Introduction
Fibroblast growth factors (FGFs) include 23 members,
which serve important roles in metabolism regulation and
development (1). FGFs have been
demonstrated to be associated with the processes of embryogenesis,
gastrulation, somitogenesis, body plan formation, organogenesis and
skin wound healing (2–7). FGF21 is a member of the FGF family that
belongs to the FGF19 subfamily, and activation of the FGF receptor
by FGF21 requires the co-receptor β-klotho (8,9).
FGF21 has been reported to preferentially express in the
liver (10). However, previous
studies have identified that FGF21 is also highly expressed in
mouse skin tissue as well as an inducible feature of FGF21
by starvation, drug administration and wounding, and its diverse
function in glucose homeostasis, liver and heart protection from
injury or skin wound healing (11–15).
Recently, FGF21 has been shown to improve hepatic insulin
sensitivity by inhibiting the mammalian target of the rapamycin
complex 1 (16).
Fasting-mediated induction of FGF21 was
regulated by the nuclear receptor peroxisome proliferator-activated
receptor α (PPARα) in the liver (17). Further studies identified that
nuclear receptor retinoic acid receptor-related receptor α (RORα)
regulates FGF21 in hepatocytes (18), whereas retinoic acid receptor β
(RARβ) regulates hepatic induction of FGF21 to promote fatty
acid oxidation (19), indicating the
complexity of transcriptional regulation of FGF21. The
canonical Wnt signaling pathway is also defined as the
Wnt/β-catenin or the β-catenin/T-cell factor (TCF) pathway
(20) that regulates diverse aspects
of biological processes (21–23). The
hallmark of the Wnt/β-catenin pathway is the stabilization of
cytosolic β-catenin. Under unstimulated conditions, β-catenin is
constantly phosphorylated by a destruction complex consisting of
glycogen synthase kinase-3β (GSK3β) and other proteins (24); phosphorylated β-catenin is
ubiquitinated by this complex and targeted for degradation by the
proteasome (24,25). Furthermore, activation of the Wnt
cascade inhibits GSK3β activity, allowing β-catenin accumulation
and subsequent relocation to the nucleus, where it associates with
TCF/lymphoid enhancer binding factor (TCF/LEF), leading to the
transcription of Wnt signaling genes that are associated with cell
survival, proliferation and differentiation (24). Genome wide array of TCF/LEF binding
sequences analysis identified that transcription regulation complex
including TCF/LEF binds to the putative cis-elements
(T/AC/GAAAG) appeared
in the promoter of downstream genes (26). However, connections between TCF/LEF
and FGF21 have not yet been reported.
In the present study, the appearance of the putative
TCF/LEF binding motifs in the promoter of FGF21 was observed
via promoter sequence analysis. Furthermore, chromatin
immunoprecipitation (ChIP) and yeast-one hybrid assays were
performed to test the possibility of transcription factor 4 (TCF4)
binding to the promoter of FGF21. In addition, β-catenin
interaction with TCF4 and transcriptional regulation of the
β-catenin/TCF4 complex to FGF21 was also examined. Finally,
the experiments transiently overexpressing TCF4 and
β-catenin or β-catenin-specific short interfering
(si) RNA were performed to test FGF21 transcription levels.
Together, these results indicate that the β-catenin/TCF4 complex
directly regulates FGF21 transcription and the present
findings may help elucidate FGF21 transcriptional
regulation.
Materials and methods
Cell culture
The mouse forestomach carcinoma (MFC) fibroblast
cell line, purchased from Bena Culture Collection (Suzhou, China;
cat. no. BNCC100581), was plated at a density sufficient to create
a confluent monolayer following 12 h of culture at 37°C in an
incubator with 5% CO2. Cells were cultured in Dulbecco's
modified Eagle's medium (cat. no. 11965-092) containing 0.5% fetal
bovine serum (cat. no. 10437-028; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Yeast-one hybrid assay
For a yeast one-hybrid assay, the 1.5-kb
FGF21 was amplified from genomic DNA of MFC fibroblast cells
by polymerase chain reaction (PCR) under the following conditions:
Initial denaturation at 98°C for 30 sec, followed by 30 cycles at
98°C for 10 sec, 65°C for 20 sec and 72°C for 2 min, with a final
extension step at 72°C for 10 min. PCR was performed using
Q5® High-Fidelity DNA Polymerase (cat. no. M0491; New
England Biolabs, Inc., Ipswich, MA, USA) and the primers listed in
Table I. Then, FGF21 promoter
sequences were cloned into the pHISi vector (cat. no.
102239; BioVector NTCC Inc., Beijing, China), in which FGF21
promoter drives histidine synthase coding sequences. The open
reading frame (ORF) sequences of TCF4 were cloned into the
pGAD424 vector (BioVector NTCC Inc., Beijing, China). The
constructed pGAD424-TCF4 or empty vector pGAD424 was
transformed into the yeast one hybrid bait strain (YM4271) using a
transformation solution (polyethylene glycol, lithium acetate,
Tris, and EDTA; STZ; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
under the following conditions: 28°C for 30 min, 42°C for 15 min
and 28°C for 5 min. After transformation, the yeast cells were
harvested at 10,000 × g for 1 min, and the supernatant was removed.
The cells were suspended in the dH2O and plated on
synthetic dropout (SD)-Leu or -His media (Takara Biotechnology Co.,
Ltd., Dalian, China) with subsequent growth at 30°C for 3 days.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequences
(5′-3′) |
|---|
| FGF21 | F:
GATGACGACCAAGACACTG |
|
| R:
CGGCCCTGTAAAGGCTCT |
| TCF4 | F:
AGAGCGACAAGCCCCAGAC |
|
| R:
ATTCGCTGCGTCTCCCATC |
| β-catenin | F:
TCGCCAGGATGATCCCAGC |
|
| R:
GCCCATCCATGAGGTCCTG |
| GAPDH | F:
GCCAAGGTCATCCATGACAACT |
|
| R:
GAGGGGCCATCCACAGTCTT |
| pFGF21 | F:
GAATTCCAGAGTTCCAGGGCCA CATCA |
|
| R:
GAGCTCCAGGGCTGCGCTCCGTTCGGGAG |
| FGF21 F1 | F:
CTAAGCAGGGGTTGGTGAGG |
|
| R:
GCGTGTCTGAGGCTTTCTTTC |
| FGF21 F2 | F:
ACACCAGCTCAGTTGCTTACAC |
|
|
ACTGAAGTCTACACTCCTGGGTCT |
| β-catenin ORF | F:
AAGCTTATGGCTACTCAAGCTGACCTGATG |
|
| R:
CTCGAGTTACAGGTCAGTATCAAACCAGGC |
| TCF4 ORF | F:
AAGCTTATGCATCACCAACAGCGAATG |
|
| R:
CTCGAGTCACATCTGTCCCATGTGATTC |
ChIP assay
ChIP assay was performed using a ChIP assay kit (cat
no. 17-295; EMD Millipore, Billerica, MA, USA) according to the
manufacturer's instructions. A total of 1 µg anti-TCF4 antibody
(cat. no. 2566; Cell Signaling Technology, Inc., Danvers, MA, USA)
and anti-β-catenin antibody (cat. no. ab32572; Abcam, Cambridge,
UK) were used for immunoprecipitation. The immunoprecipitated (IP)
DNA by the antibodies were compared with the DNA precipitated
without addition of antibodies using quantitative PCR (qPCR) with
SYBR-Green mixture (cat. no. 4472908; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. A reaction of
ChIP-PCR was performed with an initial denaturation at 95°C for 3
min, followed by 40 cycles of denaturation for 30 sec at 95°C,
annealing for 30 sec at 58°C, and extension at 72°C for 30 sec,
followed by a final extension at 72°C for 5 min. GAPDH was used as
a reference gene to normalize data. The primer sequences for qPCR
are listed in Table I. The
2−ΔΔCq method was used for the fold change calculation
(27).
Co-immunoprecipitation (Co-IP)
assay
A Nuclear-Cytosolic Protein Extraction kit (Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China) was used for protein
extraction of MFC cells. A total of 500 µg (in 500 µl) extracted
protein was incubated at 4°C with 1 µg anti-TCF4 antibody (cat. no.
2566; Cell Signaling Technology, Inc.), anti-β-catenin antibody
(cat. no. ab32572; Abcam), and horseradish peroxidase-conjugated
anti-mouse immunoglobulin G (IgG) H&L (ab6789; Abcam) for 1 h.
Subsequently, 20 µl MagnaBind Protein A Beads (cat no. 21348;
Thermo Fisher Scientific, Inc.) were added and incubated for 2 h.
Following centrifugation at 10,000 × g at 4°C for 5 min, the beads
were washed four times with extraction buffer containing 0.1%
Triton X-100 and eluted with SDS sample buffer.
Western blot analysis
Cells were lysed in an ice-cold lysis solution
containing 7 M urea, 2 M thiourea, 2%
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 40 mM
Trizma base, 40 mM dithiothreitol and 1% protease inhibitor.
Following complete lysis of the cells and centrifugation at 15,000
× g for 15 min at 4°C, the total protein concentration in the
supernatant was measured using a Bradford protein assay kit
(Bio-Rad Laboratories, Inc., Richmond, CA, USA). The proteins (40
µg/lane) were separated on 12% SDS-PAGE and electrotransferred onto
Immobilon-P transfer membranes (EMD Millipore). The membranes were
incubated in TBS containing 5% skimmed milk and 0.05% Tween-20 for
1 h and blotted with primary antibodies at 4°C overnight. Anti-TCF4
(1:1,000; cat. no. 2566; Cell Signaling Technology, Inc.),
anti-β-catenin (1:2,000; cat. no. ab32572; Abcam) and anti-GAPDH
(1:2,000; cat. no. ab8245; Abcam) antibodies were used as the
primary antibodies. GAPDH was used as the internal control. The
membranes were incubated for 1 h at 37°C with an anti-mouse or
anti-rabbit horseradish peroxidase-linked secondary antibody
(1:2,000; cat. nos. 7076 and 7074, Cell Signaling Technology,
Inc.), and the signal was visualized using an
electrochemiluminescence kit (GE Healthcare, Chicago, IL, USA).
Total RNA extraction, cDNA synthesis
and reverse transcription-qPCR (RT-qPCR)
In total, 2 µg total RNA, extracted with RNeasy Mini
kit (cat. no. 74104; Qiagen China Co., Ltd., Shanghai, China) from
MFC cells, was reverse-transcribed using a GoScript reverse
transcription kit (Promega Corporation, Madison, WI, USA) following
the manufacturer's instructions. RT-qPCR was performed with SYBR
Green mixture (cat. no. 4472908; Thermo Fisher Scientific, Inc.),
and gene expression was quantified as described previously
(27). GAPDH was used as an internal
control. The sequences of primers used in RT-qPCR are listed in
Table I.
Overexpression and RNA
interference
ORF regions of TCF4 (NM_013685.2, NCBI) and
β-catenin (NM_001165902.1, NCBI) were amplified from cDNA of
MFC cells by the gene specific primers with Q5®
High-Fidelity DNA Polymerase (cat. no. M0491; New England Biolabs,
Inc.) and cloned into the pcDNA3.1 (+) (cat. no. V79020;
Thermo Fisher Scientific, Inc.) expression vector to contract
TCF4 overexpression and β-catenin overexpressed
plasmids. The primers used for TCF4 and β-catenin are
listed in Table I. siRNA for
β-catenin (ON-TARGET plus SMART pool, L-004018) and negative
control siRNA (ON-TARGETplus si CONTROL non-targeting pool,
D-001810) were purchased from Dharmacon RNA Technologies (Chicago,
IL, USA). The MFC cells were seeded 12 h prior to transfection at
37°C with 5% CO2 and reached a density of 30–50%
confluence at the time of transfection. Subsequently, 30 nM siRNA
duplex and 2 µg TCF4 overexpression or β-catenin
overexpression plasmids were transfected on day 0 using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and
Opti-MEM I Reduced Serum Medium (Gibco; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. On day 1 (24 h
after transfection), the cells were confluent and the
overexpression or siRNA solutions were exchanged with full growth
medium. These transfected cells were subsequently used for RT-qPCR
(as described above), for which the density reached 80–90%
confluence at the time of harvest for RNA preparation on day 3 (72
h after transfection).
Statistical analysis
Data are presented as the mean ± standard error of
the mean. Each experiment was performed in triplicate. Statistical
calculations were performed with Prism 5 (GraphPad Software, Inc.,
San Diego, CA, USA). Data were analyzed using the Student's t-test
for two groups and one-way analysis of variance followed by Tukey's
test for more than two groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
TCF4 directly activates FGF21
transcription
FGF21 promoter sequences analysis
demonstrated that two putative TCF/LEF binding motifs (AGAAAG)
(26) appeared within 1.5 kb of the
promoter. The two putative binding motifs are located 1,163 and 876
bp upstream of the start codon, respectively (Fig. 1A). In order to examine whether TCF4
directly binds to the putative motifs, ChIP assays were performed
using the TCF4 antibody and the precipitation without antibody as
the control. FGF21 exhibited a higher expression level in
the MFC cells (data not shown); therefore, the MFC cells were used
in the experiments (14,15). The IP DNA fragments were amplified by
two primer pairs, which spanned the F1 and F2 regions in the
FGF21 promoter (Fig. 1B). The
ChIP-PCR data was normalized to input DNA and the results revealed
that TCF4 bound to the F1 but not F2 region in the promoter of the
FGF21 gene (Fig. 1B). To
further verify the ChIP results, a yeast-one hybrid assay was
performed by co-expressing activation domain (AD)-TCF4 and
pFGF21-His or mpFGF21-His (Fig. 1C). The results indicate that AD-TCF4
co-expressing pFGF21-His were able to grow in SD media
missing histidine, whereas the empty vector transforming or AD-TCF4
co-expressing mpFGF21-His cells failed to grow (Fig. 1D). These data indicated that TCF4
activates the FGF21 promoter in yeast.
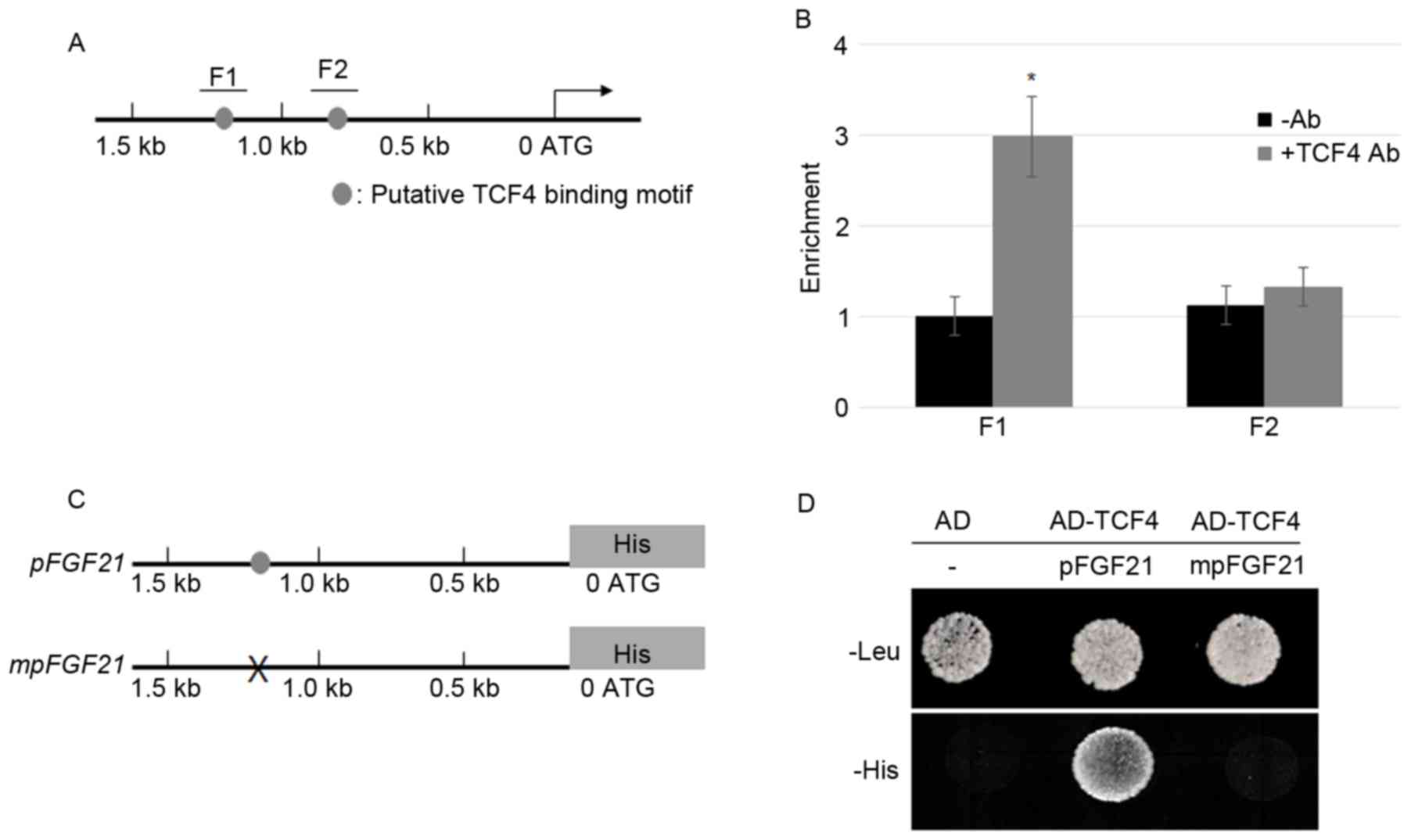 | Figure 1.TCF4 bound directly to the promoter
of FGF21. (A) The schematic diagram indicates the locations
of the putative TCF binding motifs, F1 and F2 (gray circles),
within the 1.5 kb FGF21 promoter. (B) A ChIP assay was
performed by amplifying IP DNA to detect the F1 and F2 regions in
the FGF21 promoter; relative ratios of IP DNA to input DNA
was determined by ChIP-PCR, and input DNA was used to normalize the
data. Data are presented as the mean ± standard error (n=3);
*P<0.05 vs. -Ab. (C) A 1.5 kb sequence of the FGF21
promoter with or without mutations at the TCF4 binding motif was
cloned into the pHISi vector in which His was a reporter
gene. The gray circle indicates the TCF4 binding motif and ‘X’
indicates mutation of the TCF4 binding sequences. (D) A yeast
one-hybrid assay was performed to analyze the activation of TCF4 on
the FGF21 promoter. Yeast cells harboring either the AD
vector without promoter or AD-TCF4 together with pFGF21-His
or mpFGF21-His were grown on synthetic dropout media lacking
Leu or His. TCF4, transcription factor 4; FGF21, fibroblast growth
factor 21; ChIP, chromatin immunoprecipitation; IP,
immunoprecipitated; PCR, polymerase chain reaction; Ab, antibody;
-Ab, without antibody; AD, activation domain. |
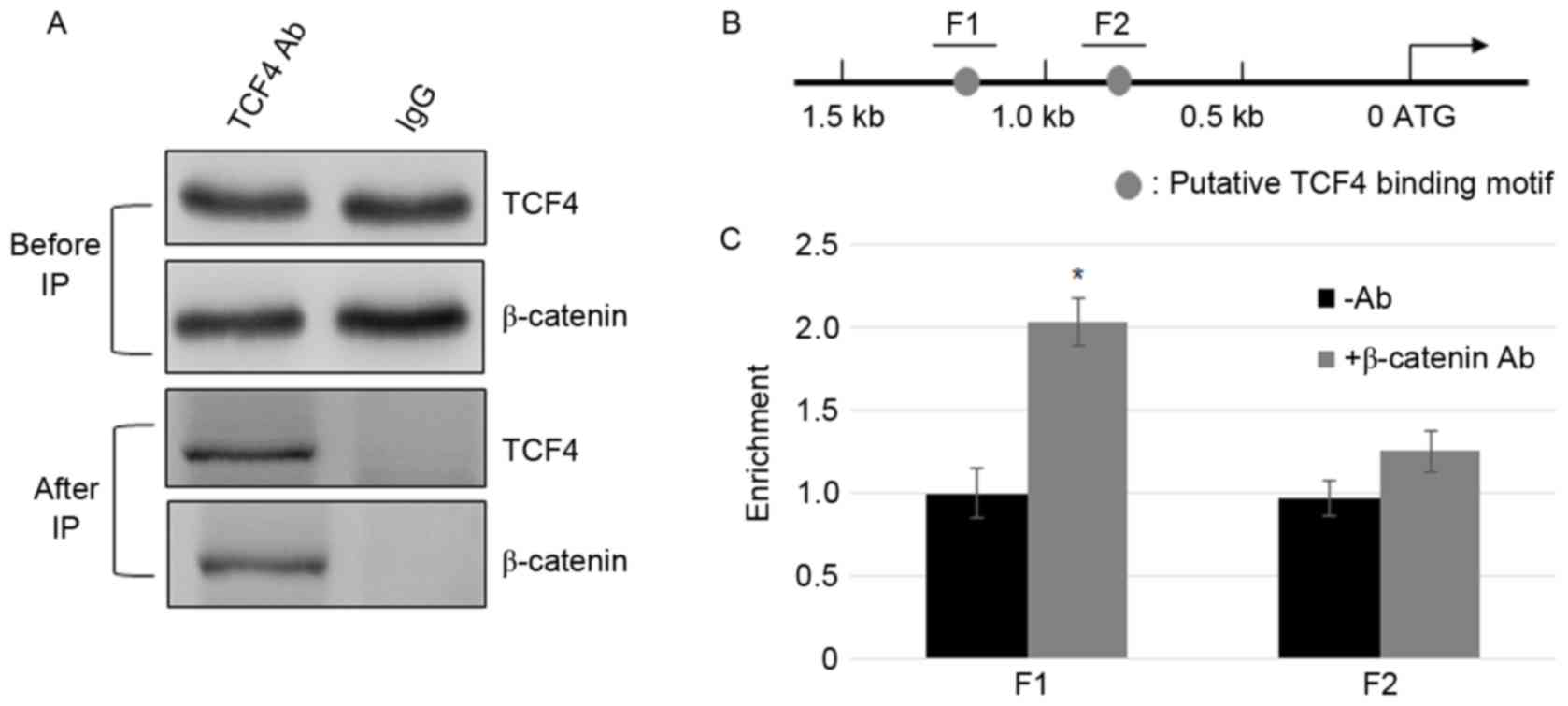
β-catenin interacts with TCF4 and
β-catenin/TCF4 complex binds to the promoter of FGF21
After β-catenin accumulates in the nucleus, it
interacts with TCF/LEF, and activates the transcription of Wnt
signaling genes (24). To test
whether TCF4 and β-catenin interact in MFC cells, Co-IP assay was
performed. The cell lysate was incubated with TCF4 or IgG
antibodies, and β-catenin and TCF4 were detected using β-catenin
and TCF4 antibodies prior to and following immunoprecipitation
(Fig. 2A). GAPDH levels in IgG and
TCF4 incubated samples were similar (Fig. 2A). The results indicate that
β-catenin and TCF4 physically interact with the MFC cells. To
further test the binding of the β-catenin complex to the promoter
of FGF21, ChIP assays were performed using β-catenin.
ChIP-PCR results indicate that similar to the TCF4, β-catenin IP
DNA was enriched in the F1 region but not in the F2 region
(Fig. 2B and C).
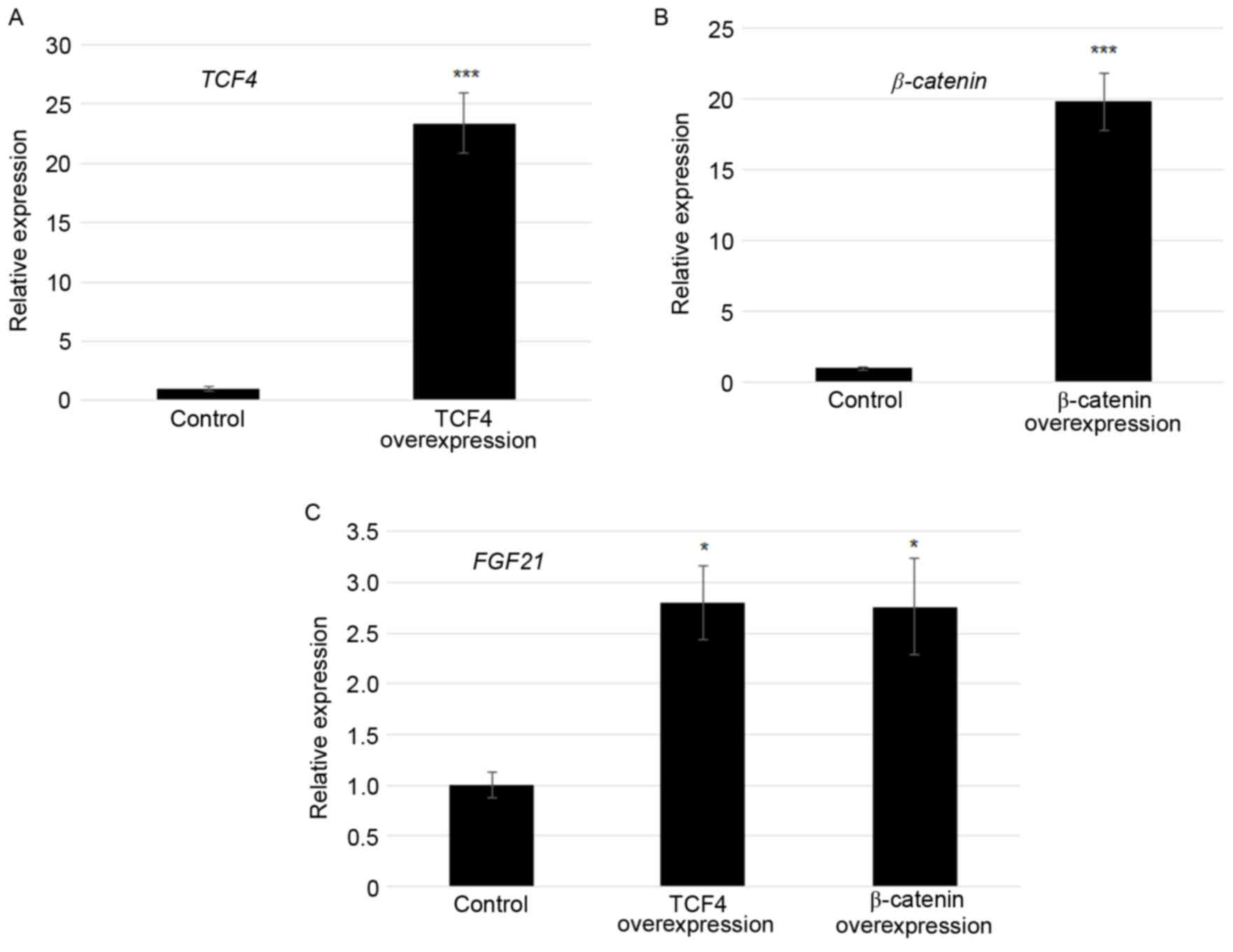
Overexpression of TCF4 and β-catenin
activated FGF21 transcription
As TCF4 and β-catenin bound to the F1 region of the
FGF21 promoter, effects of TCF4 and β-catenin on
FGF21 expression were tested. TCF4 and β-catenin modulated
transcription of FGF21 was confirmed by plasmid
(pcDNA3.1-TCF4 or pcDNA3.1-β-catenin)-mediated
transfection experiments using Lipofectamine 2000. At 24 h
following transfection, TCF4 was ~22-fold and
β-catenin was ~20-fold higher in the TCF4
overexpression and β-catenin overexpressed cells compared
with the control in which empty vector was transformed (Fig. 3A and B). Furthermore, FGF21
expression levels were monitored by RT-qPCR. The results revealed
that the FGF21 transcript was 2.7 and 2.6 fold higher in
TCF4 overexpression and β-catenin overexpressed cells
compared with the control, respectively (Fig. 3C).
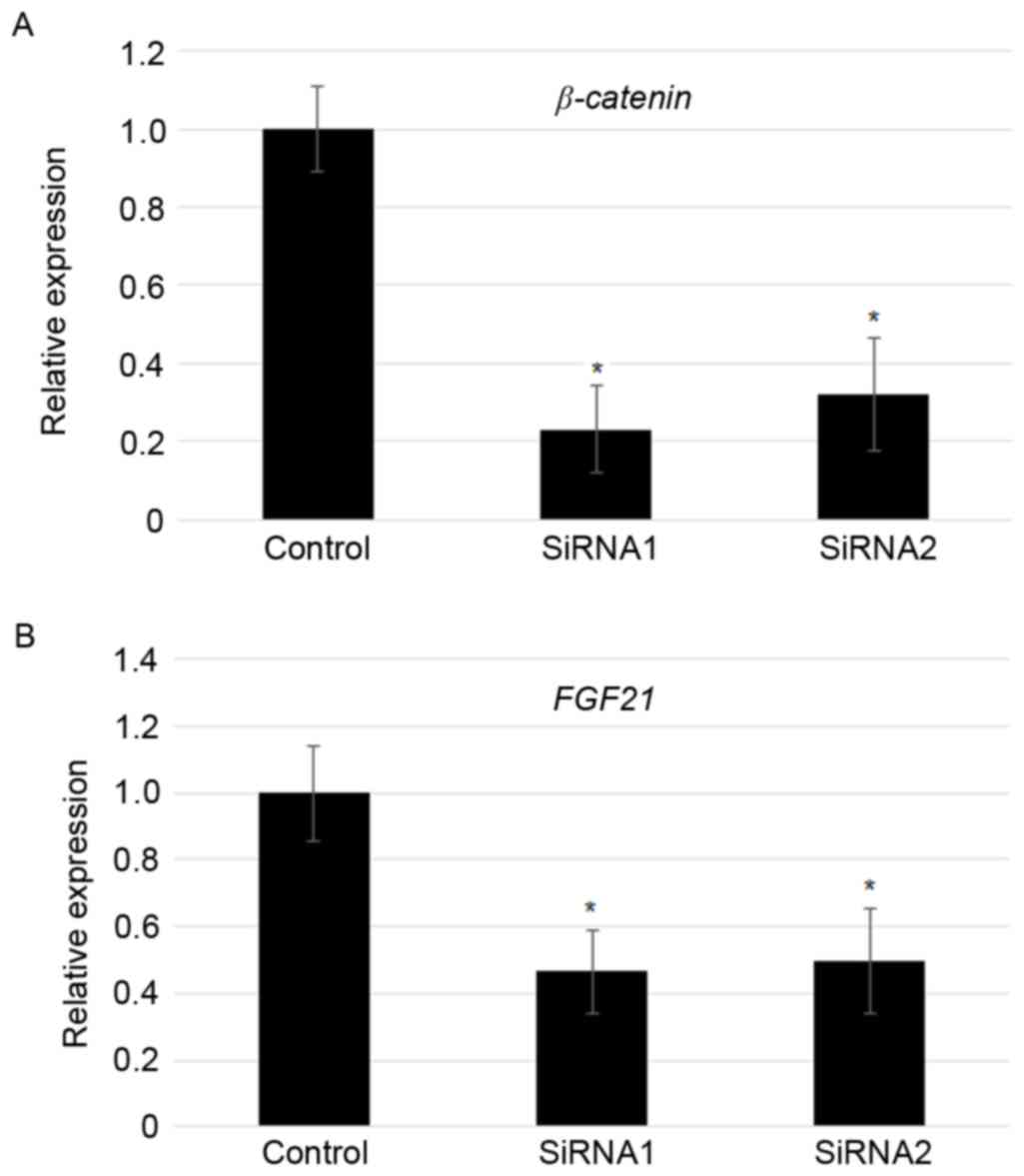
Suppression of β-catenin reduced FGF21
levels
As overexpression of β-catenin induced
FGF21 expression, FGF21 levels in the
β-catenin knockdown MFC cells was analyzed further. RT-qPCR
results indicated that two independent transfection of
β-catenin specific siRNAs significantly reduced the
expression of β-catenin (~70–80%; Fig. 4A). In addition, siRNA-mediated
knockdown of β-catenin largely contributed to the reduction
in the expression of FGF21 (~50%; Fig. 4B).
Discussion
FGF21 is sensitive to diverse physiological changes
(e.g., fasting and injury) and serves an important role in
development and metabolism, including glucose metabolism (11–15).
Transcription of FGF21 was previously identified under the
control of PPARα (17), RORα
(18), and RARβ in hepatocytes
(19). In addition, FGF21
transcription was also induced by feeding or cold treatment in
brown adipose tissue (28,29), indicating the complicated regulation
of FGF21 transcription. To elucidate transcriptional
regulation, the promoter of FGF21 was investigated for
identifying cis-element information. The putative TCF/LEF
binding motifs (26) were observed
within 1.5 kb of its promoter region. Furthermore, ChIP and
yeast-one hybrid assays confirmed that TCF4 directly bound to one
of the two putative TCF/LEF motifs in the FGF21
promoter.
TCF and β-catenin are the master transcriptional
regulators of the Wnt signaling pathway, and translocation of
β-catenin from the cytosol to the nucleus resulted in formation of
a TCF complex, including TCF and β-catenin (24). Furthermore, the complex activates a
large number of downstream genes via binding to the specific
sequences in their promoters (26).
To further identify the possibility that TCF4/β-catenin
complex-mediated regulation of FGF21 transcription, Co-IP
and ChIP assays were performed. Co-IP results indicated that TCF4
and β-catenin interact in MFC cells. Furthermore, ChIP assay data
revealed that β-catenin is associated with the FGF21
transcriptional regulator complex that may function together with
TCF4. Overexpression of TCF4 or β-catenin in MFC
induced FGF21 transcription, whereas suppression of
β-catenin via a specific siRNA repressed the FGF21
level. These results indicated that TCF4 and β-catenin activates
FGF21 by binding to the F1 region of its promoter.
Previous reports identified that Wnt/β-catenin acts
upstream of FGF2 in lung tissues to regulate proximal-distal
patterning (30). However, no direct
connection between β-catenin and FGF gene was observed. In the
present study, TCF4/β-catenin was identified to directly regulate
FGF21 in a mouse embryonic fibroblast cell line, MFC, and
further experiments are indeed required to examine whether
TCF4/β-catenin mutant mice exhibit a similar phenomenon as that
presented in FGF21 mutants, including glucose metabolism
abnormality and heart dysfunction. However, the observations of the
present study have improved the understanding of FGF21
regulation as direct connections were observed between Wnt and FGF
signaling.
Acknowledgements
The present study was funded by Wenzhou Medical
University (grant no. 025gt8972). This work was also supported by
the Doctoral Scientific Research Foundation of Xingxiang Medical
University (grant no. XYBSKYZZ201512&201513) and grants from
The Education Department of Henan Province (grant nos.
201610472040, 172102310584). The vectors and yeast strain used in
the yeast two hybrid assay were kindly donated by the Yuan lab at
Wenzhou Medical University.
References
|
1
|
Mohammadi M, Olsen SK and Ibrahimi OA:
Structural basis for fibroblast growth factor receptor activation.
CytokineGrowth Factor Rev. 16:107–137. 2005. View Article : Google Scholar
|
|
2
|
Feldman B, Poueymirou W, Papaioannou VE,
DeChiara TM and Goldfarb M: Requirement of FGF-4 for
postimplantation mouse development. Science. 267:246–249. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dubrulle J and Pourquié O: fgf8 mRNA decay
establishes a gradient that couples axial elongation to patterning
in the vertebrate embryo. Nature. 427:419–422. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun X, Meyers EN, Lewandoski M and Martin
GR: Targeted disruption of Fgf8 causes failure of cell migration in
the gastrulating mouse embryo. Genes Dev. 13:1834–1846. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martin GR: The roles of FGFs in the early
development of vertebrate limbs. Genes Dev. 12:1571–1586. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Goldfarb M: Functions of fibroblast growth
factors in vertebrate development. Cytokine Growth Factor Rev.
7:311–325. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kanazawa S, Fujiwara T, Matsuzaki S,
Shingaki K, Taniguchi M, Miyata S, Tohyama M, Sakai Y, Yano K,
Hosokawa K and Kubo T: bFGF regulates PI3-kinase-Rac1-JNK pathway
and promotes fibroblast migration in wound healing. PloS One.
5:e122282010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yie J, Wang W, Deng L, Tam LT, Stevens J,
Chen MM, Li Y, Xu J, Lindberg R, Hecht R, et al: Understanding the
physical interactions in the FGF21/FGFR/beta-Klotho complex:
Structural requirements and implications in FGF21 signaling. Chem
Biol Drug Des. 79:398–410. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Belov AA and Mohammadi M: Molecular
mechanisms of fibroblast growth factor signaling in physiology and
pathology. Cold Spring Harb Perspect Biol. 5:a0159582013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishimura T, Nakatake Y, Konishi M and
Itoh N: Identification of a novel FGF, FGF-21, preferentially
expressed in the liver. Biochim Biophys Acta. 1492:203–206. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang Q, Zhong L, Zhang J, Wang Y,
Bornstein SR, Triggle CR, Ding H, Lam KS and Xu A: FGF21 maintains
glucose homeostasis by mediating the cross talk between liver and
brain during prolonged fasting. Diabetes. 63:4064–4075. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin Z, Tian H, Lam KS, Lin S, Hoo RC,
Konishi M, Itoh N, Wang Y, Bornstein SR, Xu A and Li X: Adiponectin
mediates the metabolic effects of FGF21 on glucose homeostasis and
insulin sensitivity in mice. Cell Metab. 17:779–789. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin Z, Wu F, Lin S, Pan X, Jin L, Lu T,
Shi L, Wang Y, Xu A and Li X: Adiponectin protects against
acetaminophen-induced mitochondrial dysfunction and acute liver
injury by promoting autophagy in mice. J Hepatol. 61:825–831. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song Y, Ding J, Jin R, Jung J, Li S, Yang
J, Wang A and Li Z: Expression and purification of FGF21 in Pichia
pastoris and its effect on fibroblast-cell migration. Mol Med Rep.
13:3619–3626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song YH, Zhu YT, Ding J, Zhou FY, Xue JX,
Jung JH, Li ZJ and Gao WY: Distribution of fibroblast growth
factors and their roles in skin fibroblast cell migration. Mol Med
Rep. 14:3336–3342. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gong Q, Hu Z, Zhang F, Cui A, Chen X,
Jiang H, Gao J, Chen X, Han Y, Liang Q, et al: Fibroblast growth
factor 21 improves hepatic insulin sensitivity by inhibiting
mammalian target of rapamycin complex 1 in mice. Hepatology.
64:425–438. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inagaki T, Dutchak P, Zhao G, Ding X,
Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et
al: Endocrine regulation of the fasting response by
PPARalpha-mediated induction of fibroblast growth factor 21. Cell
Metab. 5:415–425. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Solt LA and Burris TP: Regulation
of FGF21 expression and secretion by retinoic acid receptor-related
orphan receptor alpha. J Biol Chem. 285:15668–15673. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Wong K, Walsh K, Gao B and Zang M:
Retinoic acid receptor β stimulates hepatic induction of fibroblast
growth factor 21 to promote fatty acid oxidation and control
whole-body energy homeostasis in mice. J Biol Chem.
288:10490–10504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, D'Amico M, Pestell R and Ben-Ze'ev A: The cyclin D1
gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad
Sci USA. 96:pp. 5522–5527. 1999; View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moon RT, Kohn AD, De Ferrari GV and Kaykas
A: WNT and β-catenin signalling: Diseases and therapies. Nat Rev
Genet. 5:691–701. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ,
Keller C and Rando TA: Increased Wnt signaling during aging alters
muscle stem cell fate and increases fibrosis. Science. 317:807–810.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gordon MD and Nusse R: Wnt signaling:
Multiple pathways, multiple receptors, and multiple transcription
factors. J Biol Chem. 281:22429–22433. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aberle H, Bauer A, Stappert J, Kispert A
and Kemler R: beta-catenin is a target for the ubiquitin-proteasome
pathway. EMBO J. 16:3797–3804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schuijers J, Mokry M, Hatzis P, Cuppen E
and Clevers H: Wnt-induced transcriptional activation is
exclusively mediated by TCF/LEF. EMBO J. 33:146–156. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-tie quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dutchak PA, Katafuchi T, Bookout AL, Choi
JH, Yu RT, Mangelsdorf DJ and Kliewer SA: Fibroblast growth
factor-21 regulates PPARγ activity and the antidiabetic actions of
thiazolidinediones. Cell. 148:556–567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fisher FM, Kleiner S, Douris N, Fox EC,
Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS,
Maratos-Flier E and Spiegelman BM: FGF21 regulates PGC-1α and
browning of white adipose tissues in adaptive thermogenesis. Genes
Dev. 26:271–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shu W, Guttentag S, Wang Z, Andl T,
Ballard P, Lu MM, Piccolo S, Birchmeier W, Whitsett JA, Millar SE
and Morrisey EE: Wnt/beta-catenin signaling acts upstream of N-myc,
BMP4, and FGF signaling to regulate proximal-distal patterning in
the lung. Dev Biol. 283:226–239. 2005. View Article : Google Scholar : PubMed/NCBI
|