Introduction
Lung cancer has the highest incidence and mortality
of all malignant tumors worldwide, and it severely threatens
people's health (1–3). China's cancer registration report in
2013 demonstrated that the incidence and mortality of lung cancer
ranked the first in cities and rural areas (4). Therefore, the prevention and treatment
of lung cancer has become a focus in medicine. Primary lung cancer
includes small cell lung carcinoma (SCLC) and non-SCLC (NSCLC), and
NSCLC cases account for ~85% of all lung cancer cases (2). Surgical treatment is the most effective
method for the treatment of NSCLC. Although surgical treatment has
been greatly developed in recent years, the prognosis for patients
with NSCLC remains poor, with a 5-year survival rate of only ~16%
(5). Tumor recurrence and metastasis
are the predominant causes of treatment failure in patients with
NSCLC (6); however, the molecular
mechanisms of NSCLC are not yet clear. It has been reported that
various genes are involved in the regulation of proliferation, drug
resistance, apoptosis, invasion and metastasis of NSCLC (7); therefore, molecular targeted therapy
for NSCLC has demonstrated great clinical significance (8). However, the therapeutic targets for
NSCLC remain to be further studied due to the complex molecular
mechanisms of the disease.
MicroRNA (miR), with a length of 18–25 nt, are
important regulatory factors in the occurrence and development of
tumors, and are also targets for tumor diagnosis and treatment
(9). miR bind with the
3′-untranslated region (UTR) of mRNA and regulate gene expression
by inhibiting mRNA translation (10). miR molecules may serve the roles of
oncogenes or tumor suppressor genes, and expression imbalance is a
crucial factor in the development of cancer (11). Studies have revealed that miR
molecules are involved in the proliferation, invasion and
metastasis of NSCLC. Pei et al (12) demonstrated that miR-185-5p regulates
the chemotherapy resistance of NSCLC by targeting the ABCC1 gene
(multidrug resistance-associated protein 1). Sun et al
(13) reported that miR-9600
inhibits the proliferation and metastasis of NSCLC by targeting
signal transducer and activator of transcription 3 gene expression.
Wang et al (14) indicated
that miR-509-5p inhibits the proliferation and invasion of NSCLC by
targeting YWHAG gene (14). These
studies indicate that miR molecules are involved in the occurrence
and development of NSCLC.
miR-215 is a newly discovered tumor-associated miR
molecule that has varied functions in multiple tumor types. For
example, miR-215 promotes the proliferation and metastasis of
glioma cells by targeting retinoblastoma tumor suppressor gene 1
(15). Additionally, miR-215, as an
oncogene, promotes the development of gastric cancer cells by
downregulating the expression of the Runt-related transcription
factor 1 gene (16). Li et al
(17) discovered that miR-215
directly regulates the zinc finger E-box-binding homeobox 2 (ZEB2)
gene and inhibits the epithelial-mesenchymal transition of
pancreatic cancer cells, acting as a tumor suppressor gene
(17). In addition, miR-215 has the
function of a tumor suppressor gene in NSCLC and is regulated by
its upstream transcription factor, p53. It inhibits the
proliferation and invasion and promotes the apoptosis of NSCLC
cells (18). However, the downstream
regulatory mechanism of miR-215 remains to be further studied. In
the present study, the expression of miR-215 and its mechanism of
action in NSCLC was investigated.
Materials and methods
Patients
A total of 56 patients with NSCLC (27 males and 29
females) who were subjected to resection of tumor tissues (NSCLC
group) and tumor-adjacent tissues (normal group) at Ningbo No. 2
Hospital (Ningbo, China) between January 2014 and November 2015
were included in the present study. The resected tissues were
stored at −80°C prior to use. The age range of the patients was
29–73 years, and the mean age was 46.7±1.8 years. None of the
patients received adjuvant therapy prior to surgery. Among these
patients, 32 had lymphatic metastasis (N1 group) and 24 had no
lymphatic metastasis (N0 group). According to the TNM staging
standards of the American Joint Committee on Cancer published in
2003 (19), 17 patients were at
stage I, 22 patients were at stage II, 8 patients were at stage III
and 9 patients were at stage IV. All procedures were approved by
the Ethics Committee of Ningbo No. 2 Hospital. Written informed
consent was obtained from all patients or their families.
Cells
Lung cancer A549 cell line was purchased from
Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). A549 cells were defrosted at 37°C and cultured
in 10 ml fresh Dulbecco's modified Eagle's medium (DMEM; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at 37°C and 5%
CO2 for 24 h. After 24 h of incubation, the old medium
was discarded, and 5 ml fresh high-glucose DMEM supplemented with
10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) was
added for subsequent culture. The medium was replaced every 2 days,
and the cells were passaged when a confluency of 90% was
reached.
A549 cells (1×105/well) were seeded into
24-well plates and divided into an miR-negative control (NC) group
and an miR-215 mimics group. When 70–90% confluency was reached,
1.25 µl miR-215 mimic (5′-ATGACCTATGAATTGACAGAC-3′) or miR-NC
(5′-TGACATAGACTAGCTCATACGA-3′) (20 pmol/µl; Guangzhou RiboBio Co.,
Ltd., Guangzhou, China) and 1 µl Lipofectamine 2000®
(Thermo Fisher Scientific, Inc.) were added into two individual
vials containing 50 µl Opti-Mem medium (Thermo Fisher Scientific,
Inc.), respectively. After 5 min, the liquids in the two vials were
mixed together before being left to stand for an additional 15 min.
Subsequently, the mixture was added onto the cells for an
incubation of 6 h before changing to DMEM supplemented with 10%
FBS. The cells were cultured for 48 h under normal conditions prior
to use.
For the rescue assay, the aforementioned
transfection procedure was performed with 0.5 µg NC or
pcDNA-3.1-MMP-16 plasmid DNA (Hanbio Biotechnology Co., Ltd.,
Shanghai, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Lung tissues (100 mg) were ground into powder using
liquid nitrogen prior to the addition of 1 ml TRIzol (Thermo Fisher
Scientific, Inc.) for lysis. Following lysis, total RNA was
extracted using the phenol chloroform method (20). The purity of RNA was determined by
A260/A280 using ultraviolet spectrophotometry (Nanodrop ND2000;
Thermo Fisher Scientific, Inc.). Subsequently, cDNA was obtained by
RT using a TIANScript RT kit (Tiangen Biotech Co., Ltd., Beijing,
China) from 1 µg RNA and stored at −20°C. The RT reaction mixture
was as follows: RNA template (5 µl), Oligo dT (2 µl), super pure
dNTP (2 µl) and H2O (5.5 µl). The reaction conditions
involved heating to 70°C and sudden cooling on ice for 2 min before
transient centrifugation at room temperature and 500 × g for 30
sec. Subsequently, 4 µl 5X first-strand buffer, 0.5 µl RNasin and 1
µl TIANScript M-MLV were added prior to gentle mixing with a
pipette. After being kept at 25°C for 10 min, the sample was
incubated at 42°C for 50 min, and then at 95°C for 5 min to
terminate reactions.
For reverse transcription of miR-215, Universal cDNA
Synthesis kit II (Takara, Dalian, China) was used. To determine the
expression levels of miR-215 in tissues ExiLENT
SYBR®-Green master mix (Takara Biotechnology Co., Ltd.)
was utilized, using GAPDH (forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′) as an internal reference. The
RT-qPCR reaction system (30 µl) contained 5 µl cDNA, 10 µl mix, 1
µl upstream primer (miR-215, 5′-ATGACCTATGAATTGACAGAC-3′), 1 µl
downstream universal primer (provided by the kit) and 13 µl
ddH2O. The PCR protocol was as follows: Initial
denaturation at 95°C for 3 min; followed by 40 cycles of
denaturation at 95°C for 30 sec and annealing at 60°C for 30 sec
(iQ5; Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
2−ΔΔCq method was used to calculate the relative
expression of miR-215 against GAPDH (21). Each sample was tested in
triplicate.
Cell Counting Kit (CCK)-8 assay
A549 cells in the NC and miR-215 mimic groups were
seeded into 96-well plates at a density of 5,000 cells/well in
triplicate. Every 24 h, the cells were incubated at 37°C with 20 µl
CCK-8 reagent (Beyotime Institute of Biotechnology, Shanghai,
China) for 30 min. Absorbance at 490 nm was read on a microplate
reader (168–1000; Model 680; Bio-Rad Laboratories, Inc.) at 24, 48
and 72 h, and proliferation curves were plotted using absorbance
values at each time point.
Transwell assays
Transwell chambers (8-µm pore diameter and 24 wells;
Corning Incorporated, Corning, NY, USA) were used to evaluate the
migratory ability of A549 cells. Transfected cells were collected
by trypsin digestion, and resuspended at a density of
2×105 cells/ml using DMEM. The cell suspension (200 µl)
was added into the upper chamber. In the lower chamber, 500 µl DMEM
supplemented with 10% FBS was added. Following incubation for 24 h,
the cells in the upper chamber were wiped using a cotton swab.
Subsequently, the cells in the lower chamber were fixed using 4%
formaldehyde for 10 min at room temperature, and then subjected to
Giemsa staining at room temperature for 1 min. Following three
washes with PBS, cells that migrated to the lower side of the
chamber were counted under a light microscope (five fields;
magnification, ×200) to evaluate migratory ability.
Matrigel invasion chambers (BD Biosciences, Franklin
Lakes, NJ, USA) were used to determine the invasive ability of
cells. Matrigel was first diluted with serum-free DMEM at a ratio
of 1:2. In the upper chamber (105 cells), 50 µl diluted
Matrigel was added and kept at 37°C for 1 h. In the lower chamber,
500 µl DMEM supplemented with 10% FBS was added. Following
incubation for 72 h, the cells in upper chamber were wiped with a
cotton swab. Then, the chamber was fixed using 4% formaldehyde for
10 min at room temperature, and then subjected to Giemsa staining
at room temperature for 1 min. Subsequent to three washes with PBS,
cells that moved to the lower side of the chamber were counted
under a light microscope (five fields; magnification, ×200) to
evaluate invasive ability.
Western blotting
At 48 h after transfection, cells were trypsinized
and collected by centrifugation at 12,000 × g at 4°C for 10 min.
Following this, precooled radioimmunoprecipitation assay lysis
buffer (600 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1% SDS,
1% TritonX-100, and 1% sodium deoxycholate; Beyotime Institute of
Biotechnology) and phenylmethylsulfonyl fluoride were added to the
samples. Subsequent to lysis for 5 min on ice, the mixture was
centrifuged at 12,000 × g/min at 4°C for 10 min. The supernatant
was used to determine protein concentration using a bicinchoninic
acid protein concentration determination kit [cat. no. RTP7102;
Real-Times (Beijing) Biotechnology Co., Ltd., Beijing, China].
Protein samples (50 µg) were then mixed with an equal volume of 2X
SDS loading buffer prior to denaturation in a boiling water bath
for 10 min. Afterwards, the samples (5 µl) were subject to 10%
SDS-PAGE at 100 V. The resolved proteins were transferred to
polyvinylidene difluoride membranes on ice (300 mA, 2 h) and
blocked with 5% skimmed milk at room temperature for 1 h.
Subsequently, the membranes were incubated with rabbit anti-human
MMP-16 polyclonal primary antibody (1:1,000; cat. no. ab73877;
Abcam, Cambridge, UK) and mouse anti-human GAPDH primary antibody
(1:5,000; cat. no. ab8245; Abcam) at 4°C overnight. Following
extensive washing with PBS with Tween-20 (five times, 5 min each),
the membranes were incubated, respectively, with goat anti-mouse
horseradish peroxidase (HRP)-conjugated secondary antibody for
GAPDH (1:5,000; cat. no. ab6789; Abcam) and goat anti-rabbit
HRP-conjugated secondary antibody for MMP-16 (1:2,000; cat. no.
ab205718; Abcam) for 1 h at room temperature. Following this, the
membranes were washed with PBS with Tween-20, five times for 5 min
each. The membrane was subsequently developed with an enhanced
chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for imaging. Image Lab v3.0 software (Bio-Rad
Laboratories, Inc.) was used to acquire and analyze imaging
signals. The relative content of MMP-16 protein was expressed as
the MMP-16/GAPDH ratio.
Bioinformatics
Bioinformatics prediction is a powerful tool for the
study of the functions of miR. To understand the regulatory
mechanism of MMP-16, TargetScan (release 7.1; targetscan.org) was utilized to predict miR molecules
that may regulate MMP-16.
Dual-luciferase reporter assay
The potential target genes of miR-215 were predicted
using bioinformatics. miR-215 was identified to be capable of
binding with the 3′-UTR of MMP-16 mRNA. According to bioinformatics
results, wild-type (WT) and mutant seed regions of miR-215 in the
3′-UTR of MMP-16 gene were chemically synthesized in vitro,
with the addition of the SpeI and HindIII restriction
sites, and then cloned into pMIR-REPORT luciferase reporter
plasmids (Beyotime Institute of Biotechnology). Plasmids (0.5 µg)
with WT or mutant 3′-UTR DNA sequences were co-transfected with
miR-215 mimics into A549 cells using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.). Following cultivation for 24 h,
the cells were lysed using a dual-luciferase reporter assay kit
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's manual, and fluorescence intensity was measured
using a GloMax 20/20 luminometer (Promega Corporation). Using
Renilla fluorescence activity as an internal reference, the
fluorescence value of each group of cells was measured.
Statistical analysis
The results were analyzed using SPSS 16.0 (SPSS,
Inc., Chicago, IL, USA). All measurement data were expressed as the
mean ± standard deviation. Intergroup comparison was performed
using paired Student's t-tests. The results among multiple groups
were compared using one-way analysis of variance followed by
Bonferroni's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Reduced expression of miR-215 in NSCLC
is negatively associated with lymphatic metastasis and TNM
staging
To measure the expression of miR-215 in NSCLC
tissues, RT-qPCR was employed. The data demonstrated that the level
of miR-215 in tumor tissues was significantly lower than that in
tumor-adjacent tissues (P<0.05; Fig.
1A). In addition, the expression of miR-215 in patients with
lymphatic metastasis (N1) was significantly lower than that in
patients with no lymphatic metastasis (N0) (P<0.05; Fig. 1B). Similarly, the expression level of
miR-215 in patients at TNM stages III and IV was significantly
lower than that in patients at stages I or II and all groups were
significantly different from each other (P<0.05; Fig. 1C). The results suggest that the
reduced expression of miR-215 in NSCLC is negatively associated
with lymphatic metastasis and TNM staging.
Overexpression of miR-215 inhibits the
proliferation of A549 cells in vitro
To determine the effect of miR-215 on the
proliferation of A549 cells, RT-qPCR and a CCK-8 assay were used.
The data revealed that miR-215 expression in A549 cells transfected
with miR-215 mimics was significantly higher than that in cells
transfected with miR-NC (P<0.05; Fig.
2A). In addition, A549 cells transfected with miR-215 mimics
had significantly lower absorbance at 490 nm compared with those
transfected with miR-NC at 24 and 48 h (P<0.05; Fig. 2B). The results indicate that
overexpression of miR-215 inhibits the proliferation of A549 cells
in vitro.
Upregulated expression of miR-215
inhibits the migration and invasion of A549 cells in vitro
To examine the migration and invasion of A549 cells,
Transwell assays were performed. Migration assay demonstrated that
the number of cells in the miR-215 mimics group that migrated
through the chamber membrane was significantly lower than that in
the miR-NC group (P<0.05; Fig.
3). Similarly, the invasion assay demonstrated that the number
of invaded cells in the miR-215 mimics group was significantly
smaller than that in the miR-NC group (P<0.05; Fig. 3). These results suggest that
upregulated expression of miR-215 inhibits the migration and
invasion of A549 cells in vitro.
MMP-16 is a potential target of
miR-215
In the present study, TargetScan was used to
identify target sequences of miR-215. The analysis revealed 212
transcripts with conserved sites, and it was demonstrated that
MMP-16 was a potential target of miR-215, containing two conserved
sites in the 3′-UTR. To the best of our knowledge, there have been
no previous reports claiming that miR-215 could directly regulate
MMP-16 mRNA. However, it is well established that MMP-16 serves a
crucial role in multiple cancer types and may be regulated by miR
(15). The result suggests that
MMP16 is a potential target of miR-215 (data not shown).
miR-215 may exert its biological
functions by regulating the expression of MMP-16
To determine the effect of miR-215 on the expression
of MMP-16 protein in A549 cells, western blotting was performed.
Western blotting results demonstrated that expression of MMP-16
protein in A549 cells with overexpression of miR-215 was
significantly lower than that in cells of the miR-NC group
(P<0.05; Fig. 4). This result
suggests that miR-215 may exert its biological functions by
regulating the expression of MMP-16.
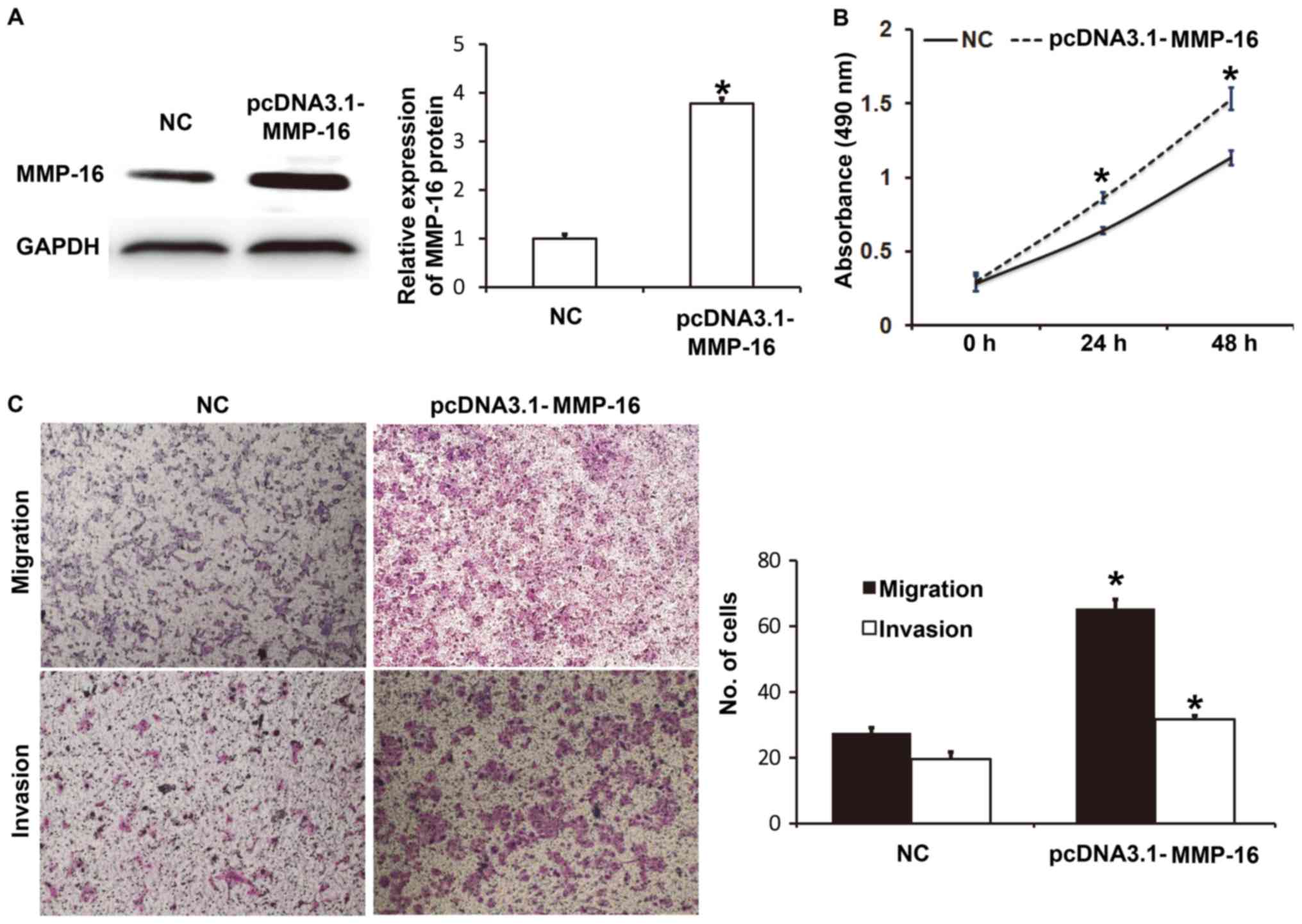
Elevated expression of MMP-16 promotes
the proliferation, migration and invasion of A549 cells
To determine the biological function of MMP-16 in
A549 cells, the cells were transfected with NC and pcDNA3.1-MMP-16
plasmids. Western blotting revealed that MMP-16 protein expression
in A549 cells transfected with pcDNA3.1-MMP-16 was significantly
higher than that in the NC group (P<0.05; Fig. 5A). CCK-8 assay demonstrated that A549
cells transfected with pcDNA3.1-MMP16 had significantly higher
absorbance at 490 nm than those transfected with NC at 24 and 48 h
(P<0.05; Fig. 5B). Transwell
assays indicated that the numbers of migrated or invaded cells in
the pcDNA3.1-MMP16 group were significantly higher than those in
the NC group (P<0.05; Fig. 5C).
These results indicate that elevated expression of MMP-16 promotes
the proliferation, migration and invasion of A549 cells.
miR-215 regulates the proliferation,
migration and invasion of A549 cells via MMP-16
To determine whether miR-215 regulates the
proliferation, migration and invasion of A549 cells via the MMP-16
protein, A549 cells were co-transfected with pcDNA3.1-MMP-16 and
miR-215 mimics. Western blotting demonstrated that MMP-16 protein
expression in A549 cells transfected with miR-215 mimics was lower
than that in the NC group, while co-transfection with miR-215
mimics and pcDNA3.1-MMP-16 increased the expression level of MMP-16
(Fig. 6A). CCK-8 assay revealed that
A549 cells transfected with miR-215 mimics had significantly lower
absorbance at 490 nm than those transfected with NC at 24 and 48 h
(P<0.05); while co-transfection with miR-215 mimics and
pcDNA3.1-MMP-16 significantly increased the absorbance at 490 nm
compared with that in the miR-215 mimics group at 24 and 48 h
(P<0.05; Fig. 6B). Transwell
assays indicated that the numbers of migrated or invaded cells in
the miR-215 mimics group were significantly lower than those in the
NC group (P<0.05); however, co-transfection with miR-215 mimics
and pcDNA3.1-MMP-16 significantly increased these numbers compared
with that in the miR-215 mimics group (P<0.05; Fig. 6C). These results suggest that miR-215
regulates the proliferation, migration and invasion of A549 cells
via MMP-16.
miR-215 binds with the 3′-UTR seed
region of MMP-16 mRNA
To identify the interaction between miR-215 and the
3′-UTR of MMP-16 mRNA, a dual-luciferase reporter assay was
performed. The fluorescence value of cells co-transfected with
miR-215 mimics and pMIR-REPORT-WT luciferase reporter plasmids was
significantly lower than that in the NC group (P<0.05). By
contrast, the fluorescence value of cells co-transfected with
miR-215 mimics and pMIR-REPORT-mutant luciferase reporter plasmids
was not significantly different from that in the NC group
(P>0.05; Fig. 7). This result
indicates that miR-215 binds with the 3′-UTR seed region of MMP-16
mRNA.
Discussion
miR-215 is a newly discovered miR molecule that is
associated with tumors. The expression of miR-215 is induced by p53
and it inhibits tumor growth by regulating cell cycle checkpoint
protein expression (22). Studies
have revealed that the expression of miR-215 is reduced in multiple
tumors, and that it is negatively associated with the metastasis
and prognosis of tumors. For example, Wang et al (23) reported that reduced expression of
miR-215 in peripheral blood of patients with acute myeloid leukemia
is associated with a poor prognosis. Zhou et al (24) demonstrated that expression of miR-215
is closely related to the prognosis of patients with breast cancer,
and the lower the expression, the lower the 5-year survival rate.
The present study indicated that expression of miR-215 in NSCLC is
significantly decreased. In the comparison between lymphatic
metastasis and TNM staging, it was revealed that the expression of
miR-215 in the group with lymphatic metastasis was lower than that
in the non-metastatic group, and the expression of miR-215 in
patients at stages III/IV was significantly lower than that of
patients at stages I/II. These results suggest that miR-215 may
have the biological functions of a tumor suppressor gene in NSCLC.
However, another report revealed that the expression of miR-215 is
upregulated in some tumors types, and promotes the proliferation,
metastasis and invasion of tumor cells. It was demonstrated that
miR-215 expression in gastric cancer cells is upregulated and
promotes the drug resistance, proliferation, migration and invasion
of gastric cancer cells (16). This
suggests that the expression and biological function of miR-215 may
be dependent on the tumor type.
Studies have indicated that miR-215 inhibits the
proliferation, metastasis and invasion of tumor cells by targeting
multiple genes in vitro. For example, Chen et al
(25) reported that miR-215 inhibits
the proliferation, migration and invasion of colon cancer cells by
targeting Yin-Yang 1 gene. Li et al (17) revealed that miR-215 suppresses the
proliferation, migration and invasion of pancreatic cancer cells by
targeting the ZEB2 gene. Additionally, Hou et al (26) discovered that miR-215 inhibits the
invasion and metastasis of NSCLC by targeting the ZEB2 gene.
Similarly, the present study demonstrated that expression of
miR-215 inhibits the proliferation, migration and invasion of lung
cancer A549 cells, and downregulated expression of miR-215 may
promote the occurrence and development of lung cancer. Notably, the
present data indicate that miR-215 serves the role of a tumor
suppressor gene by targeting MMP-16 gene expression, suggesting
that miR-215 has more than one target gene in lung cancer.
MMP-16 is a membrane-type metalloproteinase, and it
takes effect by activating proMMP-2 (gelatinase A) that is produced
by cells (27). Studies have
demonstrated that MMP-16 promotes the proliferation, metastasis and
invasion of tumors. For example, Cao et al (28) reported that MMP-16 promotes the
proliferation, invasion and metastasis of gastric cancer, and its
expression is directly related to the prognosis of patients with
gastric cancer. In addition, miR-146b-5p inhibits pancreatic cancer
progression by downregulating MMP-16 expression (29). The present data indicate that
overexpression of miR-215 downregulates the expression of MMP-16 in
A549 cells, and overexpression of MMP-16 inhibits the effects of
miR-215 on the proliferation, migration and invasion of A549 cells.
In addition, overexpression of MMP-16 itself promotes the
proliferation, migration and invasion of A549 cells.
Dual-luciferase reporter assays in the present study revealed that
miR-215 is able to directly bind to the 3′-UTR of MMP-16 to exert
its biological functions.
Taken together, the present study demonstrates that
decreased expression of MMP-16 induced by miR-215 suppresses the
proliferation, metastasis and invasion of NSCLC cells. In addition,
miR-215 may be a target for the therapy of NSCLC. In the future,
the expression of MMP-16 in NSCLC tissues compared with normal
controls will be examined, and its association with miR-215
expression and patient prognosis will be investigation. In
addition, the association between miR-215 expression and NSCLC
subtype will also be explored in future studies.
Acknowledgements
We would like to thank Dr Jie Li of the Department
of Chest Surgery, Ningbo No. 2 Hospital (Ningbo, China) for his
valuable help.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kaira K, Tomizawa Y, Imai H, Sakurai R,
Matsuura M, Yoshii A, Ochiai M, Kotake M, Ebara T, Saitoh JI, et
al: Phase I study of nab-paclitaxel plus carboplatin and concurrent
thoracic radiotherapy in patients with locally advanced non-small
cell lung cancer. Cancer Chemother Pharmacol. 79:165–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong Z, Zhao L, Lu S, Xiong J and Geng Z:
Overexpression of TSPAN8 promotes tumor cell viability and
proliferation in nonsmall cell lung cancer. Cancer Biother
Radiopharm. 31:353–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Presley CJ, Soulos PR, Tinetti M, Montori
VM, Yu JB and Gross CP: Treatment burden of medicare beneficiaries
with stage I non-small-cell lung cancer. J Oncol Pract.
13:e98–e107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiu ZL, Shen CT, Sun ZK, Wei WJ, Zhang XY,
Song HJ and Luo QY: Circulating long non-coding rnas act as
biomarkers for predicting 131I uptake and mortality in papillary
thyroid cancer patients with lung metastases. Cell Physiol Biochem.
40:1377–1390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu TC, Hsieh MJ, Wu WJ, Chou YE, Chiang
WL, Yang SF, Su SC and Tsao TC: Association between survivin
genetic polymorphisms and epidermal growth factor receptor mutation
in non-small-cell lung cancer. Int J Med Sci. 13:929–935. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma L, Qiu B, Zhang J, Li QW, Wang B, Zhang
XH, Qiang MY, Chen ZL, Guo SP and Liu H: Survival and prognostic
factors of non-small cell lung cancer patients with postoperative
locoregional recurrence treated with radical radiotherapy. Chin J
Cancer. 36:932017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei WE, Mao NQ, Ning SF, Li JL, Liu HZ,
Xie T, Zhong JH, Feng Y, Wei CH and Zhang LT: An analysis of EGFR
mutations among 1506 cases of non-small cell lung cancer patients
in Guangxi, China. PLoS One. 11:e01687952016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ghafoor Q, Baijal S, Taniere P, O'Sullivan
B, Evans M and Middleton G: Epidermal growth factor receptor (EGFR)
kinase inhibitors and non-small cell lung cancer (NSCLC)-advances
in molecular diagnostic techniques to facilitate targeted therapy.
Pathol Oncol Res. Dec 21–2017.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang T, Xu X, Xu Q, Ren J, Shen S, Fan C
and Hou Y: miR-19a promotes colitis-associated colorectal cancer by
regulating tumor necrosis factor alpha-induced protein 3-NF-κB
feedback loops. Oncogene. 36:3240–3251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Labourier E, Lloyd M, Andruss B, Adai A
and Schwarzbach A: An miRNA assay for the classification of benign
and neoplastic lesions in pancreatic fine-needle aspirates. J Clin
Oncol. 29 4 Suppl:S1632011. View Article : Google Scholar
|
|
11
|
Huang G, Pan J, Ye Z, Fang B, Cheng W and
Cao Z: Overexpression of miR-216b sensitizes NSCLC cells to
cisplatin-induced apoptosis by targeting c-Jun. Oncotarget.
8:104206–104215. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pei K, Zhu JJ, Wang CE, Xie QL and Guo JY:
MicroRNA-185-5p modulates chemosensitivity of human non-small cell
lung cancer to cisplatin via targeting ABCC1. Eur Rev Med Pharmacol
Sci. 20:4697–4704. 2016.PubMed/NCBI
|
|
13
|
Sun CC, Li SJ, Zhang F, Zhang YD, Zuo ZY,
Xi YY, Wang L and Li DJ: The novel miR-9600 suppresses tumor
progression and promotes paclitaxel sensitivity in non-small-cell
lung cancer through altering STAT3 expression. Mol Ther Nucleic
Acids. 5:e3872016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang P, Deng Y and Fu X: MiR-509-5p
suppresses the proliferation, migration, and invasion of non-small
cell lung cancer by targeting YWHAG. Biochem Biophys Res Commun.
482:935–941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei Y, Sun J and Li X: MicroRNA-215
enhances invasion and migration by targeting retinoblastoma tumor
suppressor gene 1 in high-grade glioma. Biotechnol Lett.
39:197–205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li N, Zhang QY, Zou JL, Li ZW, Tian TT,
Dong B, Liu XJ, Ge S, Zhu Y, Gao J and Shen L: miR-215 promotes
malignant progression of gastric cancer by targeting RUNX1.
Oncotarget. 7:4817–4828. 2016.PubMed/NCBI
|
|
17
|
Li QW, Zhou T, Wang F, Jiang M, Liu CB,
Zhang KR, Zhou Q, Tian Z and Hu KW: MicroRNA-215 functions as a
tumor suppressor and directly targets ZEB2 in human pancreatic
cancer. Genet Mol Res. 14:16133–16145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen B, Huang Z, Zhang Y, Chen Y and Li Z:
MicroRNA-145 suppresses osteosarcoma metastasis via targeting
MMP16. Cell Physiol Biochem. 37:2183–2193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arslan D, Bozcuk H, Gunduz S, Tural D,
Tattli AM, Uysal M, Goksu SS, Bassorgun Cİ, Koral L, Coskun HS, et
al: Survival results and prognostic factors in T4 N0-3 non-small
cell lung cancer patients according to the AJCC 7th edition staging
system. Asian Pac J Cancer Prev. 15:2465–2472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahmad J, Baig MA, Ali AA, Al-Huqail A,
Ibrahim MM and Qureshi MI: Comparative assessment of four RNA
extraction methods and modification to obtain high-quality RNA from
Parthenium hysterophorus leaf. 3 Biotech. 7:3732017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Georges SA, Biery MC, Kim SY, Schelter JM,
Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA and
Chau BN: Coordinated regulation of cell cycle transcripts by
p53-inducible microRNAs, miR-192 and miR-215. Cancer Res.
68:10105–10112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang YX, Zhang TJ, Yang DQ, Yao DM, Yang
L, Zhou JD, Deng ZQ, Ma JC, Guo H, Wen XM, et al: Reduced miR-215
expression predicts poor prognosis in patients with acute myeloid
leukemia. Jpn J Clin Oncol. 46:350–356. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou SW, Su BB, Zhou Y, Feng YQ, Guo Y,
Wang YX, Qi P and Xu S: Aberrant miR-215 expression is associated
with clinical outcome in breast cancer patients. Med Oncol.
31:2592014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Z, Han S, Huang W, Wu J, Liu Y, Cai
S, He Y, Wu S and Song W: MicroRNA-215 suppresses cell
proliferation, migration and invasion of colon cancer by repressing
Yin-Yang 1. Biochem Biophys Res Commun. 479:482–488. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R
and Zhao C: miR-215 functions as a tumor suppressor and directly
targets ZEB2 in human non-small cell lung cancer. Oncol Lett.
10:1985–1992. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang WL, Chen YF, Meng HZ, Du JJ, Luan
GN, Wang HQ, Yang MW and Luo ZJ: Role of miR-155 in the regulation
of MMP-16 expression in intervertebral disc degeneration. J Orthop
Res. 35:1323–1334. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cao L, Chen C, Zhu H, Gu X, Deng D, Tian
X, Liu J and Xiao Q: MMP16 is a marker of poor prognosis in gastric
cancer promoting proliferation and invasion. Oncotarget.
7:51865–51874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin F, Wang X, Jie Z, Hong X, Li X, Wang M
and Yu Y: Inhibitory effects of miR-146b-5p on cell migration and
invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ
Sci Technolog Med Sci. 31:509–514. 2011. View Article : Google Scholar : PubMed/NCBI
|