Introduction
Spinal cord injury (SCI), an event associated with
permanent neurologic deficit (1,2), is
reported to affect 10–40/million individuals annually in developed
countries (3). In the United States,
there are ~253,000 cases of SCI, with ~11,000 new cases diagnosed
annually (3). The economic cost of
this disease in the United States alone is estimated to be billions
of dollars (4). Violence and
accidents are the two causes of SCI, with 26% of cases associated
with violence and the remaining associated with accidents (5).
The first mention of SCI is recorded to be in 1700
BC in the Edwin Smith Papyrus, an Egyptian medical text, describing
this disease as an ‘ailment not to be treated’, which demonstrates
the severity of SCI as it was considered an untreatable ailment
(6). SCI is a severe condition, with
mortality occurring in the majority of patients prior to any
primary hospital care, while patients treated at the hospital are
prone to morbidity and mortality (5). Neurological deficit and disabilities
resulting from SCI not only affect the sensory and motor
capabilities, but also has a strong impact on the physiological
condition of the patient (7). SCI is
categorized into primary SCI, defined as the damage inflicted at
the time of trauma, and secondary SCI, which is defined as the
body's response to the initial trauma (8).
Due to the significant decrease in the quality of
life of patients following SCI, investigation of the
pathophysiology and treatment of this disease is urgent. To date,
studies have focused on preventing secondary SCI, promoting
regeneration and replacing the destroyed spinal cord tissue
(9,10). Methylprednisolone is the only current
agent approved for the treatment of primary SCI that is widely
used, although it presents selective/limited efficacy and
significant side effects (11–15). In
order to overcome the side effects of this glucocorticoid, the use
of poly (lactic-co-glycolic acid) nanoparticle formulations for the
encapsulation of methylprednisolone has been used (16). Novel therapeutic strategies are being
developed based on the basic approach involving the targeting of
the cascading mechanism that leads to secondary SCI (10). In this regard, altering the
neuro-inflammation (17), reducing
free-radical damage (18), reducing
excitotoxic damage to neurons (19),
improving the blood flow to the primary injury site (20) and countering the effect of ionic
exchange at the primary injury site (21) are the most commonly studied
therapeutic strategies.
Estrogen and its analogs have been widely
investigated as secondary SCI inhibitors, with previous studies
suggesting their anti-apoptotic effects, as well as their role in
decreasing the activation of cysteine protease, in the attenuation
of vascular endothelial growth factor and in aquaporin upregulation
(21–24). Estrogen has been reported to aid in
the improvement of SCI and promotion of the repair of the damaged
spinal cord by modulating the immune response (25). Thus, studies have been conducted on
boosting and/or modulating the immune response, and one such
approach involved the inhibition of the Ras/phosphatidylinositol
3-kinase (PI3K)/RAC α serine/threonine-protein kinase
(Akt)/serine/threonine-protein kinase mTOR (mTOR) signaling pathway
(22,26,27).
The involvement of Ras in the sympathetic neuronal
survival via the PI3K and dual specificity mitogen-activated
protein kinase kinase mek-1 signaling pathway is well established,
while its role in suppressing apoptosis by regulating the cellular
tumor antigen p53 pathway is known to benefit SCI treatment
(28–30). The majority of studies to date have
examined the Ras/PI3K/Akt/mTOR pathway as the neuroprotective and
neuroregenerative functions in SCI are regulated by this pathway,
and rapamycin inhibition of mTOR has been reported to exhibit a
positive effect on mice with SCI (31–33).
Although the role of the Ras/Raf/extracellular signal-regulated
kinase 1/2 (ERK1/2) signaling pathway in cell proliferation,
migration, differentiation and death is well documented (34–36), its
involvement in the treatment of SCI has not been widely
investigated. Studies have revealed that the U0126 inhibitor
functions as a potential drug for restoring SCI by affecting spinal
cord neurons (SCNs) via altering the Raf/ERK signaling pathway
(26,37–39).
The present study attempted to identify a novel
natural compound (Lead) involved in the alteration of the Raf/ERK
pathway for the regeneration of SCNs following SCI. The study used
the Interbioscreen (IBS) natural compound database (www.ibscreen.com) consisting of 48,531 natural
compounds, known to be largest collection of natural compounds,
their derivatives and mimetics worldwide. A multistep
structure-based virtual screening technique, followed by docking
and simulation, was employed for identifying a promising lead
compound, which is a widely used method for identifying such
compounds (40–42).
Materials and methods
Protein and compounds of natural
origin dataset preparation
The atomic coordinates for the Ras [Protein Data
Bank (PBD) ID, 4LPK] structure required for the development of a
lead compound were obtained from the PDB (rcsb.org/pdb/home/home.do). This crystal structure of
GTPase KRas is GDP-bound, and the coordinates of this Ras protein
were used. The energy-minimization of the structure was performed
by using the Swiss-PdbViewer software (version 4.1; Swiss Institute
of Bioinformatics, Lausanne, Switzerland). The root mean square
deviation (RMSD) was monitored using the GROMOS96 43b1 force field
(43). Approximately 48,531 natural
compounds from the IBS database were used for targeting the GTP
binding site of Ras.
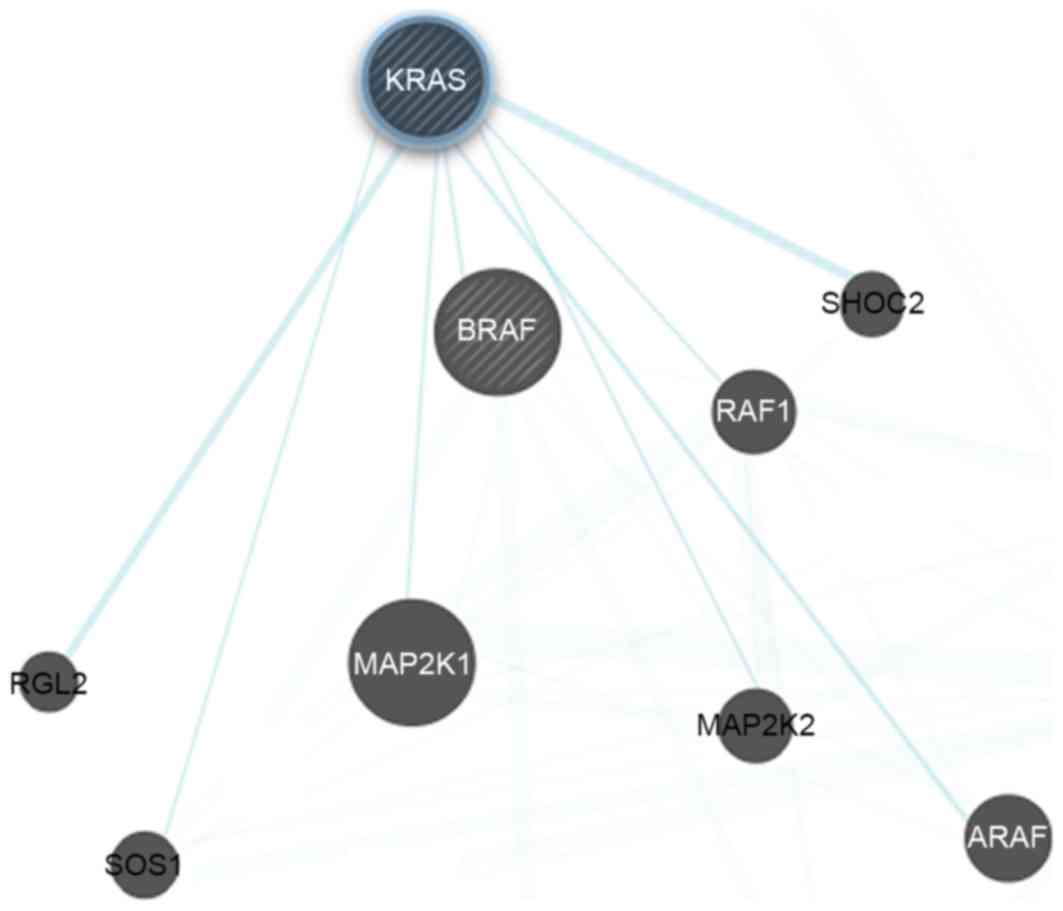
Protein network reconstruction
The Ras/PI3K/Akt/mTOR protein-protein interactions
networks were constructed using Cytoscape (version 3.5.1; National
Resource of Network Biology, San Francisco, CA, USA). The tool was
used for compiling protein functional classification, and physical
and genetic interaction networks.
Virtual screening and drug-likeliness
prediction
The ArgusLab suit was used for virtual screening of
compounds of natural origin (44).
In total, >1% (414) compounds were shortlisted according to the
binding energy (∆G) calculations. A ∆G of −7 kcal/mol was set as
the cut off to obtain the initial subset of compounds. This cut off
value was a limiting bias set in order to limit the number of
compounds, since a ∆G of −8 kcal/mol provided very few compounds
for further investigation and a value of −6 kcal/mol provided a
very large number of compounds. Thus, ∆G was set at −7 kcal/mol to
provide an acceptable number of compounds for further validation
(40–42). The number of selected compounds was
then further reduced by subjecting to the rules set by Lipinski
(45), which assesed the molecular
weight (MW), hydrogen bond acceptor (HBA), hydrogen bond donor
(HBD) and octanol/water partition coefficient (cLogP) of each
compound. Lipinski's rule of five (RO5) parameters provided five
compounds that were subjected to further analysis. Absorption,
distribution, metabolism and excretion (ADME) analysis was
performed using the PreADME/T online server
(preadmet.bmdrc.kr/adme/).
Molecular docking analysis
A structure-based drug designing method was used by
employing the AutoDock 4.2 tool for molecular docking analysis
(46). This tool calculates energy
values by classification into internal and torsional free energy.
The internal energy is the sum of the desolvation, hydrogen
bonding, van der Waals and electrostatic energies. The Lamarckian
genetic algorithm default parameters (46) were used for calculating the ∆G of
each shortlisted compound. A grid box (40×40×40 Å) was built around
the GTP binding site. Energy values were generated and the binding
mode with the GTP binding site was used to limit the investigated
compounds to a single molecule.
Molecular dynamics simulation
(MDS)
The single compound shortlisted was simulated for
100 nsec using MDS, which was conducted using the GROMACS suit
(47). From the compounds initially
identified, molecular docking identified the optimal compound as
lead3, which was then studied using MDS. To mimic in vitro
conditions, the Ras-Lead3 complex was kept in an in silico
neutral (no charge) water bath, where a water molecule was
represented by a simple point charge (SPC216). GROMOS 43a1 force
field for Ras was used for simulation, and the force field for the
compound was calculated using the PRODRG server (48). Energy minimization procedures for 1
nsec were performed using canonical [number of particles, volume
and temperature (NVT) are held constant] and isothermal-isobaric
[N, pressure and T (NPT) are held constant] ensembles. In NVT and
NPT ensembles the coupling scheme of Berendsen, SHAKE algorithm and
particle mesh Ewald method were used (49–51).
Next, the energy-minimized Ras-Lead3 complex was simulated for 100
nsec, and the trajectories generated were subjected to Molecular
Mechanics Poisson-Bolzmann Surface Area (MM-PBSA) calculations. The
g_mmpbsa tool developed for GROMACS, which was used for principal
component analysis (PCA) (52).
Snapshots of the coordinates and the total energies were obtained
after 500 psec, while 501 snapshots of the RAS-Lead3 complex were
also subjected to the MM-PBSA calculation. The binding free energy
(ΔGbind) was composed of the following species:
ΔGbind=Gcomplex–Gprotein–Gligand=ΔEMM+ΔGsol–TΔSΔEMM=ΔEval+ΔEele+ΔEvdwΔGsol=ΔGp+ΔGnpΔGnp=γSASA+β
In the aforementioned equations,
Gcomplex, Gprotein and Gligand
represent the free energy of the respective species.
ΔEMM refers to the gas phase energy, ΔGsol is
the solvation energy, TΔS represents an entropy term,
ΔEval is the sum of the internal energy of bonds, angle
and torsion, ΔEele is the electrostatic interaction,
ΔEvdw is the van der Waals interaction energy,
ΔGp is the polar salvation free energy, and finally
ΔGnp refers to the nonpolar salvation free energy.
Molecular visualization analysis
The RAS-Lead3 complex was investigated using the
visualization tools Pymol (version 1.8.6.2) (53) and Discovery Studio (version
16.1.015350) (54). For graph
construction, the Grace Program (version 5.1.22) (55) and Gnuplot (version 5.0.6) (56) were used.
Results and Discussion
A database comprised of natural compounds, their
derivatives and mimetics were considered in order to identify
compounds that would inhibit the Ras/Raf/ERK1/2 pathway by
inhibiting the GTP binding site of Ras (Fig. 1). The role of the Ras/Raf/ERK
signaling pathway in promoting apoptosis in neural cells in SCI is
well established (27). Ras
inhibitors modulate the intracellular signaling of Ras to exhibit a
pro-apoptotic or neuroprotective effect (31). The U0126 inhibitor demonstrates a
neuroprotective activity by suppressing ERK1/2 activation (57). The consistent evidence on the role of
ERK1/2 in neuronal apoptotic cell death provided an opportunity to
identify novel small molecule inhibitors using in silico
approaches (37). This signaling
cascade is triggered by the membrane receptor, which allows Ras to
swap GDP for a GTP and to become active. This activated Ras then
activates the kinase activity of Raf, which in turn phosphorylates
and activates ERKs (ERK1 and ERK2). In the present study, in order
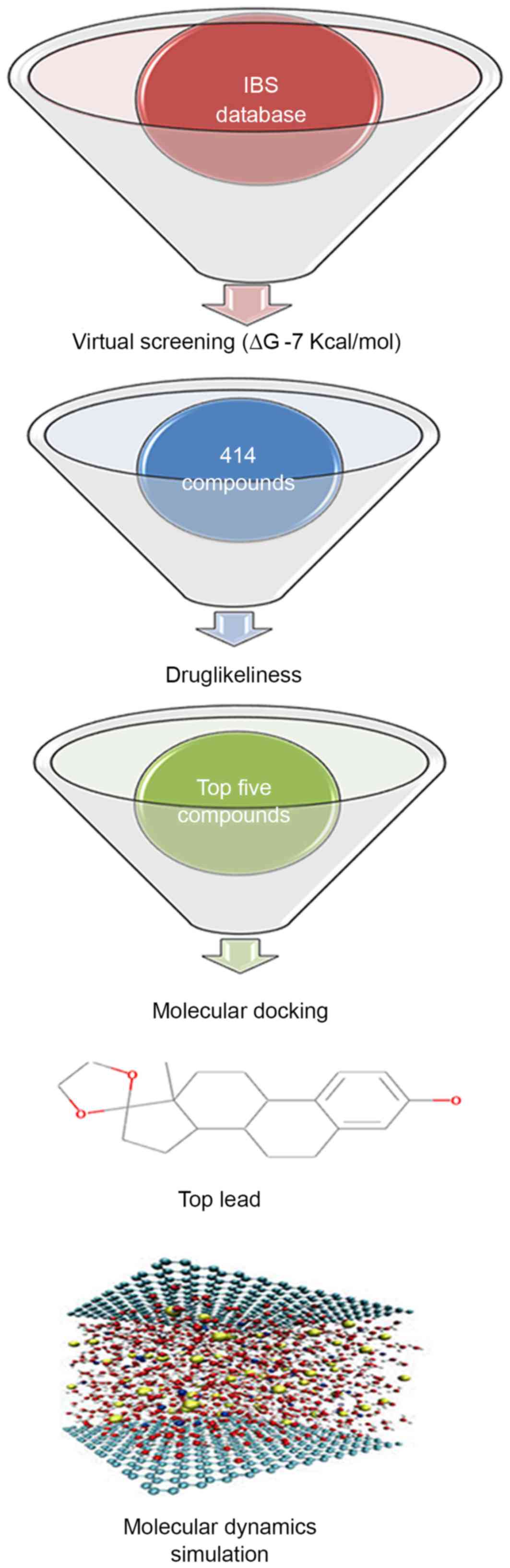
to identify a novel Ras inhibitor, virtual screening,
drug-likeliness, molecular docking analysis and MDS methods were
used, and this methodology is demonstrated in Fig. 2. Virtual screening helped to limit
the number of compounds from 48,531 natural products to 414
compounds using a limiting bias of ∆G of −7 kcal/mol.
In order to focus on compounds that may be promising
for further development, each of the top identified compounds was
examined for drug-likeliness. The drug-likeliness of the
shortlisted compounds was defined according to the mutagenic and
carcinogenic properties, Lipinski's RO5 and total polar surface
area (TPSA). The RO5 properties included the number of HBDs and
HBAs, the MW and cLogP, with the permissible range being HBD≤5,
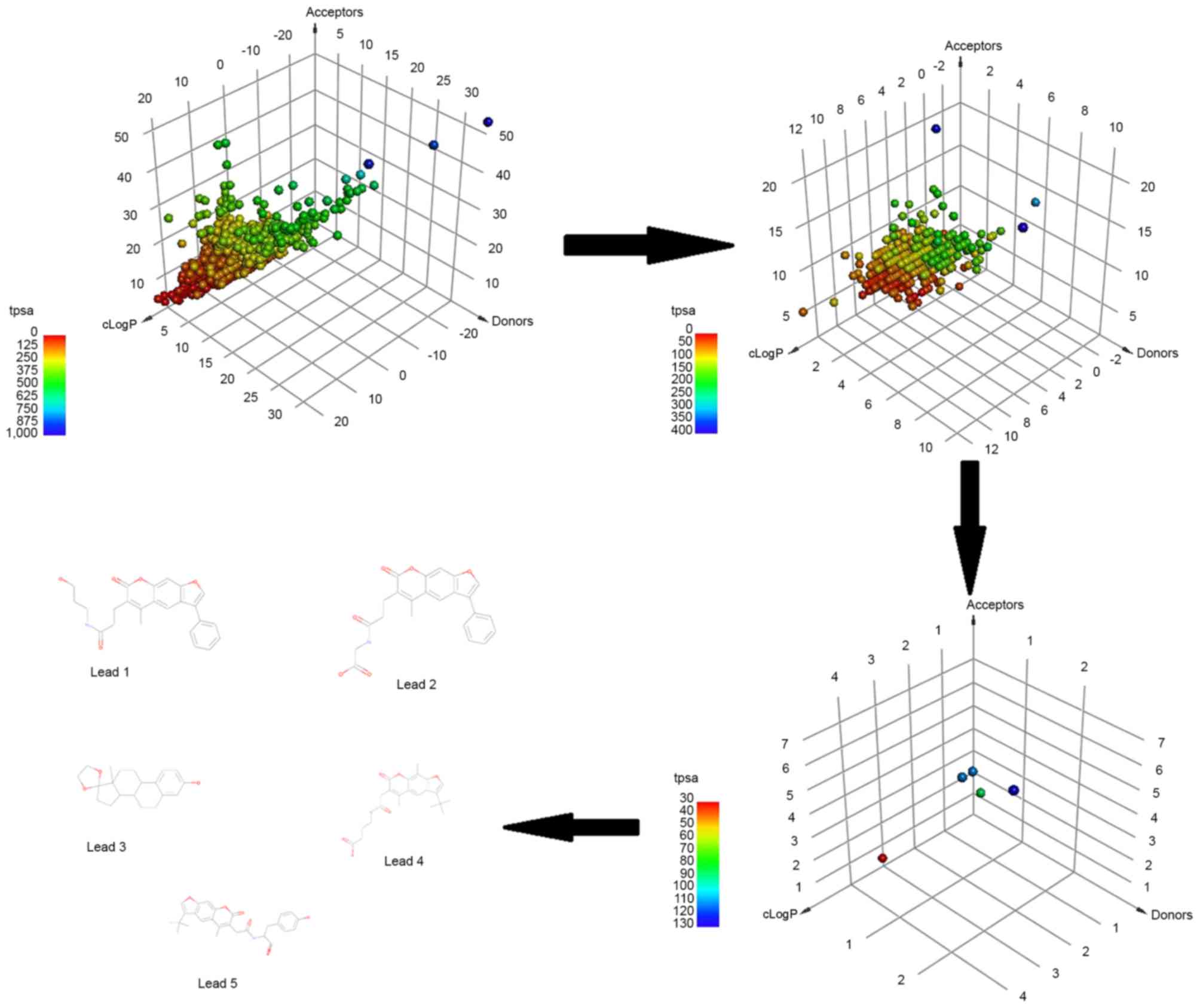
HBA≤10, MW≤500 Da and cLogP≤5. Fig.
3 represents the three-dimensional point plot of the HBA, HBD
and cLogP values. The coloring in the figure is according to the
TPSA.
Table I demonstrates
the drug-likeliness properties of the top five identified
compounds. The drug-likeliness values of these compound conformed
to the values expected from typical drugs. These five compounds
where subjected to ADME analysis. In silico methods were
used to calculate the Caco-2 cell permeability (Caco-2 p), MDCK
cell permeability, human intestinal absorption, plasma protein
binding (PPB) and blood-brain barrier (BBB), and the results
generated are shown in Table II.
The ADME properties of the shortlisted compounds were within the
acceptable range for known drugs available in the market.
 | Table I.Drug-likeliness of top five lead
compounds. |
Table I.
Drug-likeliness of top five lead
compounds.
| IBS no. | MW | cLogP | HBD, n | HBA, n | TPSA | Mutagenicity |
Carcinogenicity |
|---|
| 05678 | 314.419 | 4.41 | 1 | 3 | 38.69 | No | No |
| 46780 | 405.401 | 3.35 | 2 | 7 | 109.57 | No | No |
| 49515 | 413.183 | 3.65 | 2 | 7 | 109.75 | No | No |
| 49817 | 405.151 | 3.94 | 3 | 8 | 129.28 | No | No |
| 64118 | 405.121 | 3.12 | 2 | 6 | 96.28 | No | No |
 | Table II.Absorption, distribution, metabolism
and excretion properties calculated by in silico
approach. |
Table II.
Absorption, distribution, metabolism
and excretion properties calculated by in silico
approach.
| Name | IBS no. | Definition | Caco-2 p
(nm/sec) | MDCK p
(nm/sec) | HIA (%) | BBB (c.b/c.bl) | PPB PPB (%) |
|---|
| Lead1 | 64118 |
N-(3-hydroxypropyl)-3-(5-methyl-7-oxo-3-phenyl-7H-furo[3,2-g]chromen-6-yl)propanamide | 20.79 | 0.14 | 94.97 | 0.052 | 91.18 |
| Lead2 | 46780 |
2-(3-(5-methyl-7-oxo-3-phenyl-7H-furo[3,2-g]
chromen-6-yl)propanamido)acetic acid | 19.14 | 0.09 | 97.82 | 0.012 | 93.02 |
| Lead3 | 05678 |
(13S)-13-methyl-6,7,8,9,11,12,13,14,15,16-decahydrospiro[cyclopenta[a]phenanthrene-17,2′-[1,3]dioxolan]-3-ol | 43.61 | 56.29 | 95.62 | 3.620 | 100.00 |
| Lead4 | 49515 |
4-(2-(3-(tert-butyl)-5,9-dimethyl-7-oxo-7H-furo
[3,2-g]chromen-6-yl)acetamido)butanoic acid | 20.02 | 0.12 | 96.89 | 0.012 | 89.00 |
| Lead5 | 49817 |
(S)-2-(2-(3-(tert-butyl)-5-methyl-7-oxo-7H-furo
[3,2-g]chromen-6-yl)acetamido)-3-(4-hydroxyphenyl)propanoic
acid | 21.23 | 0.05 | 95.97 | 0.019 | 89.61 |
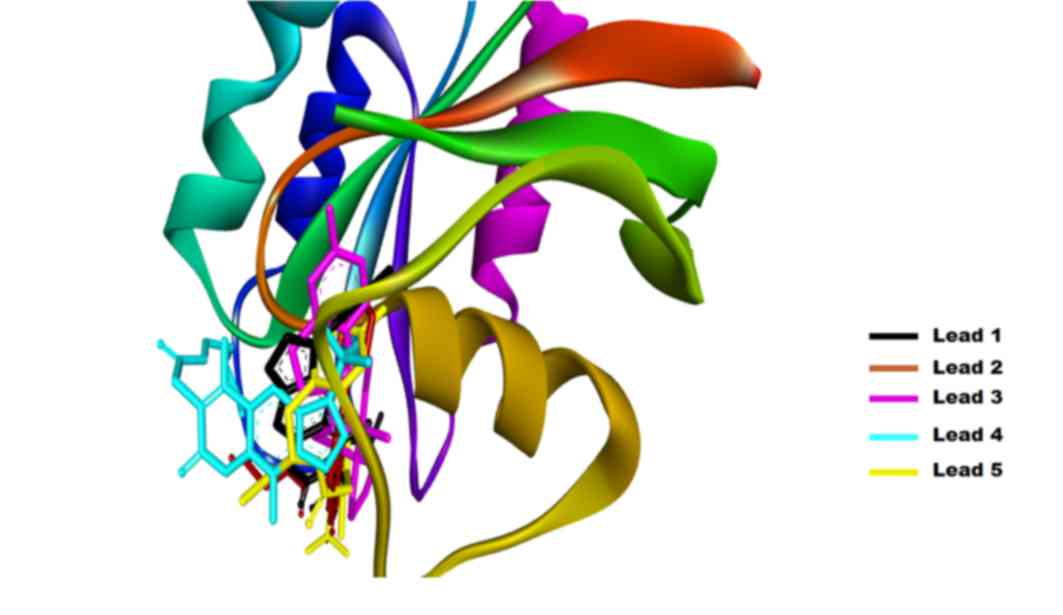
In the current study, the AutoDock tool was used for
molecular docking simulations, selecting the top binding pose based
on the ∆G for further analysis (Fig.
4). Each binding pose was analyzed using the Discovery Studio
software, and the default parameters were used to calculate all the
possible interactions. The results generated are listed in Table III, where the ∆G, binding pocket
and number of hydrogen bonds formed are demonstrated. The
interactions investigated included the van der Waals, conventional
hydrogen bond, carbon hydrogen bond, π-cation, π-donor hydrogen
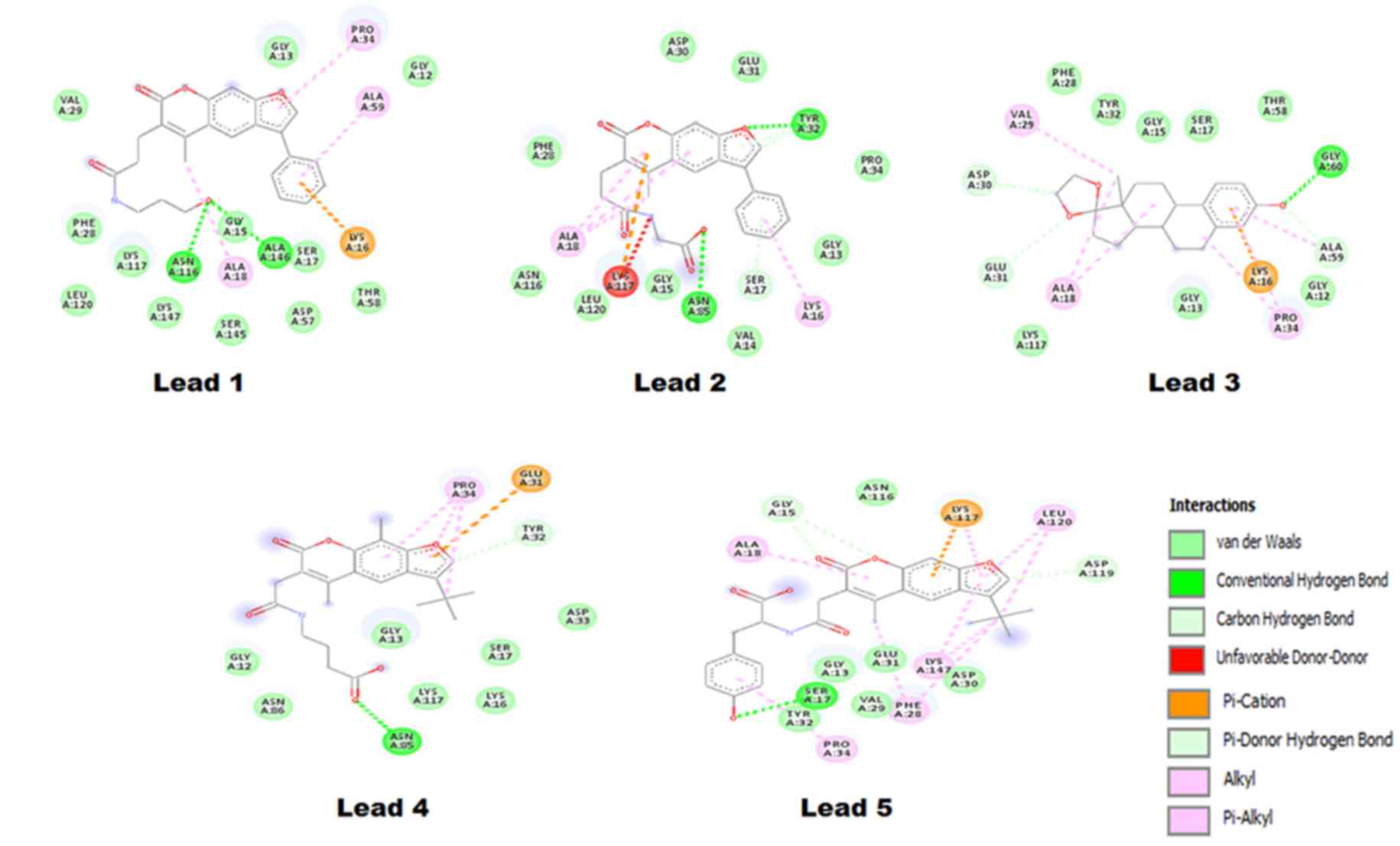
bond, alkyl and π-alkyl interactions. The Lead1-Ras complex
presented a ∆G of −6.12 kcal/mol and the interaction is shown in
Fig. 5, where Lead1 is forming two
conventional hydrogen bonds with ALA146 and ASN116 of the Ras GTP
binding site. The binding pocket of Lead1 comprises following amino
acids ALA59, GLY12, PRO34, GLY13, VAL29, PHE28, LEU120, LYS117,
ASN116, LYS147, GLY15, ALA18, SER145, ALA146, SER17, ASP57, LYS16
and THR58. The O23 atomic site of Lead1 shows hydrogen
bond interaction with ALA146 and ASN 116, with a distance between
the Lead and Ras of 2.14 and 1.97 Å, respectively.
 | Table III.Binding pose analysis of top five
identified compounds, including calculation of the ligand-binding
pocket and the hydrogen bond formation using Discovery Studio. |
Table III.
Binding pose analysis of top five
identified compounds, including calculation of the ligand-binding
pocket and the hydrogen bond formation using Discovery Studio.
| Name | IBS no. | ΔG (kcal/mol) | Ligand binding
pocket | H-bonds |
|---|
| Lead1 | 64118 | −6.27 | ALA59, GLY12,
PRO34, GLY13, VAL29, PHE28, LEU120, LYS117, ASN116, LYS147, GLY15,
ALA18, SER145, ALA146, SER17, ASP57, LYS16 and THR58 |
ALA146:HN-Lead1:O23 (2.14 Å)
ASN116:HD21-Lead1:O23 (1.97 Å) |
| Lead2 | 46780 | −7.72 | PRO34, TYR32,
GLU31, ASP30, PHE28, ALA18, ASN116, LEU120, LYS117, GLY15, ASN85,
VAL14, SER17, LYS16 and GLY13 |
TYR32:HN-Lead2:O12 (2.02 Å)
ASN85:HD21-Lead2:O25 (1.88 Å) |
| Lead3 | 05678 | −8.58 | GLY60, THR58,
SER17, GLY15, TYR32, PHE28, VAL29, ASP30, GLU31, ALA18, LYS117,
GLY13, LYS16, PRO34, GLY12 and ALA59 |
GLY60:HN-Lead3:O21 (2.11
Å) |
| Lead4 | 49515 | −5.47 | TYR32, GLU31,
PRO34, GLY12, ASN86, ASN85, GLY13, LYS117, LYS116, SER17 and
ASP33 |
ASN85:HD21-Lead4:O20 (2.07
Å) |
| Lead5 | 49817 | −6.08 | ASP119, LEU120,
LYS117, ASN116, GLY15, ALA18, TYR32, PRO34, SER17, GLY13, VAL29,
PHE28, GLU31, LYS147, PHE28 and ASP30 |
Lead5:O32-SER17:OG (2.58 Å)
SER17:HN-Lead5:O32 (1.68 Å) |
Lead2 exhibited two conventional hydrogen bond
interactions with the GTP binding pocket of Ras. Two atoms of
Lead2, O12 andO25, formed a bond with TYR32
and ASN85, with a bond length of 2.02 Å and 1.88 Å, respectively.
In addition, the binding pocket of Lead2 was comprised of 15 amino
acids, as follows: PRO34, TYR32, GLU31, ASP30, PHE28, ALA18,
ASN116, LEU120, LYS117, GLY15, ASN85, VAL14, SER17, LYS16 and
GLY13. Furthermore, Lead3 exhibited a ΔG of −8.58 kcal/mol, which
was the best reported value among the top five shortlisted
compounds. The binding pocket of the Lead3 molecule with a GTP
binding site of Ras comprises of the following amino acids: GLY60,
THR58, SER17, GLY15, TYR32, PHE28, VAL29, ASP30, GLU31, ALA18,
LYS117, GLY13, LYS16, PRO34, GLY12 and ALA59. Out of these, Lead3
formed hydrogen bond with GLY60, while the interaction of interest
in the present study was formed by the Lead3 O21
position.
Based on the ΔG and the number of interactions,
Lead4 was ranked last among the top five shortlisted compounds. Its
binding pocket was comprised of 11 amino acids, namely: TYR32,
GLU31, PRO34, GLY12, ASN86, ASN85, GLY13, LYS117, LYS116, SER17 and
ASP33. Lead4 presented a ΔG of −5.47 kcal/mol, and had a single
hydrogen bond interaction between the Lead4 O20 position
and Ras ASN85 (2.07 Å). The last Lead compound identified in the
current study was termed Lead5, which demonstrated a ΔG of −6.08
kcal/mol and formed a maximum number of interactions in the GTP
binding pocket of Ras. The binding pocket of Lead5 was comprised of
ASP119, LEU120, LYS117, ASN116, GLY15, ALA18, TYR32, PRO34, SER17,
GLY13, VAL29, PHE28, GLU31, LYS147, PHE28 and ASP30.
The analysis in the present study observed that
Lead3 was compound with the best BBB and PPB, as well as the lowest
ΔG value, and was thus selected for further analysis. In order to
explore the stability and binding mode of Lead3 with the GTP
binding site of Ras, MDS was performed for a 100 nsec run under
GROMOS 43a1 force field on the energy-minimized Lead3-Ras complex.
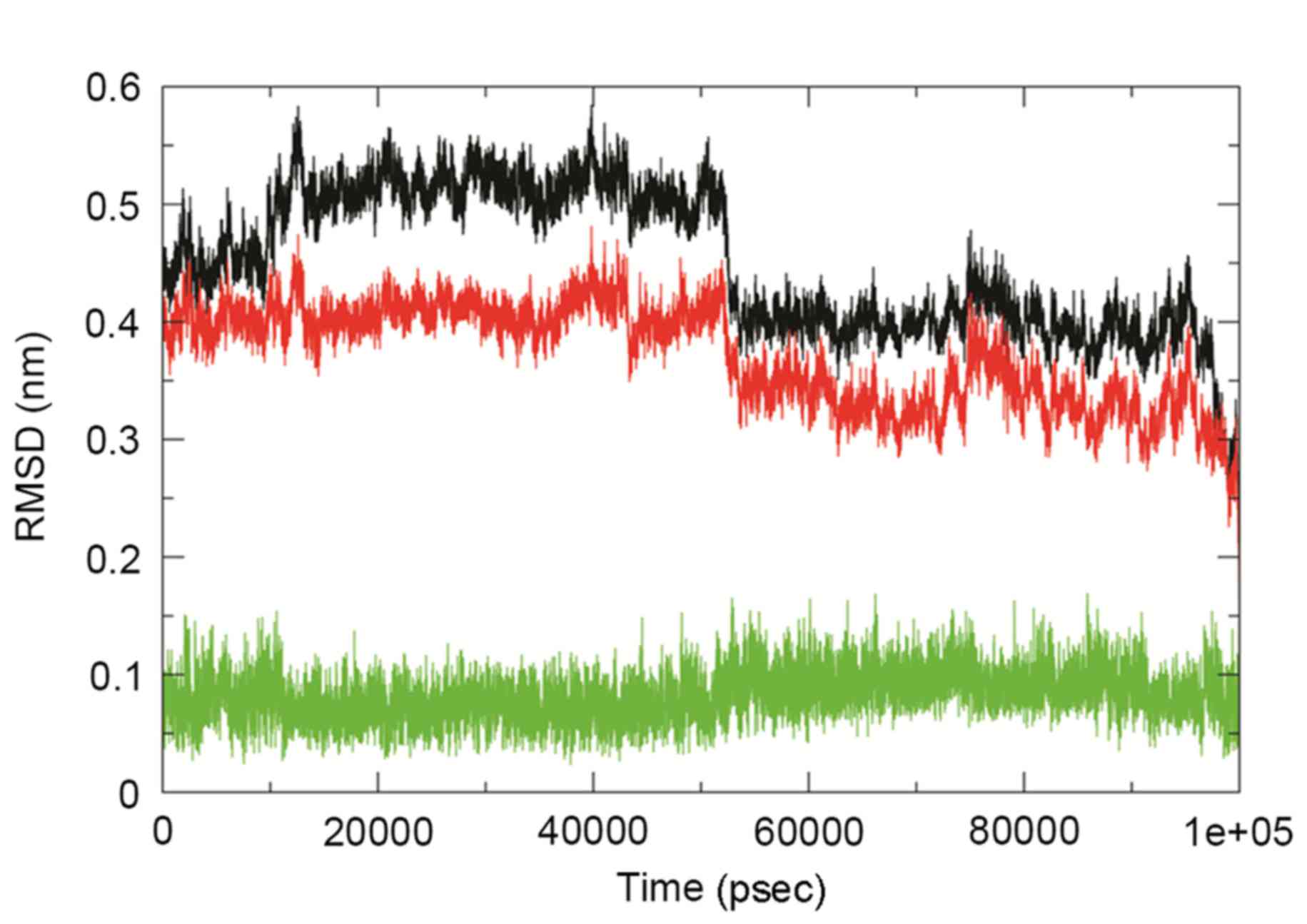
The comparative RMSD plot was analyzed to examine the stability of
this complex over time, and this plot revealed a substantial
decrease in the RMSD of the complex throughout the simulation
(Fig. 6). Furthermore, the dominant
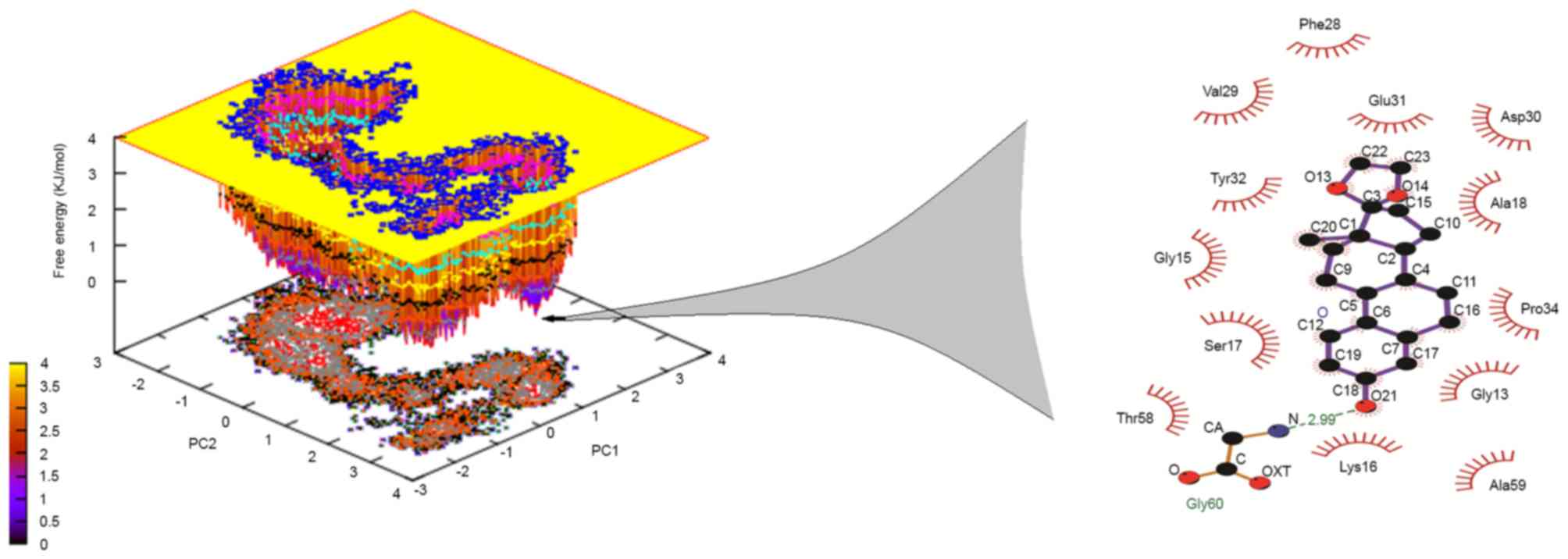
motions of the Lead3-Ras complex were also investigated using the
PCA tool. The free energy landscape was plotted in order to
retrieve the lowest energy conformer (t=67,324 psec). Subsequently,
the retrieved structure of the complex was used investigate the
interactions at that time point, as shown in Fig. 7. The MM-PBSA calculations revealed
that, in the GTP binding site, GLY60 was the best binding site with
Lead3 over time, thus revealing the inhibitory property of Lead3.
Table IV demonstrates the ∆G of the
simulated complex.
 | Table IV.Molecular mechanics poisson-bolzmann
surface area calculations. |
Table IV.
Molecular mechanics poisson-bolzmann
surface area calculations.
| Binding energy | Values
(kJ/mol) |
|---|
| Van der Waal |
−142.32±16.21 |
| Electrostatic |
−6.51±1.31 |
| Polar
solvation |
44.87±7.89 |
| Solvent accessible
volume |
−102.32±18.61 |
| Binding |
−189.33±28.22 |
In patients with SCI, the Ras/Raf/ERK1/2 signaling
pathway has been associated with the promotion of apoptosis in
neural cells (39); small molecule
inhibitors and other methods of modulating the Ras/Raf/ERK1/2
signaling pathway have become a promising target for the
development of novel treatments for traumatic injuries to the
nervous system. To the best of our knowledge, the present study is
first to use structure-based drug design to identify a novel lead
drug of natural origin as a small molecular inhibitor for
modulating Ras/Raf/ERK signaling pathway. Inhibitors, including
PD98059 and U0126, have been used in in vivo studies
targeting the Ras/Raf/ERK1/2 signaling pathway (58).
In conclusion, the in silico approach used in
the present study investigated the IBS database of ~50,000
compounds and identified a single compound that bound to the GTP
binding site of Ras, thus downregulating of the Ras/Raf/ERK1/2
signaling pathway. The inhibition of this pathway is able to
restore the neuronal migration, adhesion and dendritic spine
development. A chemical-computational approach was adopted in the
current study, focusing on the development of Ras inhibitors of a
natural origin that may prevent neuronal apoptosis in SCI, using
used computer-aided drug designing approaches. The Ras GTP
inhibitor proposed in the present study can be developed into a
drug in further studies; thus, these findings provide a starting
point for the development of a drug that may have a potential
effect in the treatment of SCI. However, further in vitro
and in vivo studies are required to translate the proposed
lead compound into a potential drug.
References
|
1
|
Schwartz G and Fehlings MG: Evaluation of
the neuroprotective effects of sodium channel blockers after spinal
cord injury: Improved behavioral and neuroanatomical recovery with
riluzole. J Neurosurg. 94(2 Suppl): 1–256. 2001.
|
|
2
|
Dobkin BH and Havton LA: Basic advances
and new avenues in therapy of spinal cord injury. Annu Rev Med.
55:255–282. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sekhon LH and Fehlings MG: Epidemiology,
demographics, and pathophysiology of acute spinal cord injury.
Spine (Phila Pa 1976). 26(24 Suppl): S2–S12. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Harvey C, Wilson SE, Greene CG, Berkowitz
M and Stripling TE: New estimates of the direct costs of traumatic
spinal cord injuries: Results of a nationwide survey. Paraplegia.
30:834–850. 1992.PubMed/NCBI
|
|
5
|
Albin MS and White RJ: Epidemiology,
physiopathology, and experimental therapeutics of acute spinal cord
injury. Crit Care Clin. 3:441–452. 1987.PubMed/NCBI
|
|
6
|
Porter R: The Cambridge illustrated
history of medicine. Cambridge Univ Press; pp. 4002001
|
|
7
|
Silva NA, Sousa N, Reis RL and Salgado AJ:
From basics to clinical: A comprehensive review on spinal cord
injury. Prog Neurobiol. 114:25–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cadotte DW and Fehlings MG: Spinal cord
injury: A systematic review of current treatment options. Clin
Orthop Relat Res. 469:732–741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Budh CN and Osteråker AL: Life
satisfaction in individuals with a spinal cord injury and pain.
Clin Rehabil. 21:89–96. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Varma AK, Das A, Wallace G IV, Barry J,
Vertegel AA, Ray SK and Banik NL: Spinal cord injury: A review of
current therapy, future treatments, and basic science frontiers.
Neurochem Res. 38:895–905. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bracken MB: Steroids for acute spinal cord
injury. Cochrane Database Syst Rev. 1:CD0010462012.PubMed/NCBI
|
|
12
|
Bracken MB, Collins WF, Freeman DF,
Shepard MJ, Wagner FW, Silten RM, Hellenbrand KG, Ransohoff J, Hunt
WE, Perot PL Jr..et al: Efficacy of methylprednisolone in acute
spinal cord injury. JAMA. 251:45–52. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bracken MB, Shepard MJ, Collins WF Jr,
Holford TR, Baskin DS, Eisenberg HM, Flamm E, Leo-Summers L, Maroon
JC, Marshall LF, et al: Methylprednisolone or naloxone treatment
after acute spinal cord injury: 1-year follow-up data. results of
the second national acute spinal cord injury study. J Neurosurg.
76:23–31. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bracken MB, Shepard MJ, Collins WF,
Holford TR, Young W, Baskin DS, Eisenberg HM, Flamm E, Leo-Summers
L, Maroon J, et al: A randomized, controlled trial of
methylprednisolone or naloxone in the treatment of acute
spinal-cord injury: Results of the second national acute spinal
cord injury study. N Engl J Med. 322:1405–1411. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tator CH: Review of treatment trials in
human spinal cord injury: Issues, difficulties, and
recommendations. Neurosurgery. 59:957–987. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chvatal SA, Kim YT, Bratt-Leal AM, Lee H
and Bellamkonda RV: Spatial distribution and acute
anti-inflammatory effects of methylprednisolone after sustained
local delivery to the contused spinal cord. Biomaterials.
29:1967–1975. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Das A, Smith JA, Gibson C, Varma AK, Ray
SK and Banik NL: Estrogen receptor agonists and estrogen attenuate
TNF-α-induced apoptosis in VSC4.1 motoneurons. J Endocrinol.
208:171–182. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bains M and Hall ED: Antioxidant therapies
in traumatic brain and spinal cord injury. Biochim Biophys Acta.
1822:675–684. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rong W, Wang J, Liu X, Jiang L, Wei F,
Zhou H, Han X and Liu Z: 17β-estradiol attenuates neural cell
apoptosis through inhibition of JNK phosphorylation in SCI rats and
excitotoxicity induced by glutamate in vitro. Int J Neurosci.
122:381–387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lutton C, Young YW, Williams R, Meedeniya
AC, Mackay-Sim A and Goss B: Combined VEGF and PDGF treatment
reduces secondary degeneration after spinal cord injury. J
Neurotrauma. 29:957–970. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ray SK, Samantaray S, Smith JA, Matzelle
DD, Das A and Banik NL: Inhibition of cysteine proteases in acute
and chronic spinal cord injury. Neurotherapeutics. 8:180–186. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee JY, Choi SY, Oh TH and Yune TY:
17β-Estradiol inhibits apoptotic cell death of oligodendrocytes by
inhibiting RhoA-JNK3 activation after spinal cord injury.
Endocrinology. 153:3815–3827. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Samantaray S, Smith JA, Das A, Matzelle
DD, Varma AK, Ray SK and Banik NL: Low dose estrogen prevents
neuronal degeneration and microglial reactivity in an acute model
of spinal cord injury: Effect of dosing, route of administration,
and therapy delay. Neurochem Res. 36:1809–1816. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang YF, Fan ZK, Cao Y, Yu DS, Zhang YQ
and Wang YS: 2-Methoxyestradiol inhibits the up-regulation of AQP4
and AQP1 expression after spinal cord injury. Brain Res.
1370:220–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Knoller N, Auerbach G, Fulga V, Zelig G,
Attias J, Bakimer R, Marder JB, Yoles E, Belkin M, Schwartz M and
Hadani M: Clinical experience using incubated autologous
macrophages as a treatment for complete spinal cord injury: Phase I
study results. J Neurosurg Spine. 3:173–181. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu D, Cao F, Sun S, Liu T and Feng S:
Inhibition of the Ras/Raf/ERK1/2 signaling pathway restores
cultured spinal cord-injured neuronal migration, adhesion, and
dendritic spine development. Neurochem Res. 41:2086–2096. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bonni A, Brunet A, West AE, Datta SR,
Takasu MA and Greenberg ME: Cell survival promoted by the Ras-MAPK
signaling pathway by transcription-dependent and -independent
mechanisms. Science. 286:1358–1362. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chan KM, Gordon T, Zochodne DW and Power
HA: Improving peripheral nerve regeneration: From molecular
mechanisms to potential therapeutic targets. Exp Neurol.
261:826–835. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mazzoni IE, Saïd FA, Aloyz R, Miller FD
and Kaplan D: Ras regulates sympathetic neuron survival by
suppressing the p53-mediated cell death pathway. J Neurosci.
19:9716–9727. 1999.PubMed/NCBI
|
|
30
|
Liu ZD, Zhang S, Hao JJ, Xie TR and Kang
JS: Cellular model of neuronal atrophy induced by DYNC1I1
deficiency reveals protective roles of RAS-RAF-MEK signaling.
Protein Cell. 7:638–650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanno H, Ozawa H, Sekiguchi A, Yamaya S,
Tateda S, Yahata K and Itoi E: The role of mTOR signaling pathway
in spinal cord injury. Cell Cycle. 11:3175–3179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Namikawa K, Honma M, Abe K, Takeda M,
Mansur K, Obata T, Miwa A, Okado H and Kiyama H: Akt/protein kinase
B prevents injury-induced motoneuron death and accelerates axonal
regeneration. J Neurosci. 20:2875–2886. 2000.PubMed/NCBI
|
|
33
|
Sun Z, Wen Y, Mao Q, Hu L, Li H, Sun Z and
Wang D: Adenosine-triphosphate promoting repair of spinal cord
injury by activating mammalian target of rapamycin/signal
transducers and activators of transcription 3 signal pathway in
rats. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 24: 165–171, 2010.
Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 24: 165–171, 2010. 24:
165–171, 2010:165–171, 2010–171, 2010. 2010.(In Chinese).
|
|
34
|
Liu A, Prenger MS, Norton DD, Mei L,
Kusiak JW and Bai G: Nerve growth factor uses Ras/ERK and
phosphatidylinositol 3-kinase cascades to up-regulate the
N-methyl-D-aspartate receptor 1 promoter. J Biol Chem.
276:45372–45379. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu T, Cao FJ, Xu Dd, Xu YQ and Feng SQ:
Upregulated Ras/Raf/ERK1/2 signaling pathway: A new hope in the
repair of spinal cord injury. Neural Regen Res. 10:792–796. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lo LW, Cheng JJ, Chiu JJ, Wung BS, Liu YC
and Wang DL: Endothelial exposure to hypoxia induces Egr-1
expression involving PKCalpha-mediated Ras/Raf-1/ERK1/2 pathway. J
Cell Physiol. 188:304–312. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stanciu M, Wang Y, Kentor R, Burke N,
Watkins S, Kress G, Reynolds I, Klann E, Angiolieri MR, Johnson JW
and DeFranco DB: Persistent activation of ERK contributes to
glutamate-induced oxidative toxicity in a neuronal cell line and
primary cortical neuron cultures. J Biol Chem. 275:12200–12206.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cao F, Zhang X, Liu T, Li XW, Malik M and
Feng SQ: Up-regulation of Ras/Raf/ERK1/2 signaling in the spinal
cord impairs neural cell migration, neurogenesis, synapse
formation, and dendritic spine development. Chin Med J (Engl).
126:3879–3885. 2013.PubMed/NCBI
|
|
39
|
Yang K, Cao F, Sheikh AM, Malik M, Wen G,
Wei H, Ted Brown W and Li X: Up-regulation of Ras/Raf/ERK1/2
signaling impairs cultured neuronal cell migration, neurogenesis,
synapse formation, and dendritic spine development. Brain Struct
Funct. 218:669–682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chikan NA, Bhavaniprasad V, Anbarasu K,
Shabir N and Patel TN: From natural products to drugs for
epimutation computer-aided drug design. Appl Biochem Biotechnol.
170:164–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Amin A, Chikan NA, Mokhdomi TA, Bukhari S,
Koul AM, Shah BA, Gharemirshamlu FR, Wafai AH, Qadri A and Qadri
RA: Irigenin, a novel lead from Western Himalayan chemiome inhibits
fibronectin-extra domain a induced metastasis in lung cancer cells.
Sci Rep. 6:371512016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao GF, Huang ZA, Du XK, Yang ML, Huang
DD and Zhang S: Molecular docking studies of traditional chinese
medicinal compounds against known protein targets to treat
non-small cell lung carcinomas. Mol Med Rep. 14:1132–1138. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van Gunsteren WF: Biomolecular Simulation:
The GROMOS96 Manual and User Guide. Biomos; Zürich: pp. 7571996
|
|
44
|
Trott O and Olson AJ: AutoDock Vina:
Improving the speed and accuracy of docking with a new scoring
function, efficient optimization, and multithreading. J Comput
Chem. 31:455–461. 2010.PubMed/NCBI
|
|
45
|
Lipinski CA: Lead- and drug-like
compounds: The rule-of-five revolution. Drug Discov Today Technol.
1:337–341. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Berendsen HJ, van der Spoel D and van
Drunen R: GROMACS: A message-passing parallel molecular dynamics
implementation. Com Phy Commun. 91:43–56. 1995. View Article : Google Scholar
|
|
48
|
Schüttelkopf AW and Van Aalten DM: PRODRG:
A tool for high-throughput crystallography of protein-ligand
complexes. Acta Crystallogr D Biol Crystallogr. 60:1355–1363. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Darden T, York D and Pedersen L: Particle
mesh Ewald: An N-log(N) method for Ewald sums in large systems. J
Chem Phy. 98:10089–10092. 1993. View Article : Google Scholar
|
|
50
|
Kholmurodov K, Smith W, Yasuoka K, Darden
T and Ebisuzaki T: A smooth-particle mesh Ewald method for DL_POLY
molecular dynamics simulation package on the Fujitsu VPP700. J Com
Chem. 21:1187–1191. 2000. View Article : Google Scholar
|
|
51
|
Hess B, Kutzner C, Van Der Spoel D and
Lindahl E: GROMACS 4: Algorithms for highly efficient,
load-balanced, and scalable molecular simulation. J Chem Theory
Comput. 4:435–447. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kumari R and Kumar R; Open Source Drug
Discovery Consortium; Lynn A: g_mmpbsa - a GROMACS tool for
high-throughput MM-PBSA calculations. J Chem Inf Model.
54:1951–1962. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
DeLano WL: The PyMOL Molecular Graphics
System. DeLano Scientific; Palo Alto, CA: 2002
|
|
54
|
Studio D: Accelrys Inc.; San Diego, CA,
USA: 2013
|
|
55
|
Turner PJ: XMGRACE, Version 5.1.19. Center
for Coastal and Land-Margin Research. Oregon Graduate Institute of
Science and Technology; Beaverton, OR: 2005
|
|
56
|
Racine J: Gnuplot 4.0: A portable
interactive plotting utility. J Appl Econome. 21:133–141. 2006.
View Article : Google Scholar
|
|
57
|
Lin B, Xu Y, Zhang B, He Y, Yan Y and He
MC: MEK inhibition reduces glial scar formation and promotes the
recovery of sensorimotor function in rats following spinal cord
injury. Exp Ther Med. 7:66–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chen J, Rusnak M, Lombroso PJ and Sidhu A:
Dopamine promotes striatal neuronal apoptotic death via ERK
signaling cascades. Eur J Neurosci. 29:287–306. 2009. View Article : Google Scholar : PubMed/NCBI
|