotemporal
dementia: Insights into the biological underpinnings of disease
through gene co-expression network analysis. Mol Neurodegener.
11:212016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Safaei A, Rezaei Tavirani M, Arefi Oskouei
A, Zamanian Azodi M, Mohebbi SR and Nikzamir AR: Protein-protein
interaction network analysis of cirrhosis liver disease.
Gastroenterol Hepatol Bed Bench. 9:114–123. 2016.PubMed/NCBI
|
|
14
|
Jin N, Wu H, Miao Z, Huang Y, Hu Y, Bi X,
Wu D, Qian K, Wang L, Wang C, et al: Network-based
survival-associated module biomarker and its crosstalk with cell
death genes in ovarian cancer. Sci Rep. 5:115662015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramadan E, Alinsaif S and Hassan MR:
Network topology measures for identifying disease-gene association
in breast cancer. BMC Bioinformatics. 17:2742016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Feser WJ, Fingerlin TE, Strand MJ and
Glueck DH: Calculating Average Power for the Benjamini-Hochberg
Procedure. J Stat Theory Appl. 8:325–352. 2009.PubMed/NCBI
|
|
17
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma X, Gao L, Karamanlidis G, Gao P, Lee
CF, Garcia-Menendez L, Tian R and Tan K: Revealing pathway dynamics
in heart diseases by analyzing multiple differential networks. PLOS
Comput Biol. 11:e10043322015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwarz E, Izmailov R, Liò P and
Meyer-Lindenberg A: Protein interaction networks link schizophrenia
risk loci to synaptic function. Schizophr Bull. 42:1334–1342. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kulshrestha A, Suman S and Ranjan R:
Network analysis reveals potential markers for pediatric
adrenocortical carcinoma. Onco Targets Ther. 9:4569–4581. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Bruijn DR, Kater-Baats E, Eleveld M,
Merkx G and Geurts Van Kessel A: Mapping and characterization of
the mouse and human SS18 genes, two human SS18-like genes and a
mouse Ss18 pseudogene. Cytogenet Cell Genet. 92:310–319. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Bruijn DR, Allander SV, van Dijk AH,
Willemse MP, Thijssen J, van Groningen JJ, Meltzer PS and van
Kessel AG: The synovial-sarcoma-associated SS18-SSX2 fusion protein
induces epigenetic gene (de)regulation. Cancer Res. 66:9474–9482.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Freed EF, Prieto JL, McCann KL, McStay B
and Baserga SJ: NOL11, implicated in the pathogenesis of North
American Indian childhood cirrhosis, is required for pre-rRNA
transcription and processing. PLoS Genet. 8:e10028922012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kmoch S, Hartmannová H, Stibůrková B,
Krijt J, Zikánová M and Sebesta I: Human adenylosuccinate lyase
(ADSL), cloning and characterization of full-length cDNA and its
isoform, gene structure and molecular basis for ADSL deficiency in
six patients. Hum Mol Genet. 9:1501–1513. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stone RL, Aimi J, Barshop BA, Jaeken J,
van den Berghe G, Zalkin H and Dixon JE: A mutation in
adenylosuccinate lyase associated with mental retardation and
autistic features. Nat Genet. 1:59–63. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fon EA, Demczuk S, Delattre O, Thomas G
and Rouleau GA: Mapping of the human adenylosuccinate lyase (ADSL)
gene to chromosome 22q13.1->q13.2. Cytogenet Cell Genet.
64:201–203. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rigo F, Hua Y, Chun SJ, Prakash TP,
Krainer AR and Bennett CF: Synthetic oligonucleotides recruit
ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol.
8:555–561. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
López-Fernández LA, Párraga M and del Mazo
J: Ilf2 is regulated during meiosis and associated to
transcriptionally active chromatin. Mech Dev. 111:153–157. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gregory RI, Yan KP, Amuthan G, Chendrimada
T, Doratotaj B, Cooch N and Shiekhattar R: The microprocessor
complex mediates the genesis of microRNAs. Nature. 432:235–240.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han C, Liu Y, Wan G, Choi HJ, Zhao L, Ivan
C, He X, Sood AK, Zhang X and Lu X: The RNA-binding protein DDX1
promotes primary microRNA maturation and inhibits ovarian tumor
progression. Cell Reports. 8:1447–1460. 2014. View Article : Google Scholar : PubMed/NCBI
|