Introduction
Colorectal cancer (CRC) is among the most common
malignant human tumors, with nearly 1.2 million new cases and more
than 600,000 CRC-related mortalities reported each year worldwide
(1,2). The treatment of CRC has greatly
developed in the past several decades, and surgery and chemotherapy
have been widely used to improve the survival rate and prognosis of
CRC patients. The 5-year relative survival rate of CRC patients is
90.1% for those with early-stage disease; however, this drops to
69.2 and 11.7% for those presenting with regional spread or distant
metastases, respectively (3). With
the use of colonoscopy, the survival rate of CRC patients has
improved in some developed countries, though there is no obvious
improvement in China (4).
To develop effective drugs for the treatment of CRC,
it is important to identify therapeutic target genes or molecules.
Coat proteins (COPs) include three main types of protein (clathrin,
COPI and COPII) and play a key role in intracellular transport by
forming transport vesicles. This role is achieved through the
coupling of two major functions. Membrane bending to generate
transport carriers from organellar membranes; and cargo binding for
the packaging of newly formed vesicles (5). The core component of the COPI complex
is a protein coatomer, a multimeric complex comprising seven
subunits: α-COP (160 kDa), β-COP (110 kDa), β'-COP (110 kDa), γ-COP
(98 kDa), δ-COP (61 kDa), ε-COP (36 kDa), and ζ-COP (20 kDa)
(6,7). Coatomer protein complex subunit β2
(COPB2) is one of the seven non-clathrin-coated vesicular coat
subunits that forms the coatomer, and plays a role in membrane
transport between the endoplasmic reticulum and Golgi apparatus
(8–11). However, to the best of our knowledge,
the relationship between the expression of COPB2 and the
development of colon cancer remains unknown. Therefore, the present
study examined the expression of COPB2 in human colon cancer
tissues, paracancerous tissues and colon cancer cell lines.
Furthermore, to investigate the physiological function of COPB2 in
colon cancer, we used a lentivirus-mediated RNAi method to suppress
the expression of COPB2 in RKO and HCT116 colon cancer cells. In
the present study, we found that COPB2 reduced the proliferation
and colony-formation abilities of colon cancer cells. Furthermore,
COPB2 silencing was associated with G0/G1 or S phase cell cycle
arrest; and after COPB2 silencing, the expression level of P21 and
P16 were significantly increased, while that of cyclin A was
reduced. Based on the present findings, we hypothesize that COPB2
is essential for the proliferation of colon cancer cells, and that
lentivirus-mediated COPB2 silencing may be a promising therapeutic
method for the treatment of colon cancer.
Materials and methods
Immunohistochemistry (IHC)
Tumor specimens and adjacent noncancerous tissues
from 35 patients who underwent surgery for colon cancer in Gansu
Provincial Hospital (Gansu, China) in 2016 were evaluated in this
study. Ethics committee approval was obtained from the Ethics
Committee of Gansu Provincial Hospital. All tumor specimens were
obtained from patients who had been newly diagnosed with colon
cancer, and none of the patients had received chemotherapy or
radiotherapy prior to sample collection. IHC was performed to
detect COPB2 protein expression by tissue microarray as described
previously (12). In brief, the
sections were deparaffinized in xylene and dehydrated with an
ethanol gradient, and then blocked with 0.3%
H2O2 at room temperature for 10 min to
inhibit endogenous peroxidase activity. Blocking with 10% normal
goat serum was performed prior to the application of primary
antibody against COPB2 (HPA036867, 1:10; Sigma-Aldrich, St. Louis,
MO, USA). Subsequently, the sections were incubated with the
anti-COPB2 antibody in PBS at 4°C overnight. The slides were then
probed for 10 min with an HRP-labeled polymer conjugated to an
appropriate secondary antibody. The immunoreactive bands were
detected by staining with DAB. The slides were reviewed and scored
in a double-blinded manner by two independent pathologists. More
than 1,000 cells were counted at ×400 magnification in ten visual
fields selected at random. The proportions of positive cells were
scored as follows: ‘0’, no staining; ‘1’, <1/3 stained; ‘2’,
1/3-2/3 stained; and ‘3’, >2/3 stained. The staining intensity
was scored as follows: ‘0’, none; ‘1’, weak; ‘2’, intermediate; and
‘3’, strong. The two scores were added to obtain the final scores,
which ranged from 0 to 6. A final score of 0–1 indicated negative
staining (−), 2–3 indicated weakly positive staining (+), 4
indicated moderately positive staining (++), and 5–6 indicated
strongly positive staining (+++) (13).
Cell culture
Six human CRC cancer cell lines, namely RKO, SW480,
HCT116, DLD1, HT-29 and SW620, were obtained from the Cell Bank of
Type Culture Collection of Chinese Academy of Sciences (Shanghai,
China). All cells were maintained in RPMI 1640 medium (Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) containing 10% FBS
(R&D Systems, Inc., Minneapolis, MN, USA), L-glutamine, and 100
U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) at 37°C in a 5% CO2
incubator.
Measurement of COPB2 expression in
colon cancer cell lines
The expression of COPB2 in the human cancer cell
lines RKO, SW480, HCT116, DLD1, HT-29, and SW620 was detected by
reverse transcription-quantitative PCR (RT-qPCR). Total RNA was
extracted from the different cell lines using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer's
instructions. The primers used to measure the expression of COPB2
were 5′-GTGGGGACAAGCCATACCTC-3′ (forward) and
5′-GTGCTCTCAAGCCGGTAGG-3′ (reverse). The primers for GAPDH, which
was used as an internal control, were 5′-TGACTTCAACAGCGACACCCA-3′
(forward) and 5′-CACCCTGTTGCTGTAGCCAAA-3′ (reverse). The reaction
system was made up as follows: 10 µl SYBR premix Ex Taq; 2 µl
forward and reverse primers (2.5 µM); 2 µl cDNA; and 4 µl
RNase-Free H2O. The conditions used for qPCR were as
follows: Initial denaturation at 95°C for 15 sec, denaturation at
95°C for 5 sec, and annealing/extension at 60°C for 30 sec (for a
total of 45 cycles). Fold changes in expression were calculated
using the 2−ΔΔCq method. The experiment was repeated 3
times.
Vector construction and lentivirus
packaging
An shRNA targeting COPB2 (shCOPB2;
AGATTAGAGTGTTCAATTA) and a random negative control shRNA (shCtrl;
TTCTCCGAACGTGTCACGT) were designed. Both shRNAs were inserted into
GV248 lentiviral vectors (Shanghai GenePharma Co., Ltd., Shanghai,
China). For lentivirus packing, HEK293T cells were transfected with
the GV248-sh COPB2 or -shCtrl vectors using Lipofectamine 2000,
according to the manufacturer's protocol.
Lentivirus-mediated silencing of COPB2
in colon cells
RKO and HCT116 cells (1×105 cells/ml)
were cultured in 6-well plates. After 24 h, GV248-COPB2-shRNA or
-control shRNA were used to infect RKO and HCT116 cells with
multiplicities of infection (MOIs) of 5 and 15, respectively. At 96
h post-infection, the cells were observed under a fluorescence
microscope (×100). The infection efficiency was determined by
counting the number of GFP-expressing cells.
RNA extraction and RT-qPCR
At 96 h post-infection, RKO and HCT116 cells were
collected and total RNA was extracted with TRIzol reagent
(Invitrogen) according to the manufacturer's instructions. RT-qPCR
was performed to determine the knockdown efficiency of the COPB2
gene in the RKO and HCT116 cells infected with shCOPB2. The primers
used to measure the expression levels of COPB2 and GAPDH, the
reaction system and the qPCR conditions were the same as above.
Fold changes in expression were calculated using the
2−ΔΔCt method, and the experiment was repeated 3
times.
Western blot analysis
Western blot analysis was performed as previously
described (14). Briefly, at 96 h
post-infection, the RKO and HCT116 cells were collected and total
protein was extracted. Protein samples were then separated on a 10%
SDS-PAGE gel and transferred to polyvinylidene difluoride (PVDF)
membranes (EMD Millipore, Bedford, UK). After blocking with 5% skim
milk, the membranes were exposed to primary antibodies against
COPB2 (Sigma-Aldrich), P16, P21, cyclin A1-A2 (Abcam, Cambridge,
UK) and GAPDH (Abcam) overnight at 4°C. After washing three times
with TBST, the membranes were incubated with fluorescently labeled
secondary antibodies at room temperature for 1 h. The bands were
visualized with an Odyssey detection system (Licor Biosciences,
Nebraska, US). The experiment was repeated 3 times.
MTT cell proliferation assay
An MTT assay was used to assess the proliferation of
the RKO and HCT116 cells infected with shCOPB2 or shCtrl, as
previously described (15,16). In brief, after being infected 96 h,
the two cell lines were seeded in 96-wells plates at a density of
1×104 cells/well, and cell density was measured at days
1, 2, 3, 4 and 5 after seeding. At each time point, 20 µl of
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
solution was added to each well. After 4 h of incubation, the MTT
was removed, 150 µl of DMSO was added, and the plate was shaken
constantly for 15 min. The absorbance was read using a microplate
reader at a wavelength of 490 nm. The experiment was repeated 3
times.
BrdU incorporation assay
A BrdU Cell Proliferation ELISA kit (11647229001;
Roche Diagnostics, Basel, Switzerland) was used to measure the
proliferation ability of cells based on DNA synthesis rate,
according to the manufacturer's protocol. After being infected 96
h, the RKO and HCT116 cells were incubated with BrdU labeling
solution (10 µl/well), and then with fixing solution (200 µl/well)
at room temperature in the dark. After 30 min, the cells were
incubated with anti-BrdU-POD (100 µl/well) for another 90 min.
Subsequently, washing buffer (200–300 µl/well) was added and the
cell culture plates were washed 3 times. Substrate solution (100
µl/well) was then added, and 30 min later, 10%
H2SO4 (50 µl/well) was added. Finally, the
absorbance was measured at a wavelength of 450 nm. The experiment
was repeated 3 times.
Colony formation assay
At 72 h post-infection, the RKO and HCT116 cells
were plated in 6-well plates at a density of 400 and 500
cells/well, respectively, and cultured in a humidified incubator at
37°C with 5% CO2. After 10 days, the natural colonies
that had formed were washed twice with PBS and then stained with
crystal violet. The numbers of colonies were counted under light
and fluorescence microscopes. The experiment was repeated 3
times.
Flow cytometry analysis
Flow cytometry was used to analyze the cell cycle
distributions of the RKO and HCT116 cells following lentiviral
infection. The infected cells were collected, fixed with 75%
ethanol, washed with PBS, and stained with propidium iodide (PI)
supplemented with RNase overnight at 4°C. Following staining, the
cells were analyzed by flow cytometry. The experiment was repeated
3 times.
Statistical analysis
SPSS 20.0 statistical software (IBM SPSS, Armonk,
NY, USA) was used for all statistical analyses. Student t-tests
were used to compare the data between two groups. To compare the
data of three or more groups, one-way ANOVA was used. The results
were presented as means ± standard deviation (SD), and P<0.05
was considered to indicate statistical significance.
Results
COPB2 is highly expressed in human
colon cancer
To investigate the relationship between COPB2
expression and human colon cancer, IHC was used to detect COPB2
expression in 35 colon cancer tissues and adjacent noncancerous
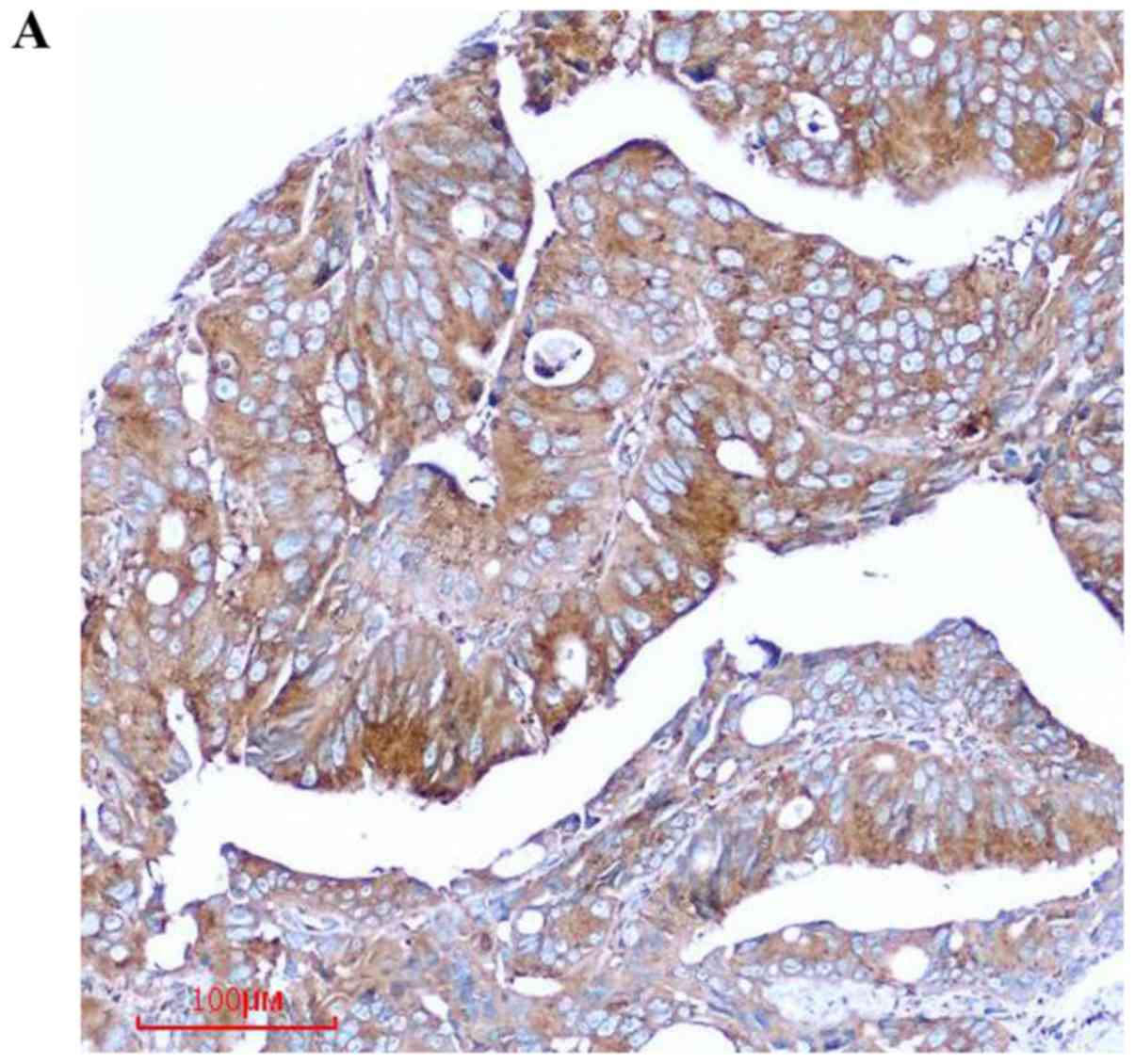
tissues. We found that COPB2 staining was markedly stronger in the
colon cancer tissues than in the paracancerous tissues (Fig. 1A). Notably, the COPB2 protein density
was determined to be 4.60±1.418 in the human colon tissues and
1.17±1.098 in the corresponding adjacent tissues (P<0.001;
Table I and Fig. 1B). These results indicated that COPB2
may be involved in the pathogenesis of human colon cancer.
 | Table I.Expression of COPB2 in human colon
cancer and paracancerous tissue by IHC staining. |
Table I.
Expression of COPB2 in human colon
cancer and paracancerous tissue by IHC staining.
| Group | Case number | Mean ± SD | P-value |
|---|
| Colon cancer | 35 |
4.60±1.418 | <0.001 |
| Paracancerous
tissue | 35 |
1.17±1.098 |
|
COPB2 is positively correlated with
pathological grading in human colon cancer
The clinicopathological parameters of 35 patients
with human colon cancer, including age, sex, tumor diameter and
pathological grade, were examined in this study. As shown in
Table II, a significant correlation
was observed between the expression of COPB2 and pathological
grade; and the expression of COPB2 was significantly higher in
stages III and IV when compared with stages I and II of human colon
cancer (P<0.01).
 | Table II.Relationship between COPB2 expression
and clinical pathological parameters by IHC staining. |
Table II.
Relationship between COPB2 expression
and clinical pathological parameters by IHC staining.
| Clinical pathological
parameters | Case number | Mean ± SD | P-value |
|---|
| Ages (years) |
|
|
|
| ≤60 | 17 |
4.530±1.546 | 0.78 |
|
<60 | 18 |
4.670±1.328 |
|
| Sex |
|
|
|
| Male | 17 |
4.820±1.510 | 0.373 |
|
Female | 18 |
4.390±1.335 |
|
| Tumor diameter
(cm) |
|
|
|
| ≤5 | 23 |
4.65±1.301 | 0.768 |
|
<5 | 12 |
4.50±1.679 |
|
| Pathological
grading |
|
|
|
| I+II | 17 |
3.94±1.391 | 0.002a |
|
III+IV | 18 |
5.17±0.707 |
|
COPB2 gene expression was detected in
six human CRC cancer cell lines
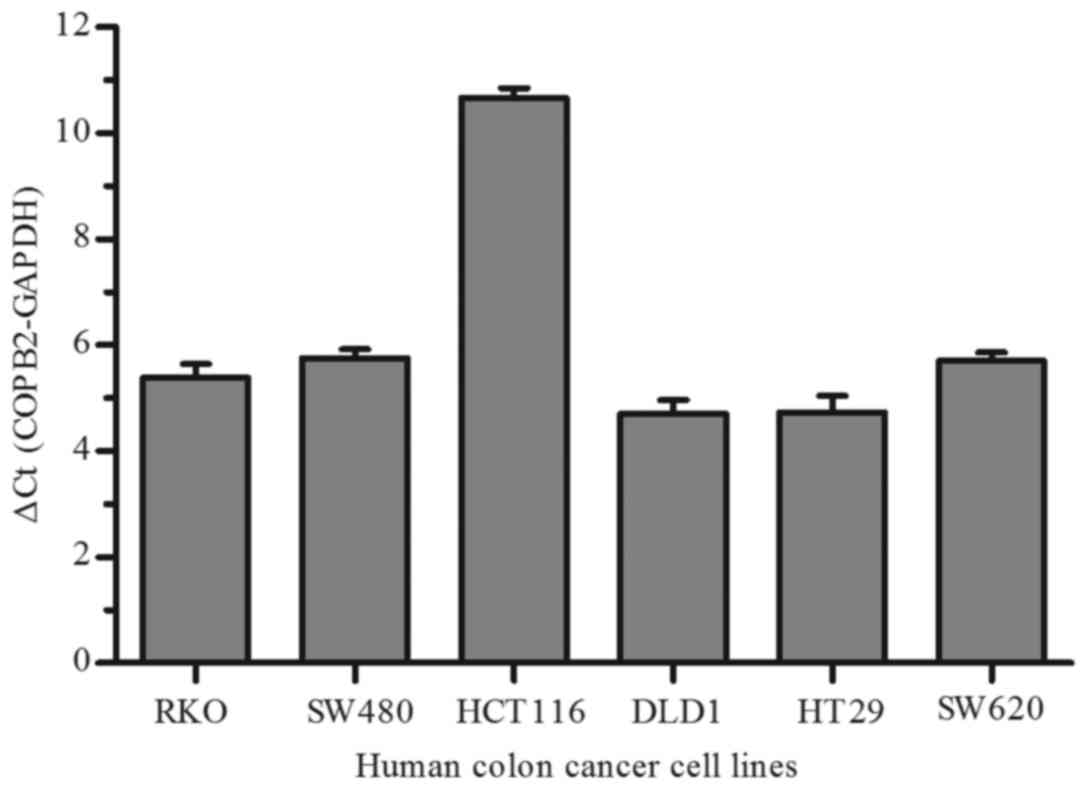
To investigate the function of COPB2 in human colon
cancer cells, we firstly detected COPB2 expression in six different
CRC cell lines (RKO, SW480, HCT116, DLD1, HT29 and SW620) by qPCR.
As depicted in Fig. 2, we found that
there was relative changes in COPB2 expression in six colon cancer
cell lines, especially in HCT116 cells. Therefore, the RKO and
HCT116 cell lines were used in subsequent assays.
COPB2 is downregulated in RKO and
HCT116 cells by lentivirus-mediated RNAi
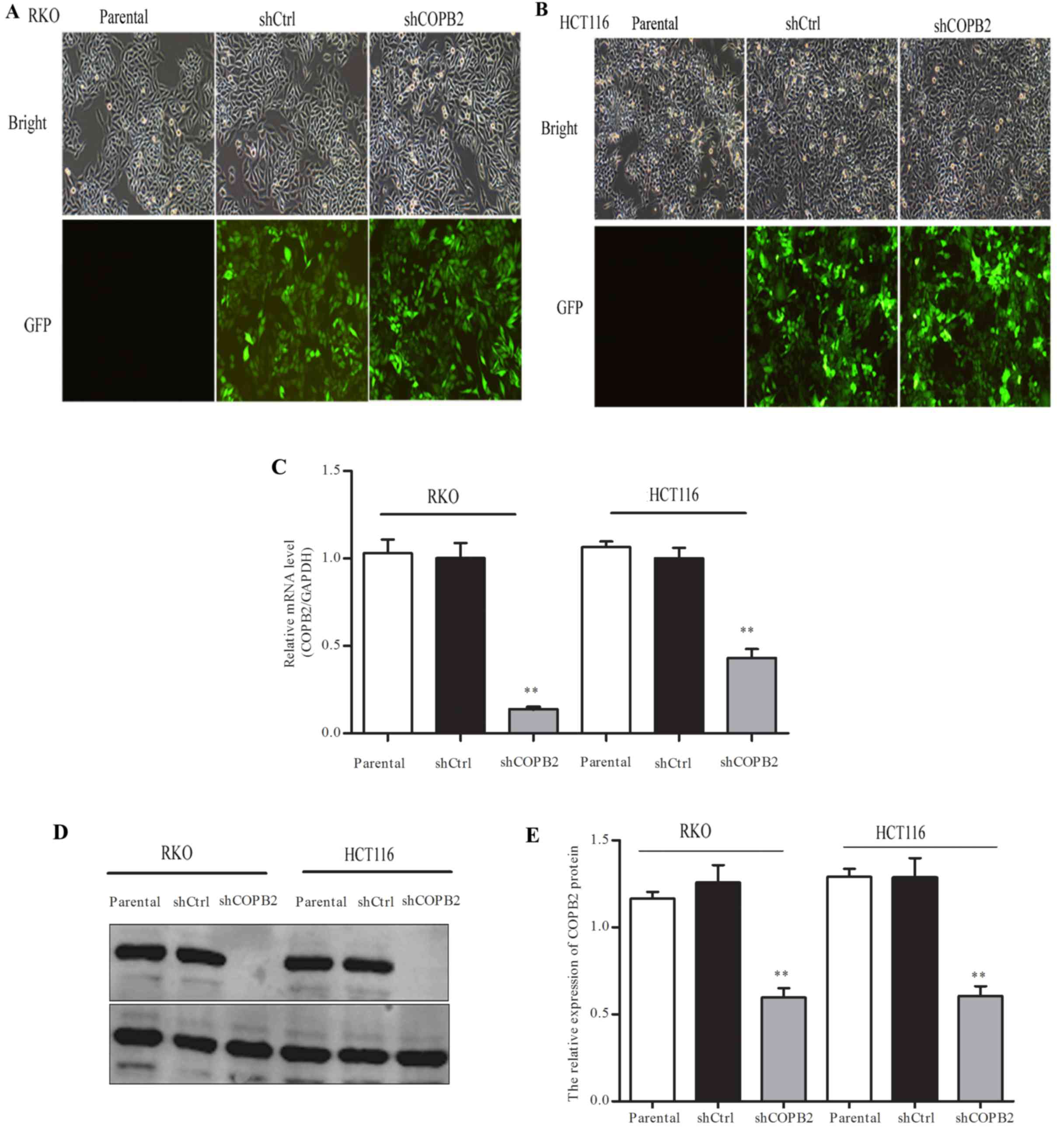
The RKO and HCT116 cells were infected with
lentiviral-shCOPB2 or -shCtrl. From the level of GFP fluorescence,
it was observed that the infection rate was >80% in each cell
line (Fig. 3A and B). Moreover,
RT-qPCR and western blotting showed that the mRNA level and protein
level of COPB2 were significantly reduced after the cells were
infected with shCOPB2 when compared with the shCtrl and parental
groups (P<0.01; Fig. 3C-E). These
results suggested that lentivirus-mediated RNAi was able to
markedly silence the expression of COPB2 in the RKO and HCT116
cells.
COPB2 silencing inhibits the
proliferation and colony formation abilities of RKO and HCT116
cells
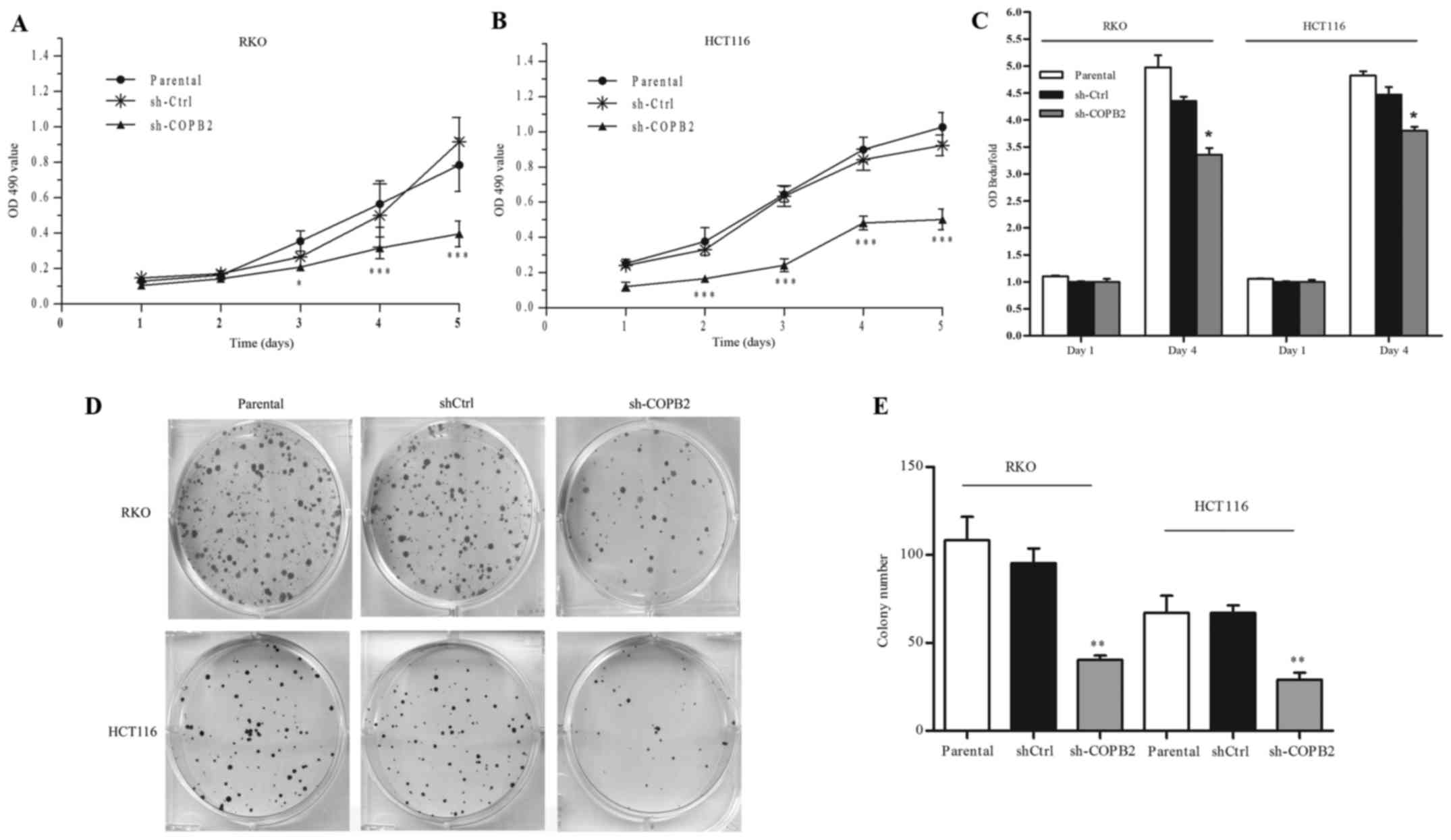
To evaluate the influence of COPB2 on the
proliferation of RKO and HCT116 cells, an MTT assay was performed.
As shown in Fig. 4A and B, we found
that cell growth was significantly reduced after infection with
lentivirus (P<0.001). Subsequently, cell proliferation ability
was assessed based on BrdU incorporation into cellular DNA. As
shown in Fig. 4C, compared with the
shCtrl and parental groups, the rates of DNA synthesis in RKO and
HCT116 cells following shCOPB2 transduction were significantly
reduced (P<0.05). Additionally, results of the colony formation
assay demonstrated that the colony numbers of RKO and HCT116 cells
were reduced following shCOPB2 transduction (P<0.01; Fig. 4D and E). Based on these results, we
concluded that COPB2 silencing inhibited the proliferation and
colony-formation abilities of the human colon cancer cells.
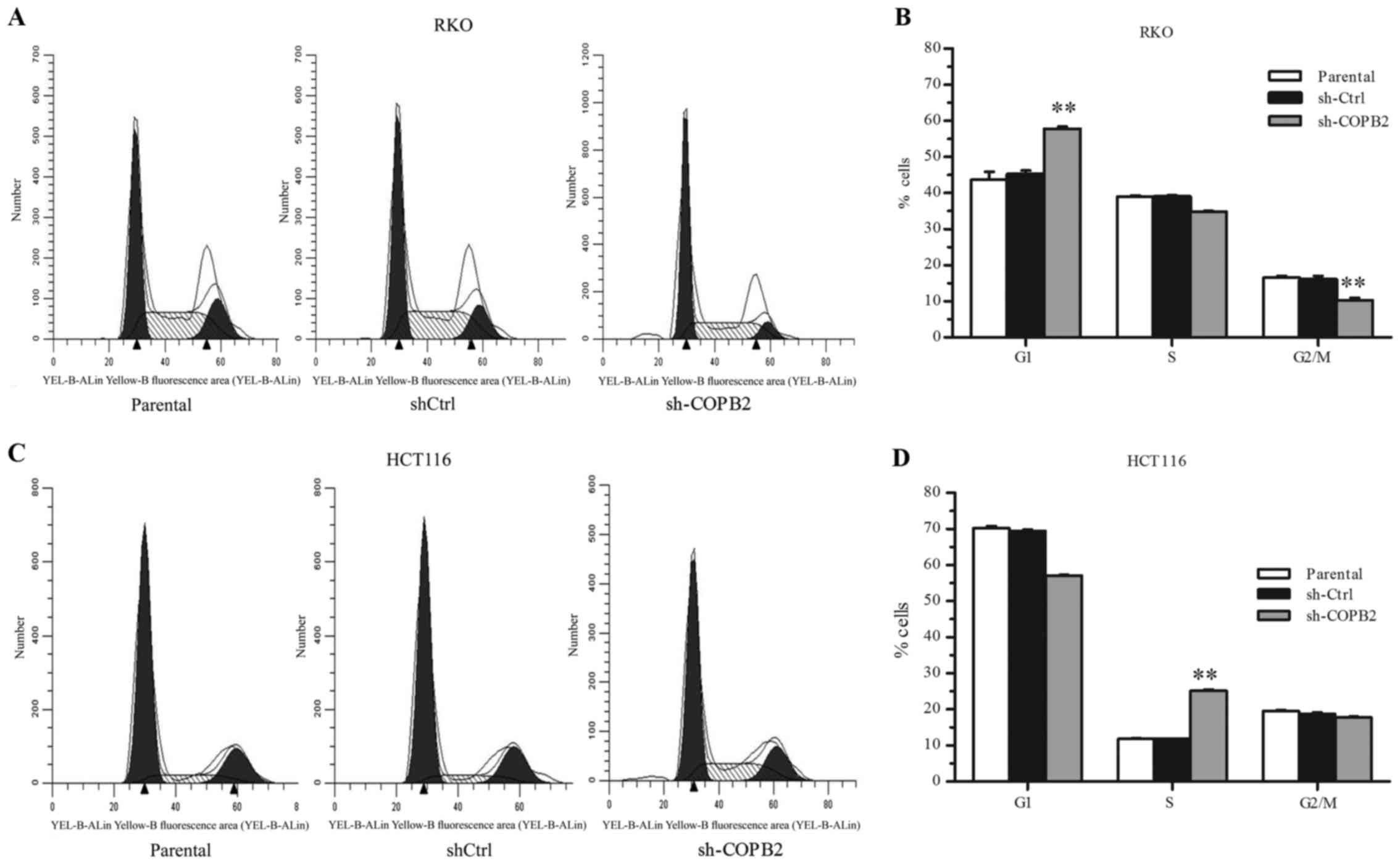
COPB2 silencing blocks cell cycle
progression in RKO and HCT116 cells
To investigate the mechanism of cell growth
inhibition mediated by COPB2 silencing, flow cytometric analysis
was performed to analyze the cell cycling of RKO and HCT116 cells
following lentivirus infection. In RKO-shCOPB2 cells, it was
observed that the percentage of cells in the G1 phase increased,
while the percentages of cells in the S phase and G2/M decreased
(P<0.01; Fig. 5A and B). These
results indicated that the silencing of COPB2 induced G1 phase
arrest and inhibited cell cycle progression in the RKO cells. By
contrast, in HCT116 cells, the percentage of cells in the S phase
significantly increased following the silencing of COPB2
(P<0.01; Fig. 5C and D). This
indicated that the HCT116 cells were arrested at the S phase
following COPB2 silencing.
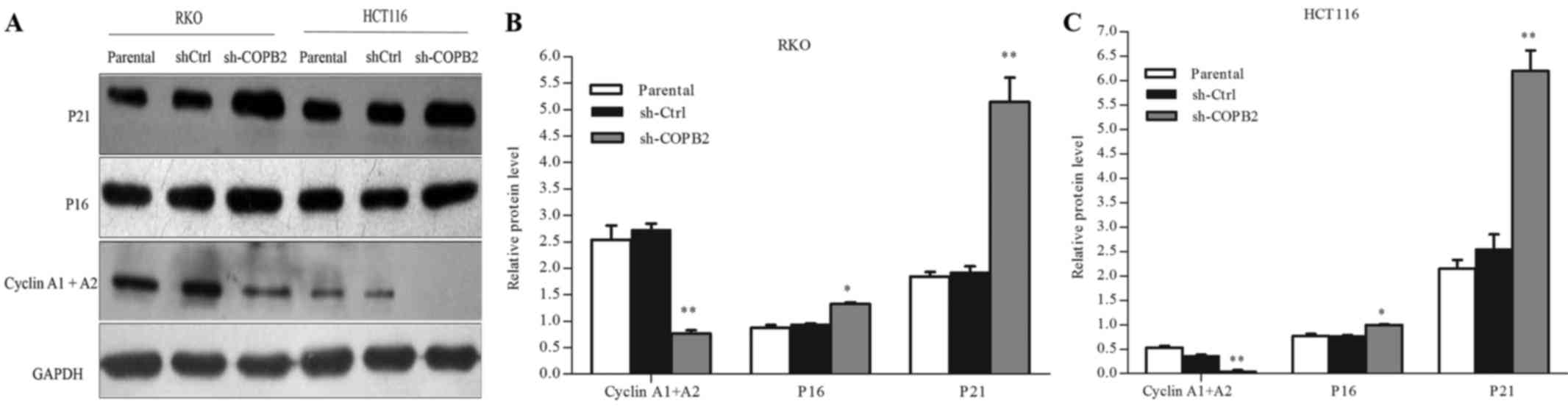
COPB2 silencing affects the expression
of cell cycle-related proteins and cyclin-dependent kinases
inhibitors
To evaluate the potential mechanism underlying the
role of COPB2 in cell cycle arrest, western blot analysis was used
to assess the protein expression levels of cell cycle-related
proteins and cyclin-dependent kinases (CDK) inhibitors. As shown in
Fig. 6A-C, silencing of COPB2
increased the protein levels of P16 (CDK inhibitor 2A) and P21 (CDK
inhibitor 1A) (P<0.01 and P<0.05, respectively), and
decreased those of cyclin A1 and A2 (P<0.01). These results
illustrated that COPB2 may be involved in regulating the expression
of cell cycle-related proteins and CDK inhibitors in RKO and HCT116
cells.
Discussion
Comprehensive tumor treatments, involving surgery,
chemotherapy, radiotherapy and biotherapy, are becoming more and
more important (17,18). Biological therapy for cancer differs
from traditional treatment methods. It comprises agents that by
virtue of their unique mechanisms of action are able to
specifically incite a response against or target malignant cells,
and is considered to be an effective method that can be used at
each stage of tumor development (19,20).
Intracellular transport is important for the functioning of cells,
and COPs play an important role in intracellular transport.
Therefore, COPs may be biological therapeutic targets in the
treatment of human colon cancer. Sudo et al (21) reported that COPA knockdown induced
apoptosis and suppressed tumor growth in a mesothelioma mouse
model, and thus we hypothesized that COPB2, which was first
reported as a novel subunit of the coatomer in 1993 (22), may be correlated with the
histogenesis and development of cancer. Mi et al (23) confirmed that COPB2 was over expressed
in human prostate cancer and promoted the proliferation of PC-3
cells. These previous findings led us to speculate that COPB2
mightv have functional impacts on other types of cancer cells. In
the present study, we first identified that COPB2 was highly
expressed in human CRC specimens and in six different colon cancer
cell lines. We subsequently investigated the biological function of
COPB2 in RKO and HCT116 cells, by using a lentivirus-medicated RNAi
method to effectively suppress the expression of COPB2 in the two
types of colon cancer cells. We confirmed that the expression of
COPB2 was associated with the proliferation and colony-formation
abilities of the RKO and HCT116 cells. Furthermore, COPB2 was
indicated to regulate the cell cycle; the knockdown of COPB2
resulted in cell cycle arrest in the tumor cells, and by western
blotting, we observed that the protein levels of Cyclin A were
reduced, with those of P16 and P21 were increased.
However, there were some unresolved factors in the
current study, which we will aim to address in future studies. In
particular, the molecular mechanism regarding the influence of
COPB2 on the proliferation of colon cancer cells, and whether the
COPB2 gene is related to tumor migration and invasion, remain
unknown and warrant further investigation. Additionally, the
function of the COPB2 gene will be further verified in
tumor-bearing mice.
In conclusion, COPB2 was indicated to be essential
for the growth of RKO and HCT116 cells through potential regulatory
effects on cell cycle progression. Lentivirus-mediated knockdown of
COPB2 could be a potential therapeutic approach for the treatment
of human CRC.
Acknowledgements
The authors would like to express their sincere
gratitude to all those who lent their support during the production
of this paper. This manuscript was supported by the National
Science Foundation of China [(grant no. 81672399(LM)].
References
|
1
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China. CA
Cancer J Clin. 66:115–132. 2015. View Article : Google Scholar
|
|
5
|
Wang S, Zhai Y, Pang X, Niu T, Ding YH,
Dong MQ, Hsu VW, Sun Z and Sun F: Structural characterization of
coatomer in its cytosolic state. Protein Cell. 7:586–600. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hara-Kuge S, Kuge O, Orci L, Amherdt M,
Ravazzola M, Wieland FT and Rothman JE: En Bloc incorporation of
coatomer subunits during the assembly of COP-coated vesicles. J
Cell Biol. 124:883–892. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuge O, Hara-Kuge S, Orci L, Ravazzola M,
Amherdt M, Tanigawa G, Wieland FT and Rothman JE: zeta-COP, a
subunit of coatomer, is required for COP-coated vesicle assembly. J
Cell Biol. 123:1727–1734. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orcl L, Palmer DJ, Amherdt M and Rothman
JE: Coated vesicle assembly in the Golgi requires only coatomer and
ARF proteins from the cytosol. Nature. 364:732–734. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Presley JF, Ward TH, Pfeifer AC, Siggia
ED, Phair RD and Lippincott-Schwartz J: Dissection of COPI and Arf1
dynamics in vivo and role in Golgi membrane transport. Nature.
417:187–193. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Beck R, Rawet M, Wieland FT and Cassel D:
The COPI system: Molecular mechanisms and function. FEBS Lett.
583:2701–2709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee MC, Miller EA, Goldberg J, Orci L and
Schekman R: Bi-directional protein transport between the ER and
Golgi. Annu Rev Cell Dev Biol. 20:87–123. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ruokun C, Yake X, Fengdong Y, Xinting W,
Laijun S and Xianzhi L: Lentivirus-mediated silencing of HSDL2
suppresses cell proliferation in human gliomas. Tumour Biol.
37:15065–15077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang D, Sun SQ, Yu YH, Wu WZ, Yang SL and
Tan JM: Suppression of SCIN inhibits human prostate cancer cell
proliferation and induces G0/G1 phase arrest. Int J Oncol.
44:161–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bie CQ, Liu XY, Cao MR, Huang QY, Tang HJ,
Wang M, Cao GL, Yi TZ, Wu SL, Xu WJ and Tang SH:
Lentivirus-mediated RNAi knockdown of insulin-like growth factor-1
receptor inhibits the growth and invasion of hepatocellular
carcinoma via down-regulating midkine expression. Oncotarget.
7:79305–79318. 2016.PubMed/NCBI
|
|
15
|
Tomuleasa C, Soritau O, Fischer-Fodor E,
Pop T, Susman S, Mosteanu O, Petrushev B, Aldea M, Acalovschi M,
Irimie A and Kacso G: Arsenic trioxide plus cisplatin/interferon
α-2b/doxorubicin/capecitabine combination chemotherapy for
unresectable hepatocellular carcinoma. Hematol Oncol Stem Cell
Ther. 4:60–66. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soritau O, Tomuleasa C, Aldea M, Petrushev
B, Susman S, Gheban D, Ioani H, Cosis A, Brie I, Irimie A, et al:
Metformin plus temozolomide-based chemotherapy as adjuvant
treatment for WHO grade III and IV malignant gliomas. J BUON.
16:282–289. 2011.PubMed/NCBI
|
|
17
|
Buck EA, Haley JD, Thomson S, Mulvihill
MJ, Epstein DM and Miglarese MR: Combination anti-cancer therapy WO
patent application 2013152252. October 10–2013
|
|
18
|
Zhang Z, Wang T, Liu Z, Tang S, Yue M,
Feng S, Hu M, Xuan L and Chen Y: Small interfering RNA targeting of
the survivin gene inhibits human tumor cell growth in vitro. Exp
Ther Med. 14:35–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y and Huang S: Discuss the strengths
and weaknesses between tumour biotherapy and chemoradiotherapy. Med
Infor. 24:3503–3504. 2011.
|
|
20
|
Arora N, Gupta A and Singh PP: Biological
agents in gastrointestinal cancers: Adverse effects and their
management. J Gastrointest Oncol. 8:485–498. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sudo H, Tsuji AB, Sugyo A, Kohda M, Sogawa
C, Yoshida C, Harada YN, Hino O and Saga T: Knockdown of COPA,
identified by loss of function screen, induces apoptosis and
suppresses tumor growth in mesothelioma mouse model. Genomics.
95:210–216. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stenbeck G, Harter C, Brecht A, Herrmann
D, Lottspeich F, Orci L and Wieland FT: beta'-COP, a novel subunit
of coatomer. EMBO J. 12:2841–2855. 1993.PubMed/NCBI
|
|
23
|
Mi Y, Yu M, Zhang L, Sun C, Wei B, Ding W,
Zhu Y, Tang J, Xia G and Zhu L: COPB2 is upregulated in prostate
cancer and regulates PC-3 cell proliferation, cell cycle, and
apoptosis. Arch Med Res. 47:411–418. 2016. View Article : Google Scholar : PubMed/NCBI
|