Introduction
Osteosarcoma is one of the most common malignant
tumors of the bone, and is typically located in the distal femur
and proximal tibia. The malignancy of osteosarcoma is high, with an
extremely poor patient prognosis. Osteosarcoma often results in
pulmonary metastasis within a number of months, with a 3–5 year
survival rate of 5–20% (1). Surgery
remains the primary treatment option for patients with
osteosarcoma; however, the incidence of relapse for these patients
is high (1). For patients with
advanced stage osteosarcoma who have missed the opportunity for
surgical therapy, comprehensive treatments, including radiotherapy,
chemotherapy and biological agents, have become the main option for
therapy (2,3). Therefore, it is important to improve
understating of the mechanisms underlying the occurrence,
progression, invasion and metastasis of osteosarcoma to provide
better treatments for this disease.
Phosphatase and tensin homolog (PTEN) is a tumor
suppressor gene that is located on chromosome 10q23.3. The
transcription product of the gene is a 515 kb long mRNA (4). PTEN belongs to the protein tyrosine
phosphatase gene family and is the first tumor suppressor with
bispecific phosphatase activity identified thus far. PTEN is
associated with the occurrence of several types of tumor, and
serves an important role in tumor cell growth, apoptosis, adhesion,
migration and invasion (5). Previous
studies have revealed that the downregulation or deletion of PTEN
activates the phosphoinositide 3-kinase/RAC-α
serine/threonine-protein kinase (PI3K/AKT) signaling pathway,
resulting in the occurrence, development, invasion and metastasis
of cancer (6,7).
MicroRNAs (miRNAs/miRs) are small RNA molecules that
are 17–25 nucleotides long and serve an important role in the
regulation of gene expression (8,9).
Although the number of miRNAs in the human genome is less than that
of protein-coding genes, they are thought to regulate the
expression of more than half of human mRNAs (10). However, few studies have reported the
role of miRNAs in osteosarcoma and their mechanism of action.
miRNA-21 was revealed to be abnormally expressed in a variety of
tumor cells and is one of the few miRNAs that is expressed in all
solid malignant tumors (10,11). A number of genes, including PTEN and
tissue inhibitor of metalloproteinase-3, have been identified as
targets of miRNA-21 (11,12).
To better understand the role and mechanism of
action of miRNA-21 in the invasion and metastasis of osteosarcoma,
in addition to cell apoptosis and proliferation, the expression of
miRNA-21 was investigated. This revealed that miRNA-21 expression
was upregulated in osteosarcoma cells. In addition, the impact of
miRNA-21 on the expression of PTEN was investigated. The findings
from the present study provide insights into the role of miRNA-21
in osteosarcoma, and reveal potential therapeutic targets for the
prevention and treatment of this disease.
Materials and methods
Cell lines and culture
The human osteosarcoma cell line, MG-63, and
osteoblast cell line, hFOB1.19, were purchased from the cell bank
of the Typical Culture Preservation Committee of the Chinese
Academy of Sciences (Beijing, China). Cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C with 5% CO2. Cell
proliferation and viability were assayed using an MTT cell
proliferation assay kit according to the manufacturer's protocol
(cat. no. C0009; Beyotime Institute of Biotechnology, Haimen,
China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from the cells using TRIzol
reagent according to the manufacturer's protocol (Invitrogen;
Thermo Fisher Scientific, Inc.), and measured for quantity and
purity using the NanoDrop™ 2000 (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Reverse transcription of the RNA was performed
using the High-Capacity RNA-to-cDNA™ kit (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. qPCR analysis was
performed using the TaqMan Gene Expression Master mix (Thermo
Fisher Scientific, Inc.) in a total volume of 20 µl containing 5 µl
cDNA products, 10 µl TaqMan® Gene Expression Master Mix
(2X), 100 µM primers and 4 µl nuclease-free water. The primer
sequences for miRNA-21 were: Forward, 5′-TGTACCACCTTGTCGGATAG-3′
and reverse, 5′-CTGCTGTTGCCATGAGAT-3′. GAPDH was used as internal
reference gene with the following primers: Forward,
5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, primer
5′-GTGCAGGGTCCGAGGGT-3′.
The thermocycling conditions were as follows: 50°C
for 2 min, 95°C for 10 min, 37 cycles of 95°C for 15 sec and 1 min
at 60°C. Samples were run in triplicate and the mean value was
calculated for each case. The data were managed using RQ Manager
software (version 1.2.1; Applied Biosystems; Thermo Fisher
Scientific, Inc.). Relative mRNA expression was calculated using
2−ΔΔCq method as previously described (13).
Transfections
The transfection of MG-63 cells was performed using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Briefly, MG-63 cells were
seeded at a concentration of 1×105 cells/well in 6-well
culture plates. Following 24 h of culturing (70% confluence), the
cells were transfected with 50 nM hsa-miR-21 mimic (cat. no.
4464066) or inhibitor (cat. no. 4464084; both Thermo Fisher
Scientific, Inc.). The transfection efficiency was checked by qPCR
and visualization of positive controls by confocal microscopy
(Olympus FV-1000 Inverted Microscope; Olympus Corporation, Tokyo,
Japan).
Cell invasion assays
For the assessment of invasion, 5×105
transfected cells in serum-free DMEM were placed into the upper
chamber of a Transwell insert coated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). For the two experiments, media containing
10% FBS were added to the lower chamber. Following 24 h of
incubation at 37°C, the cells remaining on the upper membrane were
removed with cotton wool, whereas the cells that had migrated or
invaded through the membrane were stained with 2% crystal violet in
25% methanol/PBS at room temperature for 15 min, then imaged and
counted using an EVOS XL Core inverted microscope (Thermo Fisher
Scientific, Inc.). The experiments were independently repeated
three times.
Western blot analysis
MG-63 cells were transfected with miRNA-21 mimic or
inhibitor for 48 h. Subsequently, proteins were extracted using the
ReadyPrep™ Protein Extraction kit (Total Protein) (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and quantified using the BCA
Protein Quantification kit (Vazyme, Piscataway, NJ, USA), according
to the manufacturer's protocol. A total of 10 µg per lane of
protein was separated by 12% SDS-PAGE and transferred to PVDF
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
blocked with 5% defatted milk (OriGene Technologies, Inc., Beijing,
China) at room temperature for 1 h before reacting to the secondary
antibody. Human AKT/phosphorylated (p-)AKT, PTEN and PI3K protein
expression levels were quantified using rabbit polyclonal
antibodies specific for each protein (1:1,000; Signalway Antibody
and Santa Cruz Biotechnology). The expression levels of these three
proteins were standardized to GAPDH using a mouse polyclonal
antibody directed against GAPDH (ABS16; 1:1,000; EMD Millipore) at
room temperature for 2 h. The primary antibodies were detected
using goat anti-rabbit horseradish peroxidase-conjugated secondary
antibodies (cat. no. A0208; 1:200; OriGene Technologies, Inc.) at
room temperature for 1 h. Immunoreactive bands were visualized
using Western Lighting Chemiluminescence Reagent Plus (PerkinElmer,
Inc., Waltham, MA, USA), according to the manufacturer's protocol
and then quantified by densitometry (Image J Pro Plus v1.51;
National Institutes of Health, Bethesda, MD, USA) using a
ChemiGenius Gel Bio Imaging system (Syngene, Frederick, MD,
USA).
Statistical analyses
Statistical analyses were performed using GraphPad
Prism software (version 6.0; GraphPad Software, Inc., La Jolla, CA,
USA). All experiments were repeated at least three times and
performed in triplicate. Data are expressed as the mean ± standard
deviation, and means were compared using the Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
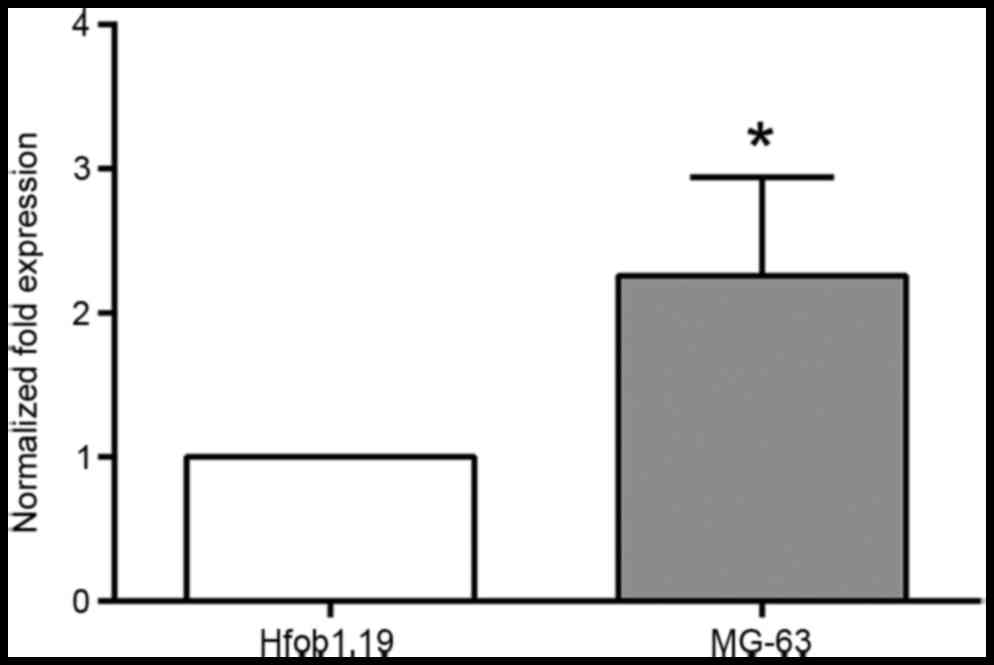
miRNA-21 is upregulated in MG-63
osteosarcoma cells
qPCR assays revealed that miRNA-21 expression was
significantly upregulated in MG-63 cells compared with hFOB1.19
cells (P<0.01; Fig. 1).
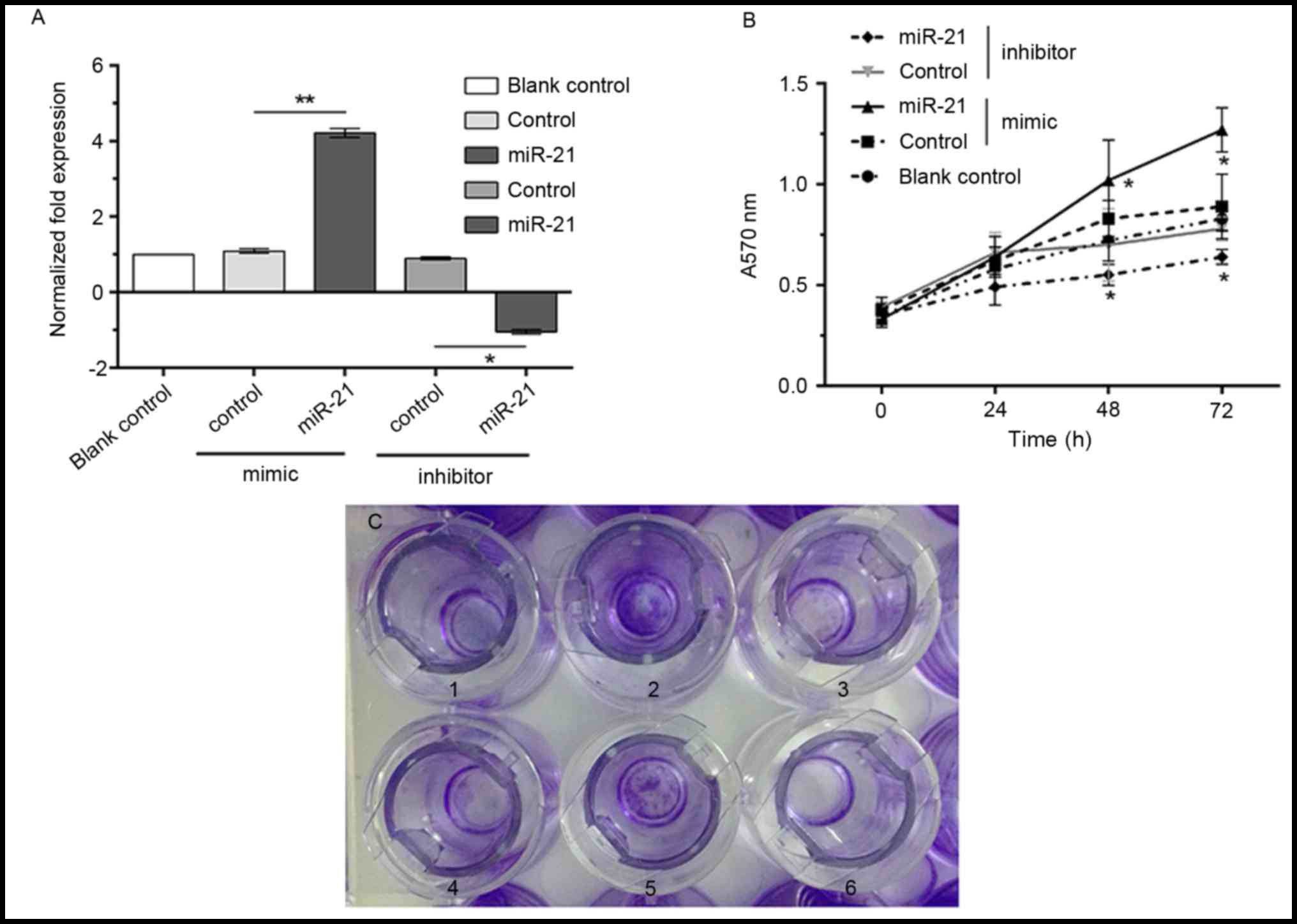
miRNA-21 increases the proliferation
and invasion of MG-63 osteosarcoma cells
To investigate the effect of miRNA-21 on the
proliferation and invasion of MG-63 cells, a miRNA-21 mimic or
inhibitor were transfected into the MG-63 cells to upregulate and
downregulate miRNA-21 expression, respectively. qPCR analyses
demonstrated that the mRNA expression level of miRNA-21 was
significantly increased or decreased following the transfection
with the mimic or inhibitor, respectively, compared with the
control group (P<0.05, Fig.
2A).
MTT assays demonstrated that at 48 and 72 h
following miRNA-21 mimic or inhibitor transfection, the
proliferation of MG-63 cells was significantly increased in cells
transfected with the mimic and decreased in cells transfected with
the inhibitor compared with their respective control groups
(P<0.05, Fig. 2B).
The Transwell assays (Fig. 2C) revealed that following
transfection with the miRNA-21 mimic, MG-63 cell invasion was
increased significantly compared with the control group (85.2±10.6
vs. 47.6±2.6), while transfection with the miRNA-21 inhibitor
resulted in a significantly reduced invasion ability compared with
the control (32.8±12.7 vs. 58.4±4.9) (both P<0.05; data not
shown).
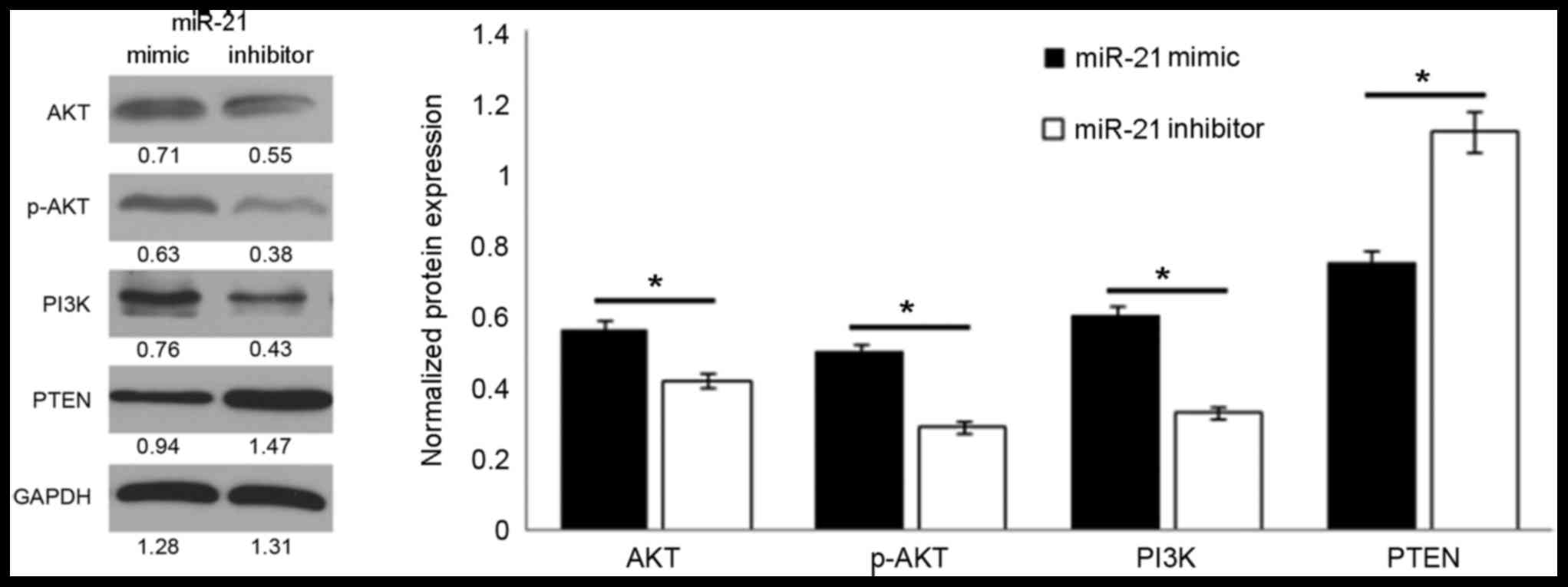
miRNA-21 increases PTEN/PI3K/AKT
signaling
The above results indicated that the expression
level of miRNA-21 impacts the proliferation and invasion of the
osteosarcoma cells. Considering this, the influence of miRNA-21 on
the expression of proteins in the PTEN/PI3K/AKT signaling pathway
was analyzed. The activation state of the PI3K/AKT signaling
pathway was determined by measuring the protein expression level of
p-AKT. The results demonstrated that the protein expression of
p-AKT was positively associated with the miRNA-21 level.
Upregulation of miRNA-21 expression significantly increased the
p-AKT level compared with treatment with the miRNA-21 inhibitor
(P<0.01; Fig. 3). In regards to
other important proteins in the PI3K/AKT signaling pathway, the
miRNA-21 mimic significantly inhibited the expression of PTEN and
increased the expression of PI3K compared with cells treated the
miRNA-21 mimic (P<0.01; Fig.
3).
Discussion
Non-coding RNA has been demonstrated to serve
important roles in numerous biological processes, and the
development and treatment of diseases (14). As a group of non-coding RNAs, miRNAs
exist in a wide variety of organisms and have been demonstrated to
have important biological functions with complicated mechanisms of
action, each having single or multiple target genes to regulate
(14–16). miRNA-21 is a ubiquitous miRNA, which
is expressed at abnormal levels in numerous types of cancer,
including colorectal cancer, bile duct carcinoma, lymphoma,
multiple myeloma and glioma, suggesting that it is an oncogene
(15–17).
At present, the pathogenesis of osteosarcoma remains
unclear. As with other solid tumors, the activation of oncogenes
and the abnormal expression of tumor suppressor genes are
considered the main reasons for the occurrence of osteosarcoma
(18). Previous studies have
revealed that PTEN inhibits the proliferation, invasion and
metastasis of tumor cells (19–21). The
mutation and deletion of the PTEN gene have been confirmed to occur
in osteosarcoma in dogs, where, in 60% of osteosarcoma cells, PTEN
gene expression was suppressed or the gene was deleted (22). It has previously been demonstrated
that in human osteosarcoma AKT, which is known to promote cell
proliferation, invasion and angiogenesis, is highly expressed and
activated (23). One of the causes
of abnormal expression and activation of AKT is the abnormal
expression of its upstream regulatory gene, PTEN (24).
The present study demonstrated that, compared with
the normal osteoblast cell line hFOB1.19, PTEN in MG-63
osteosarcoma cells was significantly downregulated, while the
levels of the downstream proteins AKT and p-AKT were significantly
elevated. This indicates that in human osteosarcoma cells PTEN is
abnormally expressed, which is consistent with the results of the
aforementioned studies.
Using a bioinformatics method, it has been predicted
that PTEN is the target gene of miRNA-21 and a number of studies
have demonstrated that miRNA-21 can regulate the mRNA expression of
PTEN (12,24). However, little is known about the
expression of miRNA-21 in osteosarcoma. Analyses in the present
study revealed that the expression of miRNA-21 in osteosarcoma
cells was upregulated in comparison with normal cells. The impact
of miRNA-21 on cell behavior and the PTEN/PI3K/AKT pathway in MG-63
osteosarcoma cells was then investigated using a miRNA-21 mimic and
inhibitor. The results of the present study demonstrated that the
downregulation of miRNA-21 in MG-63 cells resulted in decreased
proliferation, and invasion. By contrast, the upregulation of
miRNA-21 in MG-63 cells using a mimic led to increased cell
proliferation and invasion ability, suggesting that miRNA-21
promotes the growth and development of osteosarcoma.
Furthermore, analysis of the expression of proteins
in the PTEN/PI3K/AKT signaling pathway demonstrated that miRNA-21
inhibited the expression of PTEN to regulate the activity of the
PTEN/PI3K/AKT signaling pathway. The level of miRNA-21 was
positively associated with the level of AKT/p-AKT, indicating that
miRNA-21 regulates the proliferation and invasion of MG-63 cells.
Similarly, Meng et al (25,26)
revealed that miRNA-21 regulated PTEN expression in bile duct
cancer and hepatocellular cancer, and promoted the invasion of
tumor cells. Therefore, the results of the present study indicate
that in osteosarcoma cells the abnormal expression of miRNA-21 may
activate the PTEN/PI3K/AKT signaling pathway by regulating the
expression of PTEN. This suggests that miRNA-21 functions as an
oncogene. The results of the present study may provide potential
therapeutic targets for the prevention and treatment of this
disease.
References
|
1
|
Berner K, Johannesen TB, Berner A,
Haugland HK, Bjerkehagen B, Bøhler PJ and Bruland ØS: Time-trends
on incidence and survival in a nationwide and unselected cohort of
patients with skeletal osteosarcoma. Acta Oncol. 54:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ziyan W and Yang L: MicroRNA-21 regulates
the sensitivity to cisplatin in a human osteosarcoma cell line. Ir
J Med Sci. 185:85–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venkatramani R, Murray J, Helman L, Meyer
W, Hicks MJ, Krance R, Lau C, Jo E and Chintagumpala M: Risk-Based
therapy for localized osteosarcoma. Pediatr Blood Cancer.
63:412–417. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakanishi A, Kitagishi Y, Ogura Y and
Matsuda S: The tumor suppressor PTEN interacts with p53 in
hereditary cancer (Review). Int J Oncol. 44:1813–1819. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rizvi MM, Alam MS, Mehdi SJ, Ali A and
Batra S: Allelic loss of 10q23.3, the PTEN gene locus in cervical
carcinoma from Northern Indian population. Pathol Oncol Res.
18:309–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu YR, Qi HJ, Deng DF, Luo YY and Yang SL:
MicroRNA-21 promotes cell proliferation, migration, and resistance
to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal
cancer. Tumour Biol. 37:12061–12070. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xue R, Lei S, Xia ZY, Wu Y, Meng Q, Zhan
L, Su W, Liu H, Xu J, Liu Z, et al: Selective inhibition of PTEN
preserves ischaemic post-conditioning cardioprotection in
STZ-induced Type 1 diabetic rats: Role of the PI3K/Akt and
JAK2/STAT3 pathways. Clin Sci (Lond). 130:377–392. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu L, Liu J and Cheng G: Role of miRNAs
in schistosomes and schistosomiasis. Front Cell Infect Microbiol.
4:1652014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu X, Fan Q, Xu L, Li L, Yue Y, Xu Y, Su
Y, Zhang D and Wang L: Ursolic acid attenuates diabetic mesangial
cell injury through the up-regulation of autophagy via
miRNA-21/PTEN/Akt/mTOR suppression. PLoS One. 10:e01174002015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu X, Luo F, Ling M, Lu L, Shi L, Lu X,
Xu H, Chen C, Yang Q, Xue J, et al: MicroRNA-21 activation of ERK
signaling via PTEN is involved in arsenite-induced autophagy in
human hepatic L-02 cells. Toxicol Lett. 252:1–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng Y, Fan Y, Huiqing C, Zicai L and Quan
D: The emerging landscape of long non-coding RNAs. Yi Chuan.
36:456–468. 2014.(In Chinese). PubMed/NCBI
|
|
15
|
Wagner A, Mayr C, Bach D, Illig R,
Plaetzer K, Berr F, Pichler M, Neureiter D and Kiesslich T:
MicroRNAs associated with the efficacy of photodynamic therapy in
biliary tract cancer cell lines. Int J Mol Sci. 15:20134–20157.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Chen L, Xu Y, Li R and Du X:
microRNA-195 promotes apoptosis and suppresses tumorigenicity of
human colorectal cancer cells. Biochem Biophys Res Commun.
400:236–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34(Database Issue):
D140–D44. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Anderson ME: Update on survival in
osteosarcoma. Orthop Clin North Am. 47:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gildea JJ, Herlevsen M, Harding MA,
Gulding KM, Moskaluk CA, Frierson HF and Theodorescu D: PTEN can
inhibit in vitro organotypic and in vivo orthotopic invasion of
human bladder cancer cells even in the absence of its lipid
phosphatase activity. Oncogene. 23:6788–6797. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chu EC and Tarnawski AS: PTEN regulatory
functions in tumor suppression and cell biology. Med Sci Monit.
10:RA235–RA241. 2004.PubMed/NCBI
|
|
21
|
Maehama T, Okahara F and Kanaho Y: The
tumour suppressor PTEN: Involvement of a tumour suppressor
candidate protein in PTEN turnover. Biochem Soc Trans. 32:343–347.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Levine RA, Forest T and Smith C: Tumor
suppressor PTEN is mutated in canine osteosarcoma cell lines and
tumors. Vet Pathol. 39:372–378. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu ZH, Tao ZH, Zhang J, Li T, Ni C, Xie J,
Zhang JF and Hu XC: MiRNA-21 induces epithelial to mesenchymal
transition and gemcitabine resistance via the PTEN/AKT pathway in
breast cancer. Tumour Biol. 37:7245–7254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
25
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Schmittgen TD and Patel T: Involvement of
human micro-RNA in growth and response to chemotherapy in human
cholangiocarcinoma cell lines. Gastroenterology. 130:2113–2129.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|