Introduction
Nonalcoholic fatty liver disease (NAFLD) refers to a
type of complex disease with excessive fat accumulation in
hepatocytes that are the results of a variety of external and
internal causes (1). Diabetes
mellitus (DM) is closely related to NAFLD and can seriously
threatens people's health. The number of NAFLD patients with DM is
increasing yearly, however, its molecular mechanism of pathogenesis
remains unclear. NAFLD complicated with DM has been considered a
global health problem (2–4). Atherosclerosis manifests due to
abnormal blood lipid metabolism, and current studies show that it
is closely related to NAFLD complicated with DM (5). Previous studies have found that a
subtype of peroxisome proliferator activated receptors (PPARs),
peroxisome proliferator-activated receptor-α (PPAR-α), have the
effect of regulating glucose and lipid metabolism, thus
participating in the occurrence and regulation of hyperglycemia and
hyperlipidemia (6). A large number
of previous studies have shown that DNA methylation plays an
important role in metabolic diseases (7). There have been no report on whether
PPAR-α methylation plays a role in the pathogenesis of
atherosclerosis. In this study, NAFLD patients with DM were used as
subjects to determine the relationship between PPAR-α methylation
levels and atherosclerosis, providing a theoretical and
experimental basis for clarifying the pathogenesis of NAFLD
complicated with DM and finding new targets for the treatment of
atherosclerosis.
Materials and methods
Sample selection
Patients that received physical examination at Qilu
Hospital of Shandong University from May 2002 to April 2016 were
enrolled as the subjects of study, and their blood samples were
obtained. Subjects were divided into either the healthy control
group (group N, n=50), including 28 males and 22 females aged
between 32–51 years, or the NAFLD complicated with DM group (group
M, n=50), including 30 males and 20 females aged 38–62 years. All
subjects had complete clinical data. The study was approved by the
Ethics Committee of Qilu Hospital of Shandong University and
informed consents were signed by the patients and/or guardians.
Main reagents
The total cholesterol (TC), triglyceride (TG),
high-density lipoprotein (HDL) and low-density lipoprotein (LDL)
detection kits were obtained from Nanjing Jiancheng Bio-Engineering
Institute Co., Ltd. (Nanjing, China). The TRIzol total RNA
extraction kit and the polymerase chain reaction (PCR) reverse
transcription kit was obtained from Tiangen Biotech Co., Ltd.
(Beijing, China). The mouse anti-human PPAR-α monoclonal antibody
(dilution, 1:300; cat. no. sc-130640) and bovine anti-mouse IgG-HRP
secondary polyclonal antibody (dilution, 1:1,000; cat. no. sc-2371)
was obtained from Santa Cruz Biotechnology (Philadelphia, PA,
USA).
General data collection
The general information of the subjects, including
sex, age, height, weight, past histories of drinking and
hyperlipidemia, and a history of DM, hypertension and other
disorders were collected using medical records combined with a
questionnaire survey. The levels of TC, TG, HDL, LDL and liver
functions were detected.
Determination of biochemical
indexes
A total of 3 ml of fasting peripheral venous blood
were drawn in the early morning, placed in a coagulation-promoting
tube, and centrifuged for 6 min at 2,680 × g after natural
solidification. TC, TG, HDL, LDL and other indexes of the subjects
were detected.
Extraction of DNA in blood cells
The fasting whole blood was drawn in the early
morning from the patients, and the anticoagulant tube was used to
avoid hemolysis. After being placed at room temperature for 1 h,
the whole blood samples were transferred to the polypropylene EP
tube. All samples were uniformly encoded and stored in a cryogenic
refrigerator at −80°C. The blood cell sample library was built for
the PCR detection of PPAR-α.
PCR
A PCR instrument was used to amplify the target
gene's forward primer 5′-AGTAGGGGCGGGTATGGTTTTTG-3′, and reverse
primer 5′-ACCTCCTCAATAAACCCAACTCTACTACTC-3′, and the target
fragment size was 133 bp. Amplification conditions were as follows:
Preheating at 94°C for 5 min, 94°C, 58°C and 72°C for 30 sec, a
total of 40 cycles, and extension at 72°C for 10 min. The optical
density ratio of PPAR-α to the corresponding internal control,
β-actin, was detected using an agarose gel electrophoresis, and the
relative content of PCR products was determined.
Immunofluorescent staining
After hepatic tissues in N and M groups were fixed
with 10% formaldehyde for 48 h, they were embedded into paraffin
and prepared into 5 µm-thick section slides. Paraffin sections were
dewaxed with xylene, dehydrated with alcohol at a gradient
concentration and repaired with antigens. Then, the sections were
rinsed with 0.01 M polybutylene succinate (PBS) (pH 7.4) 3 times (5
min/time). Then, the sections were stored in a 10% BSA wet box for
30 min at 37°C. The appropriately diluted (1:70)
fluorescence-labeled antibodies was dropped onto the sections and
placed in a wet box for incubation overnight at 4°C. After being
washed with PBS 3 times (5 min/time), the fluorescence secondary
antibodies (diluted at 1:100) were dropped and incubated for 2 h in
a wet box at 37°C. Finally, sections were sealed using buffered
glycerinum, followed by observation under the fluorescent
microscope (Olympus, Tokyo, Japan).
Statistical analysis
The experimental data were presented as mean ±
standard deviation (mean ± SD). The experimental results were
analyzed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA) statistical
software. The means between both groups were compared using the
t-test. The mean among the groups were compared using a one-way
analysis of variance (ANOVA). The p-test was used for pairwise
comparison. P<0.05 was considered to indicate statistically
significant differences.
Results
Comparison of subjects
The height and body weight of group M were on
average 162.00±6.12 and 64.21±7.31, respectively. Subjects in group
N had a height and body weight of 165.22±8.21 and 62.97±9.21,
respectively. Differences between the two groups were not
statistically significant (P>0.05). BMI in group M was
24.69±2.93, which was significantly higher than that in group N
(23.31±2.14) (P<0.05).
Biochemical indexes
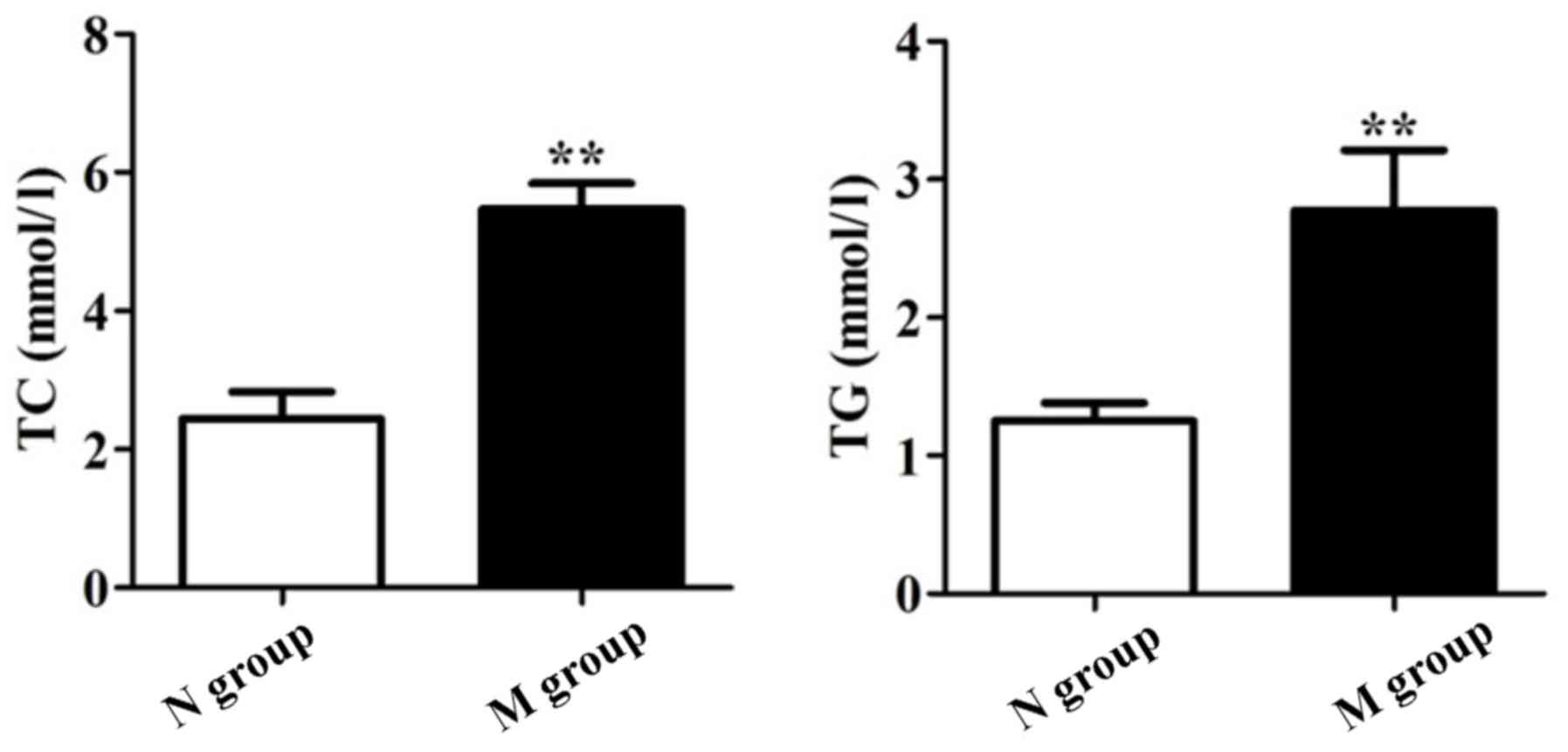
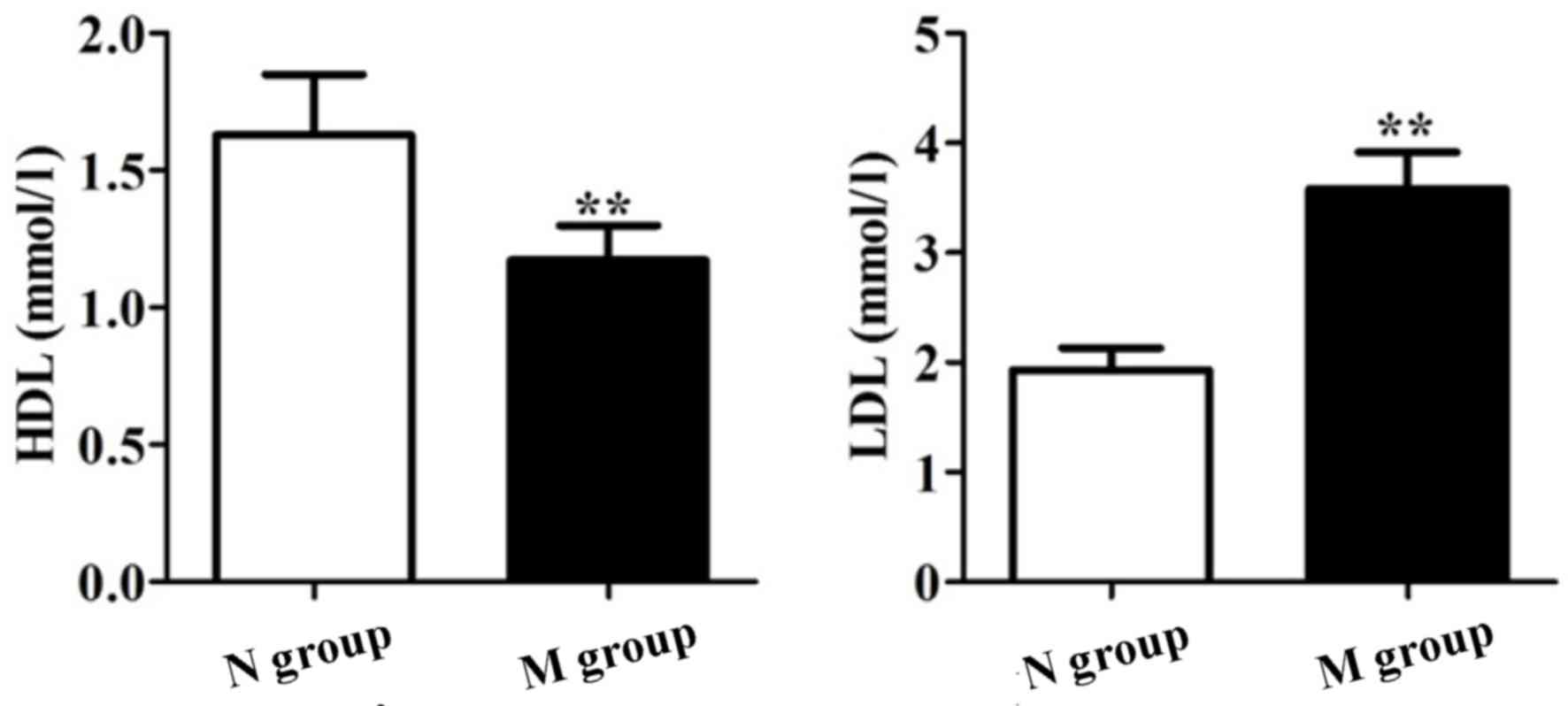
As shown in Figs. 1
and 2, TC, TG, HDL and LDL were
significantly different between groups N and M (P<0.05). Our
results showed that patients in group M suffered from dyslipidemia,
and NAFLD patients with DM suffered from atherosclerosis compared
to group N.
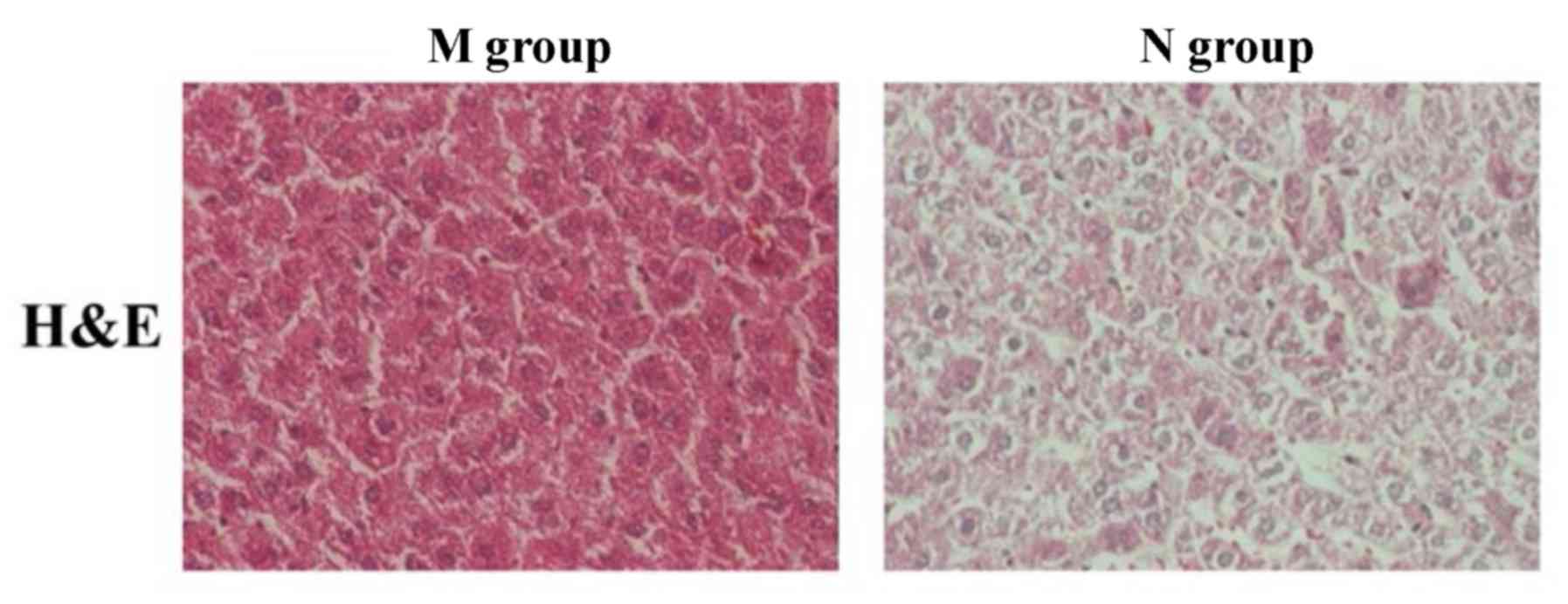
Observation of pathological situation
via H&E staining
Hematoxylin and eosin (H&E) stained sections of
normal liver tissues and liver tissues of NAFLD patients with DM
were used to determine the pathological differences between each
sample. Compared to normal liver tissue sections, substantial fatty
degeneration occurred, lipid droplets were visible in the liver,
liver tissue structures were changed and a large number of
hepatocytes had swelling and injury in group M. As shown in
Fig. 3, there was a significant
difference in the histopathology between normal liver tissues and
liver tissues of NAFLD patients with DM.
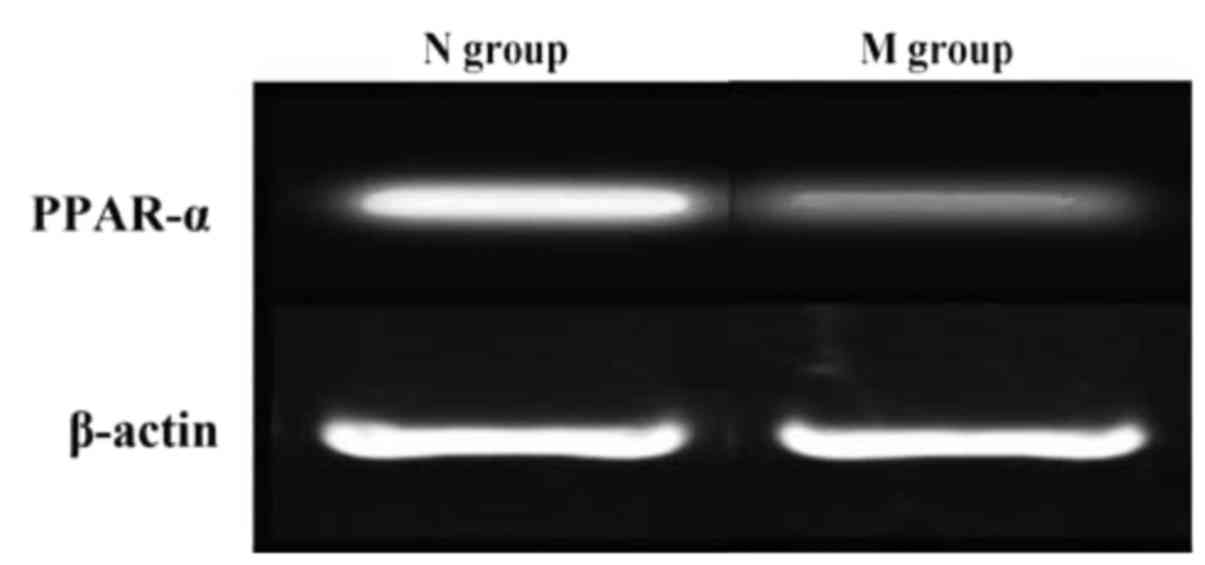
Reverse transcription-polymerase chain
reaction results
RNA was extracted from normal liver tissues and
liver tissues of NAFLD patients with DM. PCR amplification showed
that the expression of PPAR-α in normal liver tissue was
significantly higher than that in liver tissues of NAFLD patients
with DM (Fig. 4).
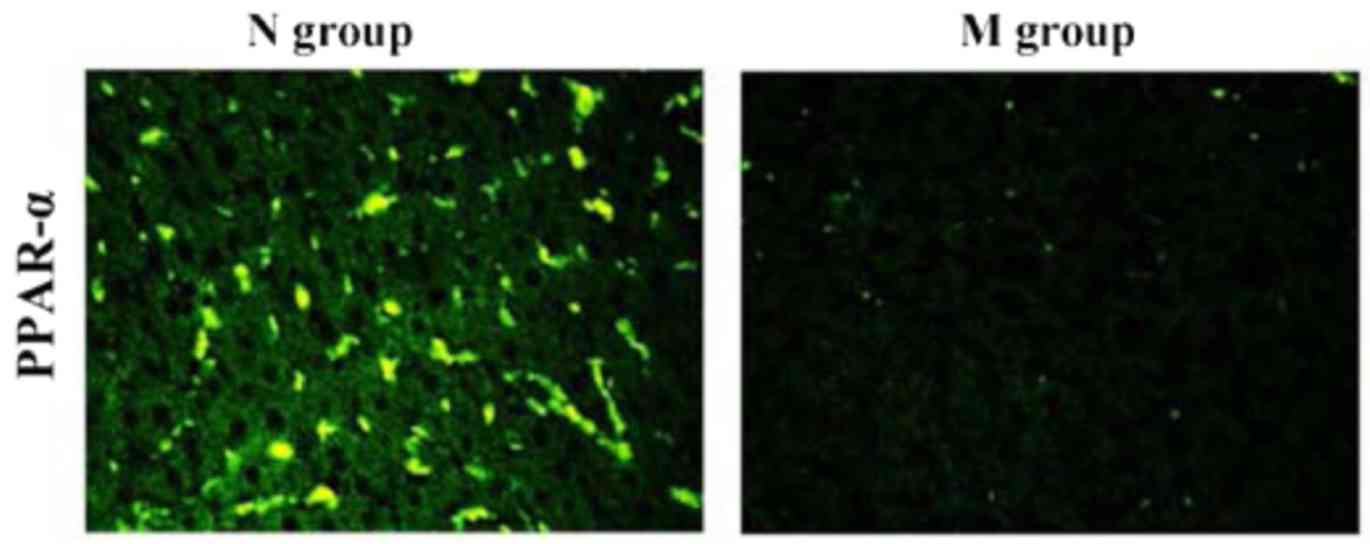
Immunofluorescence staining
results
As shown in Fig. 5,
PPAR-α was highly expressed in normal liver tissues, but rarely
expressed in liver tissues of NAFLD patients with DM. Our results
suggested that PPAR-α plays an important role in the development
and progression of atherosclerosis induced by NAFLD complicated
with DM.
Discussion
Improvement in living standards in previous years
have led to changes in the dietary structure and lifestyle.
Currently, the incidence rate of NAFLD and DM have also shown a
significantly rising trend (8,9). NAFLD
complicated with DM is a complex disease that affects fat
metabolism and glucose metabolism due to a variety of factors,
including external and internal causes (10). It may gradually develop into more
serious diseases or even threaten life and health if left
untreated. Therefore, research on prevention and treatment of NAFLD
complicated with DM has increasingly become a research hotspot and
focus for all of society and the medical community. NAFLD patients
with DM suffer from hyperglycemia, hyperlipidemia, high
cholesterol, insulin resistance and other pathological symptoms
(11). In the long term, the
cardiovascular system may suffer, and NAFLD complicated with DM is
closely related to the occurrence of atherosclerosis (12). However, the therapeutic approach of
NAFLD complicated with DM remains to be studied, and the molecular
mechanism of its pathogenesis is also unclear. Therefore, more
studies are needed to explore this field.
PPAR is a class of transcription factors that are
activated by a ligand, which belong to the nuclear receptor
superfamily (13). There are three
subtypes of PPARs that are encoded by different genes, with
differing structures and functions. PPAR-α mainly exists in the
fat, liver, heart, kidney, stomach and duodenal mucosa. It is also
highly expressed in the pancreas islet (14). Previous findings have shown that
PPAR-α binds to a ligand and plays a role in a variety of
biological effects. PPAR-α has the effect of regulating
glucose-lipid metabolism, inflammation, immunity and cell
differentiation. PPAR-α-mediated fatty acid oxidation and fat
metabolism are particularly important (15). PPAR-α target gene is related to lipid
transport and metabolic pathways, which can adjust the expression
of fat absorption and metabolism-related genes with a close
relationship with the occurrence and development of a variety of
metabolic diseases (16). PPAR-α is
involved in lipid metabolism and regulation of inflammation and
cell differentiation through a variety of mechanisms. In this
manner, PPAR-α may play an important regulatory role in the
pathogenesis of NAFLD complicated with DM.
DNA methylation is one of the most characteristic
markers of epigenetics, as well as the most in-depth mechanism of
epigenetic research (17). The
active methyl is transferred to the C5 of the cytosine by catalysis
of DNA methyltransferase to form methylcystein. Therefore,
methylation is generally associated with gene silencing, whereas
demethylation can often re-activate silencing genes (18,19).
Previous findings have shown that DNA methylation is important in
metabolic diseases. Based on the findings, our results suggest that
methylation of PPAR-α plays an important role in the occurrence and
development of NAFLD complicated with DM (20). In the present study, the levels of
TC, TG, HDL and LDL in the diseases and control populations were
determined, and PCR and immunofluorescence were used to detect
PPAR-α expression in both groups. The role of PPAR-α methylation in
the pathogenesis of NAFLD complicated with DM and its correlation
with atherosclerosis were clarified, providing theoretical support
for targeted therapy of NAFLD complicated with DM.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vernon G, Baranova A and Younossi ZM:
Systematic review: The epidemiology and natural history of
non-alcoholic fatty liver disease and non-alcoholic steatohepatitis
in adults. Aliment Pharmacol Ther. 34:274–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Charlton M: Nonalcoholic fatty liver
disease: A review of current understanding and future impact. Clin
Gastroenterol Hepatol. 2:1048–1058. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lazo M, Hernaez R, Bonekamp S, Kamel IR,
Brancati FL, Guallar E and Clark JM: Non-alcoholic fatty liver
disease and mortality among US adults: Prospective cohort study.
BMJ. 343:d68912011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nestel PJ and Mensink RP: Perspective:
Nonalcoholic fatty liver disease and cardiovascular risk. Curr Opin
Lipidol. 24:1–3. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bhatia LS, Curzen NP, Calder PC and Byrne
CD: Non-alcoholic fatty liver disease: A new and important
cardiovascular risk factor. Eur Heart J. 33:1190–1200. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barger PM and Kelly DP: PPAR signaling in
the control of cardiac energy metabolism. Trends Cardiovasc Med.
10:238–245. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee SST, Pineau T, Drago J, Lee EJ, Owens
JW, Kroetz DL, Fernandez-Salguero PM, Westphal H and Gonzalez FJ:
Targeted disruption of the α isoform of the peroxisome
proliferator-activated receptor gene in mice results in abolishment
of the pleiotropic effects of peroxisome proliferators. Mol Cell
Biol. 15:3012–3022. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Targher G, Day CP and Bonora E: Risk of
cardiovascular disease in patients with nonalcoholic fatty liver
disease. N Engl J Med. 363:1341–1350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pacana T and Fuchs M: The cardiovascular
link to nonalcoholic fatty liver disease: A critical analysis. Clin
Liver Dis. 16:599–613. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu H, Liu H, Hu F, Zou L, Luo S and Sun L:
Independent association between nonalcoholic fatty liver disease
and cardiovascular disease: A systematic review and meta-analysis.
Int J Endocrinol. 2013:1249582013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Villanova N, Moscatiello S, Ramilli S,
Bugianesi E, Magalotti D, Vanni E, Zoli M and Marchesini G:
Endothelial dysfunction and cardiovascular risk profile in
nonalcoholic fatty liver disease. Hepatology. 42:473–480. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akabame S, Hamaguchi M, Tomiyasu K, Tanaka
M, Kobayashi-Takenaka Y, Nakano K, Oda Y and Yoshikawa T:
Evaluation of vulnerable coronary plaques and non-alcoholic fatty
liver disease (NAFLD) by 64-detector multislice computed tomography
(MSCT). Circ J. 72:618–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Djouadi F, Weinheimer CJ, Saffitz JE,
Pitchford C, Bastin J, Gonzalez FJ and Kelly DP: A gender-related
defect in lipid metabolism and glucose homeostasis in peroxisome
proliferator-activated receptor α-deficient mice. J Clin Invest.
102:1083–1091. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aoyama T, Peters JM, Iritani N, Nakajima
T, Furihata K, Hashimoto T and Gonzalez FJ: Altered constitutive
expression of fatty acid-metabolizing enzymes in mice lacking the
peroxisome proliferator-activated receptor α (PPARalpha). J Biol
Chem. 273:5678–5684. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leone TC, Weinheimer CJ and Kelly DP: A
critical role for the peroxisome proliferator-activated receptor
alpha (PPARalpha) in the cellular fasting response: The
PPARalpha-null mouse as a model of fatty acid oxidation disorders.
Proc Natl Acad Sci USA. 96:pp. 7473–7478. 1999; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kersten S, Seydoux J, Peters JM, Gonzalez
FJ, Desvergne B and Wahli W: Peroxisome proliferator-activated
receptor α mediates the adaptive response to fasting. J Clin
Invest. 103:1489–1498. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Niculescu MD and Zeisel SH: Diet, methyl
donors and DNA methylation: Interactions between dietary folate,
methionine and choline. J Nutr. 132:2333–2335. 2002. View Article : Google Scholar
|
|
19
|
Oakes CC, La Salle S, Robaire B and
Trasler JM: Evaluation of a quantitative DNA methylation analysis
technique using methylation-sensitive/dependent restriction enzymes
and real-time PCR. Epigenetics. 1:146–152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Contreras AV, Torres N and Tovar AR:
PPAR-α as a key nutritional and environmental sensor for metabolic
adaptation. Adv Nutr. 4:439–452. 2013. View Article : Google Scholar : PubMed/NCBI
|