Introduction
Gastric cancer (GC), one of the leading causes of
cancer-associated mortality, is a malignant epithelial cancer that
poses a serious threat to public health, particularly in China
(1,2). Current treatments for GC depend on the
tumor-node-metastasis stage, the type of cancer tissue (intestinal
or diffuse) and the patient's physical condition (3,4). It has
been demonstrated that patients with early-stage GC have a longer
survival time, due to them receiving prompt improved gastrectomy
and radical therapy (4,5). However, the overall prognosis for the
disease remains poor, the 5-year survival rate is ~30% for patients
not with early-stage GC at the time of their initial diagnosis
(6–8). Therefore, developing a predictive
biomarker for the diagnosis and prognosis of GC may improve the
targeted therapeutic options available to treat GC.
The dysregulation of microRNAs (miRNAs/miRs) may
serve oncogenic or tumor suppressor functions in tumorigenesis, as
miRNAs regulate cell proliferation, apoptosis, migration and
invasion (9). Previous studies have
suggested that miRNAs may be used as molecular biomarkers in the
diagnosis and prognosis of GC (10–12).
Oncogenic miRNAs, which are generally upregulated in GC, exert
their function by silencing the expression of genes that inhibit
the cell cycle and/or genes associated with apoptotic pathways
(13–15). By contrast, miRNAs that are
downregulated act as tumor suppressors during GC progression. These
miRNAs generally exert their function via anti-proliferative and
pro-apoptotic associated genes (14–16).
Additionally, decreased levels of miR-144 in GC tissue may be
associated with poor patient prognosis and it was demonstrated that
the reintroduction of miR-144 increased the susceptibility of
cancer cells to chemotherapy (17,18). It
has been reported that GATA4, a key regulator of cardiogenesis via
cell autonomous or non-autonomous mechanisms, may be involved in
the activation of miR-144 (19,20).
Furthermore, the ability of miR-144 to inhibit the proliferation
and metastasis of tumor cells is dependent on the target genes
involved (21). Thus, the present
study aimed to determine the role of miR-144 in GC tissues.
COX-2 is a rate-limiting enzyme, which converts
arachidonic acid into prostaglandin. It has been demonstrated in
vivo and in vitro that the expression of COX-2 is
associated with the proliferation and metastasis of tumor cells
(22). In addition, COX-2 has been
proposed as a potential pharmacologic target to decrease the growth
of certain human tumors (23).
Previous studies identified that the expression of COX-2 was
upregulated in GC and may be targeted by miR-143 (24,25),
which suggests that targeted COX-2 by miRNA may serve as a novel
approach for the treatment of GC.
The present study demonstrated that the expression
of miR-144 and GATA4 were downregulated in GC tissues and cell
lines. Treatment with 5-aza-2′-dexoxycytidine (5-aza) upregulated
the expression of miR-144 and GATA4 in MGC-803 cells.
Overexpression of miR-144 and GATA4 was able to increase MGC-803
cell viability and colony formation, and miR-144 directly targeted
and suppressed cyclooxygenase-2 (COX-2) expression. These results
may improve understanding of the molecular mechanism of
anti-oncogenic miRNAs in the initiation and progression of GC and
provide evidence supporting the use of miRNAs to treat patients
with GC.
Materials and methods
Cell lines and human cancer tissue
samples
The human GC cell lines AGS, BGC-823, SGC-7901,
MGC-803 and the normal gastric epithelium cell line GES-1 were
obtained from the Institute of Biochemistry and Cell Biology at the
Chinese Academy of Sciences (Shanghai, China). Cells were
maintained in RPMI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 1%
penicillin/streptomycin, and cultured at 37°C in a humidified
chamber supplemented with 5% CO2. A total of 21 human GC
tissue samples were obtained from patients undergoing surgical
resection at Harbin Medical University Cancer Hospital (Harbin,
China) between January 2014 and July 2015. There were 11 males and
10 females and the mean age of the patients was 46. Matched normal
tissue samples were obtained from a segment of resected specimens
that were >5 cm from the edge of the tumor. Informed consents
were obtained from all patients and the Research Ethics Committee
of the Affiliated Tumor Hospital of Harbin Medical University
approved the study.
Vector construction and
transfection
A wild-type 3′-untranslated region (UTR) fragment of
human COX-2 mRNA was amplified and cloned downstream of the
pMIR-REPORT vector (Ambion; Thermo Fisher Scientific, Inc.) using
HindIII and SpeI restriction sites to generate the
pMIR-COX-2-3′-UTR luciferase vector (COX-2 3′-UTR). The mutation of
the 3′-UTR fragment of human COX-2 mRNA was performed using the
QuikChange mutagenesis kit (Stratagene; Agilent Technologies, Inc.,
Santa Clara, CA, USA) and cloned into the pMIR-REPORT vector to
form the pMIR-COX-2-3′UTR-mut (COX-2-3′UTR-mut). To construct
pcDNA3-GATA4, the fragment of GATA4 mRNA was amplified and cloned
downstream of the pcDNA3.1 (Invitrogen; Thermo Fisher Scientific,
Inc.). The miR-144 mimics (5′-UACAGUAUAGAUGAUGUACU-3′), miR-144
antisense oligonucleotides (ASO; 5′-AGUACAUCAUCUAUACUGUA-3′), COX-2
small interfering (si)RNA and their corresponding controls
[miRNA-negative control (NC), ASO NC and siRNA NC] were synthesized
by Shanghai GenePharma Co., Ltd. (Shanghai, China). Transfections
of miRNA mimics, ASO, siRNA and corresponding controls into MGC-803
cells were conducted using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions using a final concentration of 50 nM
for RNA mimics, 100 nM for ASO, 100 nM for siRNA and their
respective NCs. The COX-2 siRNA target sequence was
5′-GCTGGGAAGCCTTCTCTAA-3′; the siRNA sense strand was
5′-GCUGGGAAGCCUUCUCUAAdTdT-3′ and the antisense strand was
5′-dTdTCGACCCUUCGGAAGAGAUU-3′.
Luciferase reporter assay
For the luciferase assay, MGC-803 cells
(1×105 cells/well) were cultured in triplicate in
48-well plates and then co-transfected with COX-2
3′UTR/COX-2-3′UTR-mut and miR-144 mimics/miRNA NC at the indicated
concentrations using Lipofectamine 2000. After 48 h, a
dual-luciferase reporter assay kit (Promega Corporation, Madison,
WI, USA) was used to determine luciferase intensity. The pRL-TK and
pMIR-vectors were used as internal controls and the results were
expressed as the relative luciferase activity normalized to the
Renilla luciferase acitivity.
5-Aza-CdR (5-Aza) treatment of GC cell
line MGC-803
MGC-803 cells were treated with 3 µM 5-Aza (cat. no.
A3656; Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) in RPMI 1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (Hyclone; GE Healthcare Life Sciences) and
1% penicillin/streptomycin for 48 h at 37°C in a humidified chamber
supplemented with 5% CO2. And the RT-qPCR was performed
as follows.
RNA preparation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from tissues and cells using the
mirVana miRNA isolation kit (Ambion; Thermo Fisher Scientific,
Inc.) following the manufacturer's protocol. The separated and
purified RNAs were detected by 1.5% agarose gel electrophoresis.
Subsequently, 5 µg RNA was reversely transcribed into cDNA using
Moloney Murine Leukemia Virus Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.). 5× First-Strand Buffer and dNTPs
were purchased from Invitrogen (Thermo Fisher Scientific, Inc.).
The reverse transcription temperature protocol was 16°C for 30 min;
42°C for 30 min; 85°C for 5 min, then hold at 4°C. qPCR analysis
was performed using the SYBR Premix Ex Taq™ kit (Takara
Biotechnology Co., Ltd., Dalian, China) following the
manufacturer's protocol. The PCR thermocycling conditions were as
follows: 30 cycles of denaturation at 94°C for 30 sec, annealing at
58°C for 30 sec and extension at 72°C for 30 sec. U6 (for miR-144)
and GAPDH (for GATA4 and COX-2) were used an as internal control
and the 2−ΔΔCq method (26) was used for quantification. The
experiments were performed three times. The primers used are as
follows: miR-144 forward, 5′-ATCCAGTGCGTGTCGTG-3′ and reverse,
5′-TGCTTATACAGTATAGATG-3′. U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse 5′-AACGCTTCACGAATTTGCGT-3′. GATA4 forward,
5′-CTGGCCTGTCATCTCACTACG-3′ and reverse
5′-GGTCCGTGCAGGAATTTGAGG-3′. COX-2 forward, 5-CCTGTGCCTGATGATTGC-3
and reverse 5-CTGATGCGTGAAGTGCTG-3. GAPDH forward,
5-AATCCCATCACCATCTTCCAGG-3′ and reverse
5-GAGTGGGTGTCGCTGTTGAAGT-3′.
Cell viability assay
An MTT assay was used to investigate the effect of
GATA4 and miR-144 overexpression on cell viability. Briefly, cells
were seeded at 1,000 cells/well and incubated at 37°C for 24 h.
Cells were then co-transfected with miR-144 mimics or pcDNA3-GATA4.
Cells were harvested at following 2 h incubation with MTT (0.5
mg/ml) and dimethyl sulfoxide. The optical density was determined
at 570 nm on a Tecan SpectraFluor microplate reader (Tecan Group
Ltd., Männedorf, Switzerland).
Colony formation assay
For the colony formation assay, cells transfected
with miR-144 mimics or pcDNA3-GATA4 were seeded at 1,000 cells/well
in a 6-well plate and incubated for 2 weeks at 37°C in a humidified
chamber supplemented with 5% CO2. The culture medium was
replenished every 3 days and experiments were performed three
times. The media was subsequently removed and fixed with 75%
ethanol for 30 min at room temperature, 0.2% crystal violet stain
(Beyotime Institute of Biotechnology, Haimen, China) was added to
the wells for 20 min at room temperature, the plate was washed with
running water and air dried. Then the colonies in each well were
counted under a light microscope (magnification, ×400; Olympus
Corporation, Tokyo, Japan) following.
Cell cycle analysis
A total of 48 h after transfection, cells
transfected with miR-144 mimics or pcDNA3-GATA4 were subjected to
cell cycle analysis. Cells were obtained by trypsinization followed
by centrifugation at 300 × g for 5 min. The cells were fixed with
70% ethanol on ice for 2 h followed by centrifuging for 5 min at
300 × g at 4°C. Subsequently, 0.05 mg/ml propidium iodide (PI;
Sigma-Aldrich; Merck KGaA) and 0.1 mg/ml RNAse A (Sigma-Aldrich;
Merck KGaA) were added to the samples for 30 min. Cells were
examined using a BD FACSCalibur flow cytometer (BD Biosciences, San
Jose, CA, USA) and CellQuest software version 3.3 (BD Biosciences).
The experiments were repeated three times.
Apoptosis assay
Annexin V-fluorescein isothiocyanate and PI staining
were used to determine the effect of miR-144 mimics or pcDNA3-GATA4
on cell apoptosis. The adherent cells were harvested 48 h after
transfection. Subsequently, cells were washed with PBS and stained
with Annexin-V in combination with PI at room temperature in the
dark for 15 min (Annexin V-PI kit; Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China). Cells were subsequently analyzed using a BD
FACSCalibur flow cytometer (BD Biosciences) and CellQuest software
version 3.3. A total of 10,000 events were counted for each
sample.
Western blot analysis
Proteins from transfected cells were extracted using
radioimmunoprecipitation assay lysis buffer (cat. no. R0278;
1:1,000; Sigma-Aldrich; Merck KGaA) and the protein concentration
was determined using BCA assay kit (Thermo Fisher Scientific,
Inc.). Equal amounts of protein (30 µg/lane) were separated by 12%
SDS-PAGE followed by a transfer onto a nitrocellulose membrane.
Then membranes were blocked in 5% non-fat milk in TBS with 0.05%
Tween-20 (TBST) at room temperature for 1 h and incubated with
diluted primary antibodies [anti-COX-2 (cat. no. 4842; 1:1,000;
Cell Signaling Technology, Inc., Danvers, MA, USA) and anti-GAPDH
(cat. no. G9545; 1:1,000; Sigma-Aldrich; Merck KGaA)] overnight at
4°C and were subsequently washed three times in TBST, followed by
incubation with horseradish peroxidase conjugated secondary
antibodies (cat. no. AC111P; 1:1,000; Sigma-Aldrich; Merck KGaA)
for 1 h at room temperature. The protein bands were detected with
the Western Blotting Luminol reagent (cat. no. sc-2048; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and quantified by
densitometric analysis of digitized autoradiograms using Quantity
One version 4.6.2 software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Bioinformatics method, 3′-UTR datasets
and microRNA datasets
Mir-144 target sites were predicted by
computer-aided algorithms obtained from Targetscan Release 7.1
(targetscan.org/). The 3′-UTR sequences of COX-2
were retrieved using the Ensembl database (ensembl.org). Human miRNA sequences of the miR-144
registry were downloaded from the miRBase website (mirbase.org).
Statistical analysis
Statistical analysis was performed using a
two-tailed Student's t-test on SPSS 17.0 software (SPSS Inc.,
Chicago, IL, USA). For comparison of multiple groups, one-way
analysis of variance followed by a Tukey's post hoc multiple
comparisons test was performed. All results are expressed as the
mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
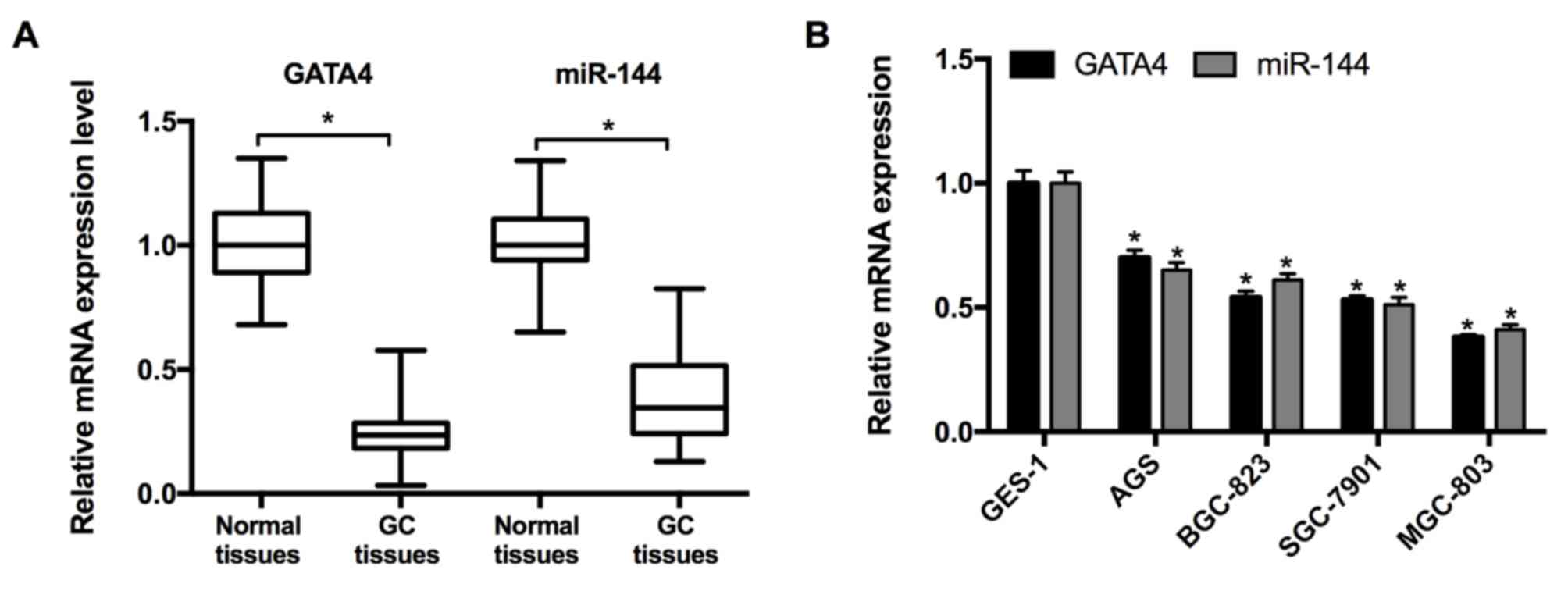
miR-144/GATA4 is downregulated in
human GC tissues and cell lines
RT-qPCR was performed to measure the expression of
miR-144 and GATA4 in 21 pairs of GC and adjacent normal tissues
taken from patients with GC. The results revealed that the
expression of miR-144 and GATA4 were significantly decreased in GC
tissues compared with the adjacent normal tissues (Fig. 1A). Subsequently, the expression of
miR-144 and GATA4 in the GC cell lines was determined. The results
from RT-qPCR indicated that the expression of miR-144 and GATA4
were significantly downregulated in the GC cell lines (MGC-803,
SGC-7901, BGC-823 and AGS) relative to the normal cell line, GES-1
(Fig. 1B). As the expression of
miR-144 was expressed at lowest level in the MGC-803 cell line, it
was selected for miR-144 overexpression and associated assays.
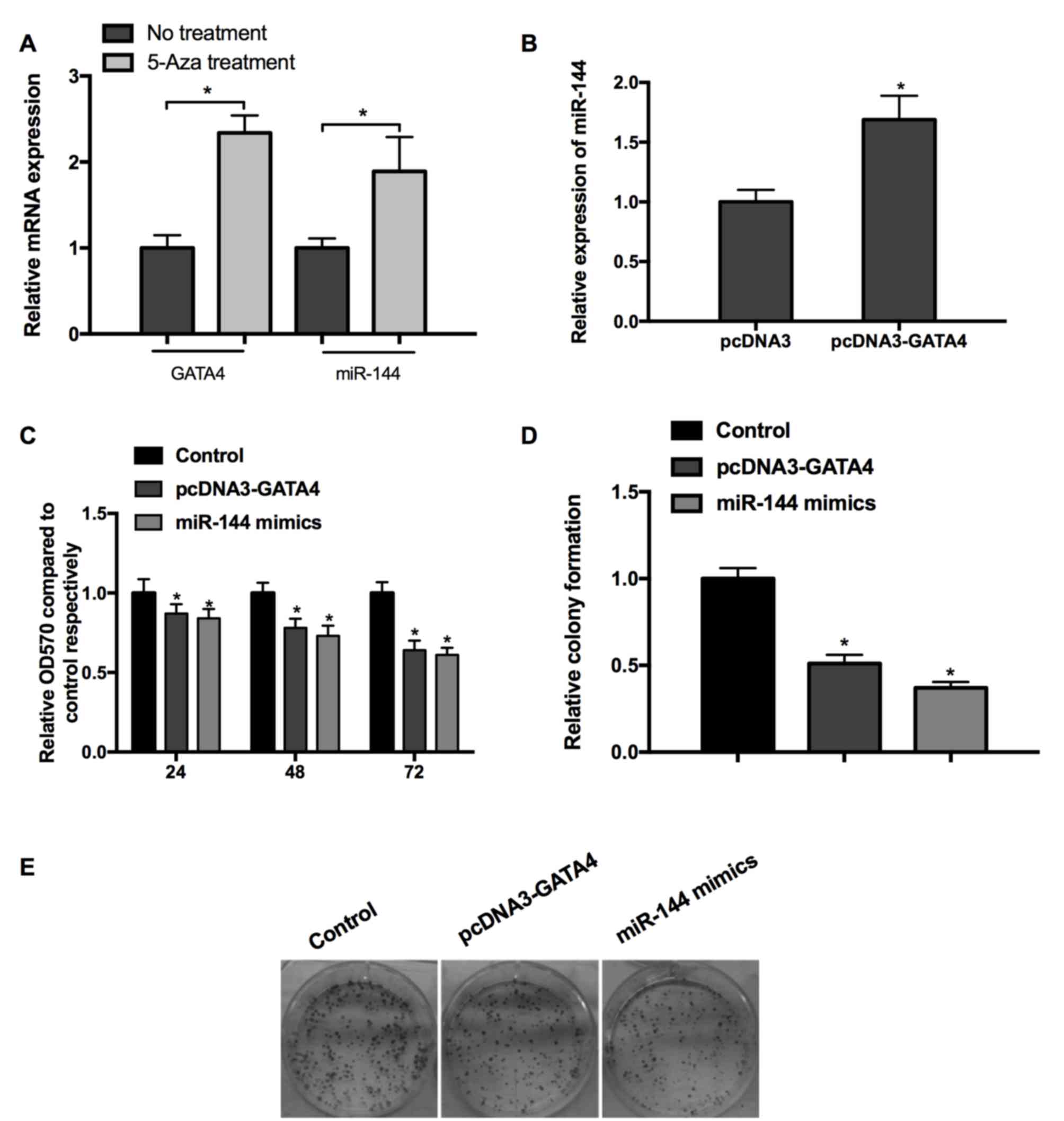
Synergistic effects of GATA4 and
miR-144 on cell viability and growth
Studies have indicated that GATA4 is able to
transcriptionally activate miR-144; additionally GATA4 may
potentially be involved in promoting the hypermethylation of tumor
suppressor genes underlying tumorigenesis (19,27,28). The
results of the present study implied that DNA demethylation
following treatment with 5-aza induced the upregulation of GATA4
and miR-144 expression compared with the cells without 5-aza
treatment (Fig. 2A). Subsequently,
miR-144 mimics/miRNA NC were transfected into cells and the
expression of miR-144 was determined by RT-qPCR. The results
confirmed that the overexpression of GATA4 upregulated miR-144
expression (Fig. 2B). In order to
test the effects of GATA4 and miR-144 on cell viability,
pcDNA3-GATA4 or miR-144 mimics were transfected into MGC-803 cells.
It was revealed that overexpression of GATA4 and miR-144 induced
time-dependent inhibition of cell viability (Fig. 2C). Furthermore, the results of the
cell colony assay exhibited a similar inhibition on cell
proliferation (Fig. 2D and E). This
evidence indicates that GATA4 and miR-144 are potentially
hypermethylated in GC, and that GATA4 and miR-144 inhibit the
viability of GC cells.
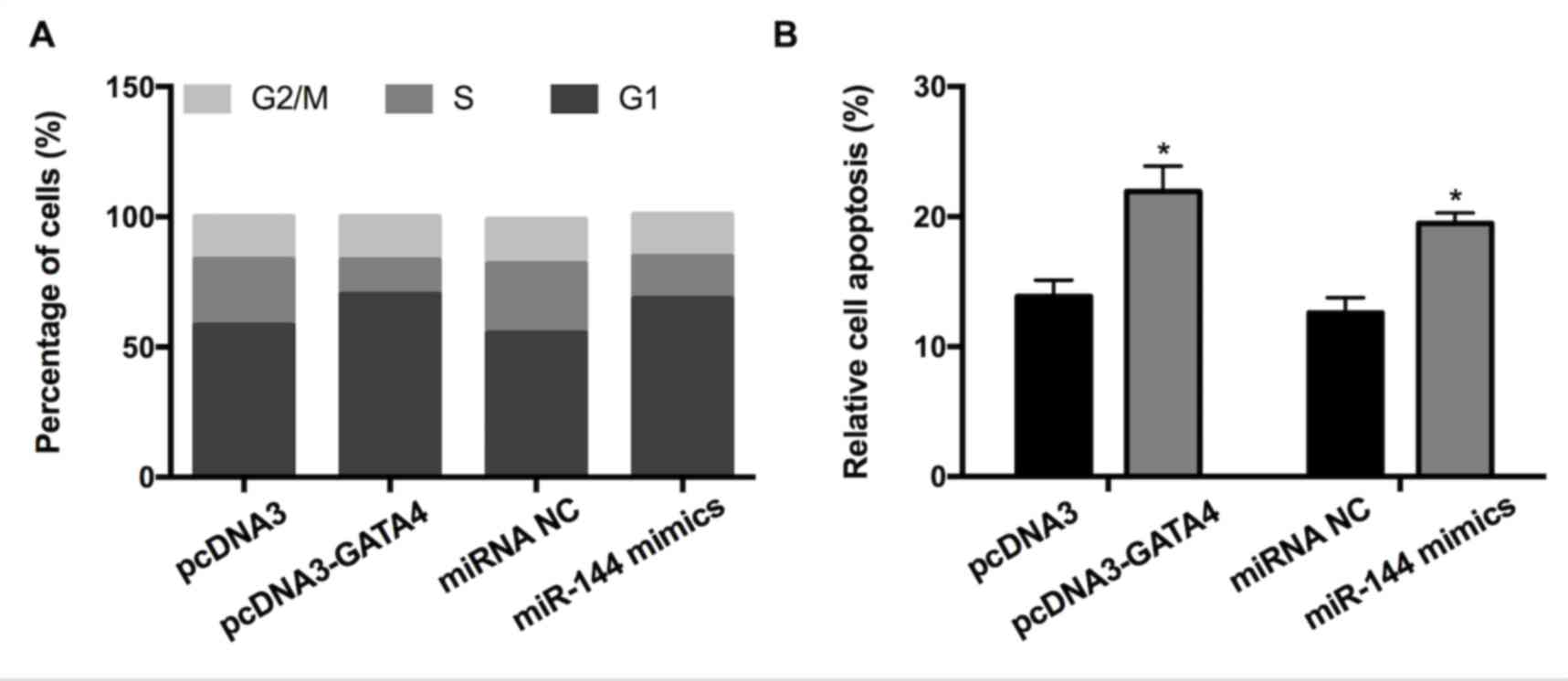
GATA4 and miR-144 induce GC cell cycle
arrest and apoptosis
To investigate whether GATA4 and miR-144 affect the
cell cycle and apoptosis of GC cells, fluorescence-activated cell
sorting and a cell apoptosis assay were performed. The results
revealed that overexpression of GATA4 increased the proportion of
cells in the G1 phase and concurrently decreased the proportion of
cells in the S-phase compared with the control (Fig. 3A). Transfection with miR-144 mimic
had a similar effect on the cell cycle. In accordance with this,
elevated expression of GATA4/miR-144 mimics significantly promoted
apoptosis (Fig. 3B). Altogether, the
results indicated that the elevated expression of GATA4/miR-144 may
induce GC cell cycle arrest and apoptosis.
miR-144 negatively regulates COX-2,
which inhibits GC cell viability and growth
Although it has been demonstrated that miR-144
targets COX-2 in esophageal squamous cell cancer (29), there have been few studies
investigating whether miR-144 regulates COX-2 in GC. Bioinformatics
prediction using Targetscan Release 7.1 (targetscan.org/) was performed and COX was confirmed
to be a direct target of miR-144 (Fig.
4A). The luciferase intensity of MGC-803 cells transfected with
miR-144 mimics+COX-2-3′-UTR was significantly lower than the other
three groups (miRNA NC+COX-2-3′-UTR, miRNA NC+COX-2-3′-UTR-mut and
miR-144+COX-2-3′-UTR-mut), indicating that miR-144 may directly
bind to COX-2 3′-UTR (Fig. 4B). The
possibility that miR-144 serves a functional role in the regulation
of endogenous COX-2 expression in GC cells was also assessed. The
results of RT-qPCR revealed that cells transfected with miR-144
mimic exhibited a significantly decreased expression of endogenous
COX-2 compared with the miRNA NC group (Fig. 4C). The results of the western blot
analysis also exhibited a similar inhibitory effect (Fig. 4D). To further explore the role of
COX-2 in the proliferation of GC cells, the expression of COX-2 was
inhibited following transfection of COX-2 siRNA or siRNA NC. The
results obtained from the MTT assay revealed that the viability of
MGC-803 cells decreased in a time-dependent manner (Fig. 4E). The results of the colony
formation assay further determined that inhibition of COX-2 reduced
cell viability (Fig. 4F). These
results indicate that miR-144 directly targets COX-2 3′UTR to
suppress its expression and inhibit GC cell viability and
growth.
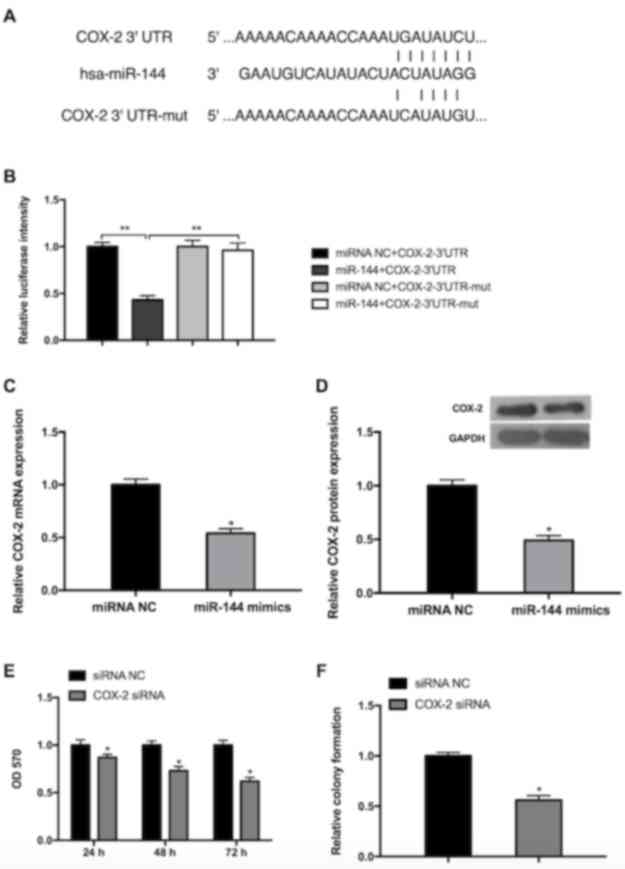 | Figure 4.miR-144 targets COX-2, which inhibits
the viability and growth of GC cells. (A) A schematic of the
bioinformatics prediction in the 3′-UTR of COX-2 as well as mutant
3′-UTR used in the present study. (B) The results of the luciferase
reporter assay demonstrated that COX-2 is a direct target of
miR-144. **P<0.01. The expression of COX-2 was reduced in cells
transfected with miR-144 mimics, as determined by (C) reverse
transcription-quantitative polymerase chain reaction and (D)
western blot analysis. (E) Knockdown of COX-2 reduced cell
viability in a time-dependent manner, as determined by the results
of an MTT assay. (F) Knockdown of COX-2 inhibited cell growth, as
determined by the results of a cell colony formation assay.
Differences were assessed by one-way analysis of variance followed
by Tukey's multiple comparison test. *P<0.05 vs. corresponding
control. miR-144, microRNA-144; COX-2, cyclooxygenase-2; UTR,
untranslated region; mut, mutant; NC, negative control; siRNA,
small interfering RNA; OD, optical density. |
Synergistic effects of GATA4 and
miR-144 on COX-2 expression in GC cells
RT-qPCR and western blotting were performed to
further investigate whether GATA4 and miR-144 are able to regulate
COX-2 expression. The results revealed that the overexpression of
GATA4 (pcDNA3-GATA4+ASO NC) significantly reduced the expression of
COX-2 mRNA and protein, whereas miR-144 ASO (pcDNA3+miR-144 ASO)
promoted the expression of COX-2 mRNA and protein (Fig. 5). Additionally, the reduction induced
by pcDNA3-GATA4 can be partially rescued by miR-144 ASO
(pcDNA3-GATA4+miR-144 ASO) compared with the pcDNA3+ASO NC group
(Fig. 5). These results indicate
that GATA4 and miR-144 exhibit a synergistic effect on the
inhibition of COX-2 expression in GC cells.
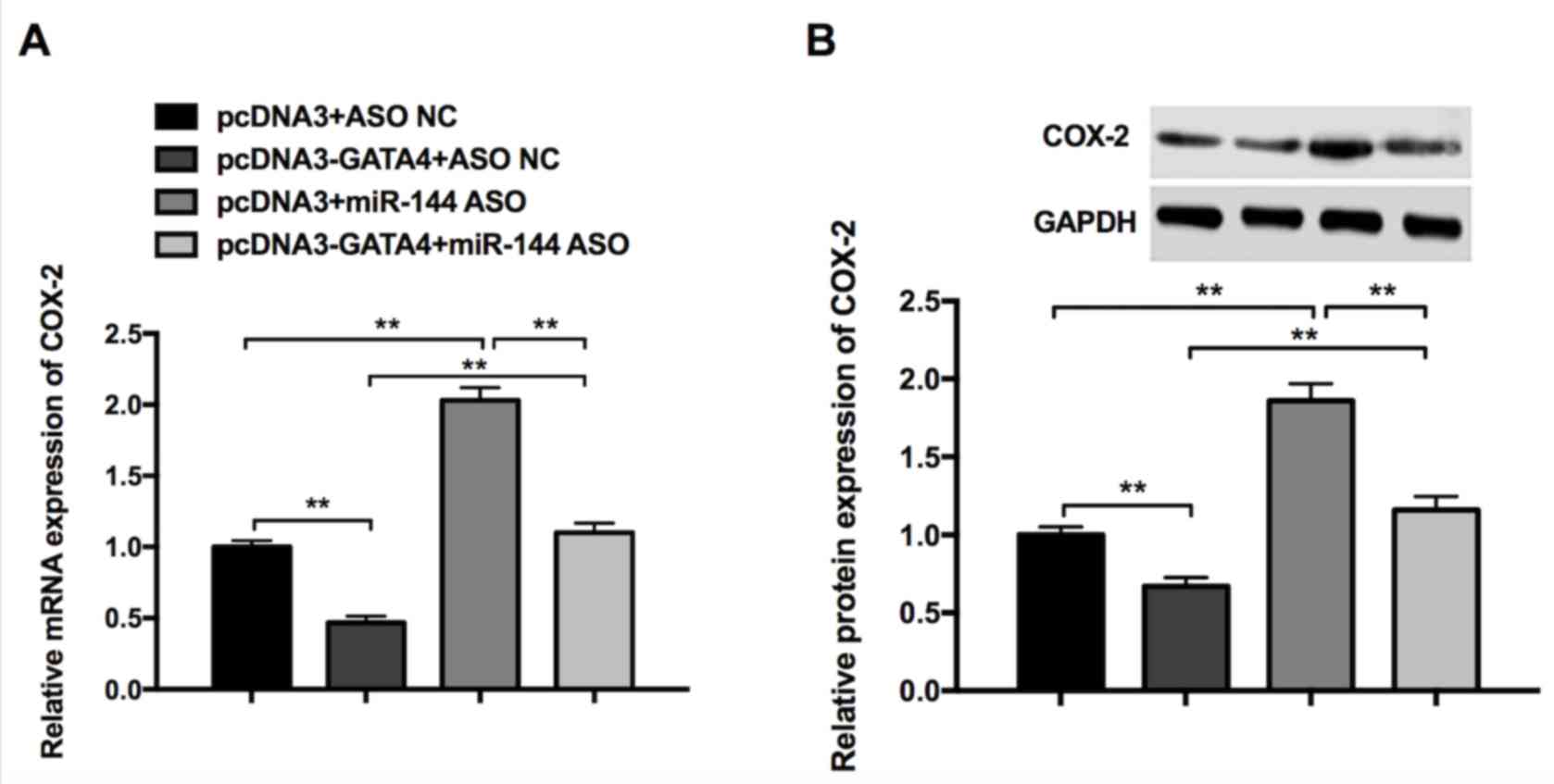 | Figure 5.Synergistic effects of GATA4 and
miR-144 on COX-2 expression in gastric cancer cells. (A) Expression
of COX-2 in cells transfected with pcDNA3+ASO NC, pcDNA3-GATA4+ASO
NC, pcDNA3+miR-144 ASO, pcDNA3-GATA4+miR-144 ASO, as determined by
reverse transcription-quantitative polymerase chain reaction. (B)
Expression of COX-2 in cells transfected with pcDNA3+ASO NC,
pcDNA3-GATA4+ASO NC, pcDNA3+miR-144 ASO, pcDNA3-GATA4+miR-144 ASO,
as determined by western blot analysis. Differences were assessed
using one-way analysis of variance followed by Tukey's multiple
comparison test. **P<0.01. miR-144, microRNA-144; COX-2,
cyclooxygenase-2; ASO, antisense oligonucleotides; NC, negative
control. |
Discussion
The mortality rate of patients with GC is increasing
in developing countries, particularly in East Asian countries
(30). The overall 5-year survival
rate remains low; ranging between 20–30% for patients past the
early stage of the disease at the time of their initial diagnosis
(31). Furthermore, the
dysregulation of miRNAs is reported to be involved in the
pathogenic processes of GC tumorigenesis by regulating the
expression of the miRNA's target genes. It has been revealed that
the regulation of miRNAs participating in GC tumorigenesis is
associated with cell proliferation and apoptosis-related genes
(32). Therefore, numerous studies
have suggested that miRNAs, including miR-10b, miR-106a and
miR-107, may be used as biomarkers in GC (14,33,34). In
addition, it has been demonstrated that gene silencing is tightly
associated with epigenetic mechanisms, particularly promoter
hypermethylation of tumor suppressor genes, which may serve a
critical role in the mechanisms underlying tumorigenesis (27,28).
In the present study, it was revealed that the
expression of GATA4 and miR-144 was repressed in GC tissues and
cell lines compared with relative controls. It was hypothesized
that the downregulation of GATA4 and miR-144 that occurs in GC is
induced by hypermethylation. Consistent with this, the results of
the present study indicated that cells treated with 5-aza exhibited
increased expression of GATA4 and miR-144. It has been demonstrated
that dysregulated cell proliferation and the induction of cell
cycle arrest are key mechanisms involved in tumor progression
(35). Furthermore, miR-144 and
GATA4 may function by modulating the expression of genes that
suppress tumor growth (36–38). The results of the MTT and colony
formation assays validated this and revealed that miR-144 and GATA4
have a synergistic effect on cell proliferation, apoptosis and the
cell cycle.
To further identify the molecular mechanism of
miR-144 in the regulation of cancer progression, a bioinformatics
analysis was performed, which indicated that COX-2 is a potential
target of miR-144. COX-2 is the rate-limiting enzyme involved in
the biosynthesis of prostaglandins and it has been suggested that
COX-2 is involved in the development and progression of tumors
(39,40). In the present study, knockdown of
COX-2 expression by siRNA in MGC-803 cells decreased cell viability
and colony formation. Furthermore, the results of the luciferase
reporter assay validated that miR-144 targets the 3′UTR of COX-2
and GATA4 and that miR-144 has a synergistic effect on the
regulation of COX-2 in GC cells.
In conclusion, the results of the present study
revealed that GATA4 and miR-144 are downregulated in GC and that
this may be attributed to hypermethylation. Furthermore, GATA4 and
miR-144 had a synergistic effect on cell proliferation and
apoptosis, potentially by targeting COX-2. These results indicate
that GATA4 and miR-144 inhibit the proliferation of GC cells and
that they may be used as a potential therapeutic target in the
treatment of GC.
Acknowledgements
The present study was supported by the Health and
Life Committee Fund of Heilongjiang Province (grant no. 2016-111)
and the Applied Technology Research and Development Project of
Harbin, Heilongjiang Province (grant no. 2016RAQXJ152).
References
|
1
|
Heise K, Bertran E, Andia ME and Ferreccio
C: Incidence and survival of stomach cancer in a high-risk
population of Chile. World J Gastroenterol. 15:1854–1862. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi C, Liu B, Yan J, Liu H, Pan Z, Yao W,
Yan F and Zhang H: Gastric cancer: Preoperative TNM staging with
individually adjusted computed tomography scanning phase. J Comput
Assist Tomogr. 40:160–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu B, El Hajj N, Sittler S, Lammert N,
Barnes R and Meloni-Ehrig A: Gastric cancer: Classification,
histology and application of molecular pathology. J Gastrointest
Oncol. 3:251–261. 2012.PubMed/NCBI
|
|
5
|
Smyth EC and Cunningham D: Gastric cancer
in 2012: Defining treatment standards and novel insights into
disease biology. Nat Rev Clin Oncol. 10:73–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dassen AE, Lemmens VE, van de Poll-Franse
LV, Creemers GJ, Brenninkmeijer SJ, Lips DJ, Vd Wurff AA, Bosscha K
and Coebergh JW: Trends in incidence, treatment and survival of
gastric adenocarcinoma between 1990 and 2007: A population-based
study in the Netherlands. Eur J Cancer. 46:1101–1110. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu CW, Lo SS, Shen KH, Hsieh MC, Lui WY
and P'eng FK: Surgical mortality, survival and quality of life
after resection for gastric cancer in the elderly. World J Surg.
24:465–472. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yalçin S: The increasing role of
pharmacogenetics in the treatment of gastrointestinal cancers.
Gastrointest Cancer Res. 3:197–203. 2009.PubMed/NCBI
|
|
9
|
Melo SA and Esteller M: Dysregulation of
microRNAs in cancer: Playing with fire. FEBS Lett. 585:2087–2099.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang YY, Li L, Ye ZY, Zhao ZS and Yan ZL:
MicroRNA-10b promotes migration and invasion through Hoxd10 in
human gastric cancer. World J Surg Oncol. 13:2592015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
12
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li X, Zhang Y, Shi Y, Dong G, Liang J, Han
Y, Wang X, Zhao Q, Ding J, Wu K and Fan D: MicroRNA-107, an
oncogene microRNA that regulates tumour invasion and metastasis by
targeting DICER1 in gastric cancer. J Cell Mol Med. 15:1887–1895.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang H, Yu WW, Wang LL and Peng Y:
miR-130a acts as a potential diagnostic biomarker and promotes
gastric cancer migration, invasion and proliferation by targeting
RUNX3. Oncol Rep. 34:1153–1161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang
Q, Chen L, Pang X, Leng W and Bi F: Plasma miR-122 and miR-192 as
potential novel biomarkers for the early detection of distant
metastasis of gastric cancer. Oncol Rep. 31:1863–1870. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meder B, Backes C, Haas J, Leidinger P,
Stähler C, Großmann T, Vogel B, Frese K, Giannitsis E, Katus HA, et
al: Influence of the confounding factors age and sex on microRNA
profiles from peripheral blood. Clin Chem. 60:1200–1208. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Imaoka H, Toiyama Y, Okigami M, Yasuda H,
Saigusa S, Ohi M, Tanaka K, Inoue Y, Mohri Y and Kusunoki M:
Circulating microRNA-203 predicts metastases, early recurrence and
poor prognosis in human gastric cancer. Gastric Cancer. 19:744–753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Akiyoshi S, Fukagawa T, Ueo H, Ishibashi
M, Takahashi Y, Fabbri M, Sasako M, Maehara Y, Mimori K and Mori M:
Clinical significance of miR-144-ZFX axis in disseminated tumour
cells in bone marrow in gastric cancer cases. Br J Cancer.
107:1345–1353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu J, Xue H, Zhang J, Suo T, Xiang Y,
Zhang W, Ma J, Cai D and Gu X: MicroRNA-144 inhibits the metastasis
of gastric cancer by targeting MET expression. J Exp Clin Cancer
Res. 34:352015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grépin C, Nemer G and Nemer M: Enhanced
cardiogenesis in embryonic stem cells overexpressing the GATA-4
transcription factor. Development. 124:2387–2395. 1997.PubMed/NCBI
|
|
22
|
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu
WT, Jegga AG and Fan GC: Synergistic effects of the GATA-4-mediated
miR-144/451 cluster in protection against simulated
ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell
Cardiol. 49:841–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yin J, Liu B, Li B, Liu Z, Xie X, Lv Z,
Gao S and Guang J: The cyclooxygenase-2 inhibitor celecoxib
attenuates hepatocellular carcinoma growth and c-Met expression in
an orthotopic mouse model. Oncol Res. 19:131–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q,
Dan ZL, Tian DA and Zhang P: MicroRNA143 suppresses gastric cancer
cell growth and induces apoptosis by targeting COX-2. World J
Gastroenterol. 19:7758–7765. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Chen JQ and Liu JL: COX-2
inhibitors and gastric cancer. Gastroenterol Res Pract.
2014:1323202014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lynch J, Meehan MH, Crean J, Copeland J,
Stallings RL and Bray IM: Metastasis suppressor microRNA-335
targets the formin family of actin nucleators. PLoS One.
8:e784282013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma J, Hong L, Chen Z, Nie Y and Fan D:
Epigenetic regulation of microRNAs in gastric cancer. Dig Dis Sci.
59:716–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shao Y, Li P, Zhu ST, Yue JP, Ji XJ, Ma D,
Wang L, Wang YJ, Zong Y, Wu YD and Zhang ST: MiR-26a and miR-144
inhibit proliferation and metastasis of esophageal squamous cell
cancer by inhibiting cyclooxygenase-2. Oncotarget. 7:15173–15186.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsu KW, Hsieh RH, Huang KH, Fen-Yau Li A,
Chi CW, Wang TY, Tseng MJ, Wu KJ and Yeh TS: Activation of the
Notch1/STAT3/Twist signaling axis promotes gastric cancer
progression. Carcinogenesis. 33:1459–1467. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu JW, Wu JG, Zheng LH, Zhang B, Ni XC, Li
XQ and Jiang BJ: Influencing factors and clinical significance of
the metastatic lymph nodes ratio in gastric adenocarcinoma. J Exp
Clin Cancer Res. 28:552009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi
HC, Lin YH, Lu PH and Lin KH: Potential diagnostic, prognostic and
therapeutic targets of MicroRNAs in human gastric cancer. Int J Mol
Sci. 17:E9452016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hersey P and Zhang XD: Treatment
combinations targeting apoptosis to improve immunotherapy of
melanoma. Cancer Immunol Immunother. 58:1749–1759. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xiao B, Guo J, Miao Y, Jiang Z, Huan R,
Zhang Y, Li D and Zhong J: Detection of miR-106a in gastric
carcinoma and its clinical significance. Clin Chim Acta.
400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Z, Kong Y, Yang W, Ma F, Zhang Y, Ji
S, Ma EM, Liu H, Chen Y and Hua Y: Upregulation of microRNA-34a
enhances the DDP sensitivity of gastric cancer cells by modulating
proliferation and apoptosis via targeting MET. Oncol Rep.
36:2361–2397. 2016.
|
|
37
|
Song B, Yan J, Liu C, Zhou H and Zheng Y:
Tumor suppressor role of miR-363-3p in gastric cancer. Med Sci
Monit. 21:4074–4080. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li D, Li Z, Xiong J, Gong B, Zhang G, Cao
C, Jie Z, Liu Y, Cao Y, Yan Y, et al: MicroRNA-212 functions as an
epigenetic-silenced tumor suppressor involving in tumor metastasis
and invasion of gastric cancer through down-regulating PXN
expression. Am J Cancer Res. 5:2980–2997. 2015.PubMed/NCBI
|
|
39
|
Adhim Z, Matsuoka T, Bito T, Shigemura K,
Lee KM, Kawabata M, Fujisawa M, Nibu K and Shirakawa T: In vitro
and in vivo inhibitory effect of three Cox-2 inhibitors and
epithelial-to-mesenchymal transition in human bladder cancer cell
lines. Br J Cancer. 105:393–402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Elmets CA, Ledet JJ and Athar M:
Cyclooxygenases: Mediators of UV-induced skin cancer and potential
targets for prevention. J Invest Dermatol. 134:2497–2502. 2014.
View Article : Google Scholar : PubMed/NCBI
|