Introduction
Osteoarthritis (OA) is a common disease of the
skeletal system. As the most frequent form of joint disease, OA can
occur in any joint but primarily affects knees, hips, hands and the
spine, thus constituting a leading cause of musculoskeletal
disability worldwide (1,2). Degradation of hyaline articular
cartilage and remodeling of the subchondral bone with sclerosis are
the main features of OA (3,4). At first, patients with OA present with
joint stiffness and soreness (5).
The articulation may show some swelling caused by an effusion and
synovitis. When the bare bony surfaces grate against each other,
locking may also occur, which will cause more serious pain
(6) and will result in symptoms that
decrease quality of life. A previous epidemiological survey showed
that approximately 10% of the world's population aged over 60 years
may present with symptomatic OA (7,8).
Moreover, according to the World Health Organization (WHO), OA has
been the fourth major cause of disability for many years, with an
incidence rate of 40% in the 55–64 age group (9). As the global population mages, the
incidence of OA increases year after year. In clinical practice,
diagnosis of OA is based mainly on radiological examination and
clinical assessment, and a key point for OA diagnosis is to
identify early stage onset and progression (10). Additionally, OA is considered to be a
polygenic and multifactorial illness (11–13).
Studies have shown that age, sex, obesity, genetics, ethnicity,
behavioral influences, occupation and environmental factors, and
particularly genetic factors strongly affect the occurrence and
development of OA (14,15). Exploring the genetic factor mechanism
would prove helpful in the early diagnosis of OA.
The asporin (ASPN) gene has been reported as a
predisposing gene for OA in several previous studies and encodes a
cartilage extracellular protein belonging to the small leucine-rich
proteoglycan (SLRP) family (3).
These proteins can regulate chondrogenesis by binding to cartilage
transforming growth factor-β (TGF-β) and inhibiting the expression
of the TGF-β1-induced gene in cartilage (16–18). In
normal cartilage, ASPN is expressed at low levels but in OA
articular cartilage, it is abundantly expressed, thus playing a key
role in cartilage metabolism (19).
The D14 allele of ASPN has been reported as a gene potentially
contributing to knee OA (KOA), and there is a significant
difference in the allele frequency of ASPN D13 between female OA
patients and controls (20). A
number of studies have examined the association between ASPN
polymorphisms and OA, but their results and conclusions have been
contradictory. Therefore, there is not yet strong evidence of the
association between OA and ASPN. Therefore, it is necessary to
conduct additional studies to confirm the relationship between OA
and ASPN. In the present study, an updated, more comprehensive and
more detailed cumulative meta-analysis was performed based on the
newly published original studies evaluating the association between
ASPN polymorphism and OA susceptibility, to investigate whether the
D-repeat polymorphism is associated with susceptibility to OA and
whether it can provide a reference for the early diagnosis of
OA.
Materials and methods
Search strategy
To perform this meta-analysis, the Cochrane Library,
PubMed and Excerpta Medica databases (EMBASE) were used to
comprehensively search the relevant literature. The following
keywords were used: ‘(Asporin or ASPN), (Osteoarthritis or OA), and
(Asporin and ASPN or Osteoarthritis and OA)’. In addition, the
Chinese electronic databases: VIP, CNKI and Wan Fang were used to
search the relevant literature. All articles that were selected for
analysis focused on ASPN gene polymorphisms and OA susceptibility
and had been published from January 1st, 1915 through February 1st,
2017. By using the aforementioned search strategy, two authors
independently completed screening the records and then assessed the
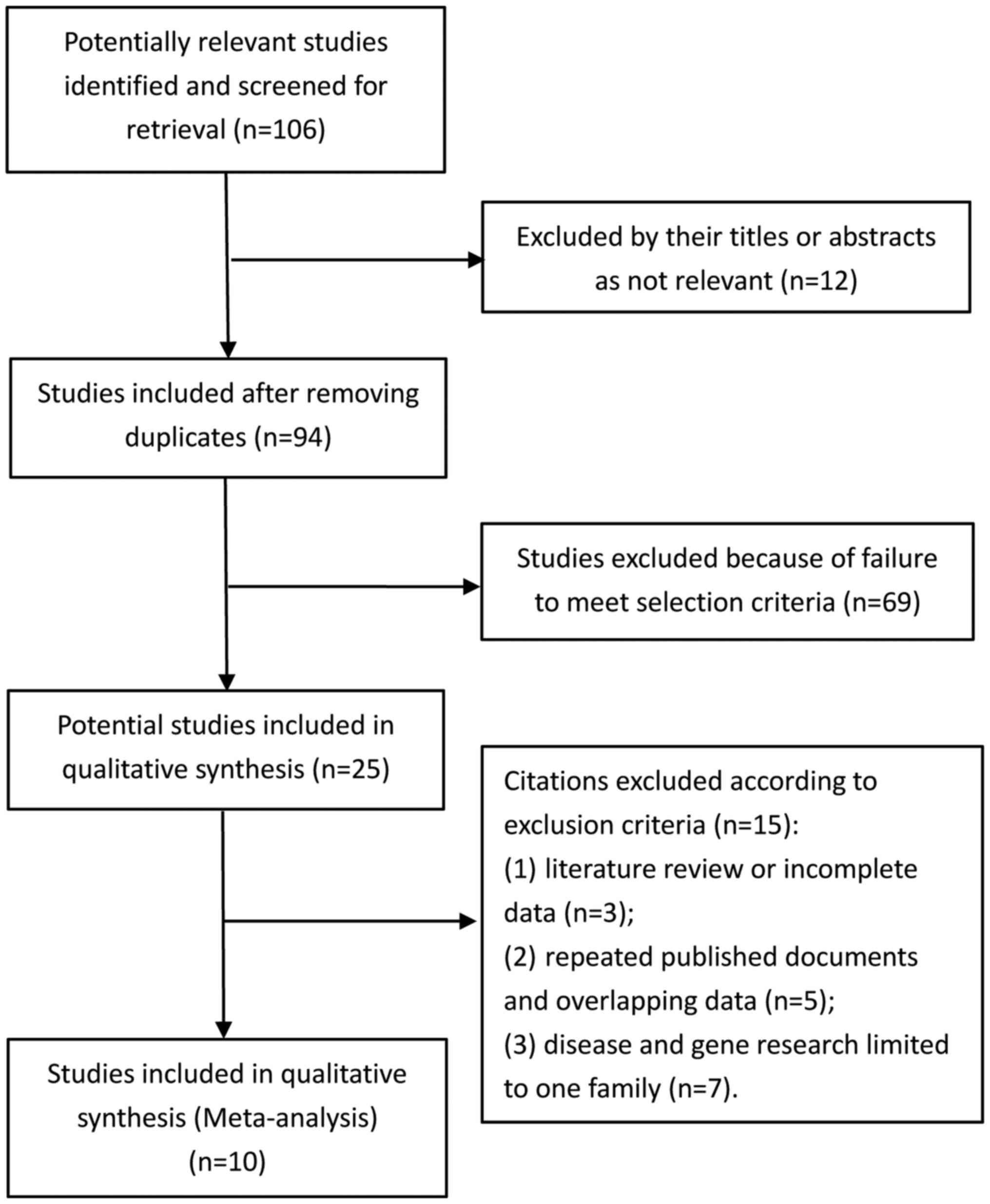
final results. The study selection flowchart is shown in Fig. 1.
Inclusion and exclusion criteria
To be included in the meta-analysis, studies had to
meet the following criteria: i) They had to be a case-control study
(or cohort study); ii) studies had to provide data on the number of
studies and ratios, and OA diagnosis had to be performed using a
standardized method; and iii) they had to contain a study of
genotype and the ASPN allele, the relevant odds ratio (OR) and the
95% confidence interval (CI). Exclusion criteria were: i)
Literature review or incomplete data; ii) repeated published
documents and overlapping data; and iii) disease and gene research
limited to one family. There were no restrictions on the country
and language under study.
Data extraction
Two reviewers (Jing Wang and Rongqiang Zhang)
independently reviewed and extracted data from all qualified
studies. Data extraction from original studies included first
author's name, year of publication, total sample size and age of OA
patients and controls, diagnostic criteria, country where the study
was performed, ethnicity of the participants, OA sites and genotype
frequencies of the two groups. The continent of origin was
categorized as follows: Europe or America (=1) and Asia (=2). Total
sample sizes of OA patients and controls were categorized as
<1,000 (=1) and ≥1,000 (=2). Moreover, OA sites were categorized
as KOA (=1) and hand or hip OA (=2). The two investigators
carefully checked the data extraction information and reached
consensus to ensure accuracy of the extracted data. In the case of
disagreement, the two reviewers double-checked the original data
together and then discussed them in order to reach an agreement. If
they failed to reach an agreement, a third reviewer (Aimin Yang)
was invited to the discussion.
Methodological quality assessment
The methodological quality of the included studies
was independently assessed by two reviewers (Jing Wang and
Rongqiang Zhang), according to our revised Scale for Quality
Assessment (Table I) based on the
scales of Thakkinstian et al (21) and Qin et al (22). The revised scale covered criteria
such as representativeness of cases, credibility of controls,
ascertainment of OA, genotyping examination, Hardy-Weinberg
equilibrium (HWE) and association assessment. Any disagreement
related to methodological quality was resolved by consensus. Scores
ranged from 0 to 12. Articles that were assigned a score <8 were
considered ‘low-quality’ studies, whereas papers with a score ≥8
were considered ‘high-quality’ studies.
 | Table I.Scale for quality assessment. |
Table I.
Scale for quality assessment.
| Criteria | Score |
|---|
| Representativeness
of cases |
|
|
Selected from population | 2 |
|
Selected from any OA/surgery
service | 1 |
|
Selected without clearly
defined sampling frame or with extensive inclusion/exclusion
criteria | 0 |
| Credibility of
controls |
|
|
Population-or
neighbor-based | 3 |
| Blood
donors or volunteers | 2 |
|
Hospital-based (Joint
diseases-free patients) | 1 |
| Healthy
volunteers, but without total description | 0.5 |
| Not
described | 0 |
| Ascertainment of
OA |
|
|
Clinical and X-ray or MRI
confirmation | 2 |
|
Diagnosis of OA by patient
medical record | 1 |
| Not
described | 0 |
| Genotyping
examination |
|
|
Genotyping done under
‘blinded’ condition | 1 |
|
Unblinded or not
mentioned | 0 |
| Hardy-Weinberg
equilibrium |
|
|
Hardy-Weinberg equilibrium in
controls | 2 |
|
Hardy-Weinberg disequilibrium
in controls | 1 |
| No
checking for Hardy-Weinberg disequilibrium | 0 |
| Association
assessment |
|
| Assess
association between genotypes and OA with appropriate statistics
and adjustment for confounders | 2 |
| Assess
association between genotypes and OA with appropriate statistics
without adjustment for confounders | 1 |
|
Inappropriate statistics
used | 0 |
Statistical analysis
RevMan software v5.2 (The Cochrane Collaboration,
Oxford, UK) was used to perform the meta-analysis. When the
selected original studies met the criteria for homogeneity
(I2<50% or P>0.05), a fixed-effects model
was chosen to conduct the meta-analysis. If they met the criteria
for heterogeneity (I2>50% or P<0.05), a
random-effects model was chosen to conduct the meta-analysis.
Univariate and multivariate meta-regression analyses were then
performed to explore the source of heterogeneity between studies
using Stata 13.0 software (Stata Corp, College Station, TX, USA).
In meta-regression analyses, LogOR (where OR stands for the
relevant OR of cases vs. control) was taken as the dependent
variable. The sample size, OA sites and continent of origin were
taken as independent variables. If necessary, a subgroup analysis
was conducted according to the source of heterogeneity. Publication
bias was analyzed with a funnel plot and Begg's test.
Results
Literature search and study
selection
A total of 34,084 relevant articles were searched
from the Cochrane Library, PubMed, the Excerpta Medica database,
VIP, CNKI and Wan Fang. Finally, 10 articles including 11 studies
(Kizawa's research included both an independent case-control study
and an independent cohort study) were included in our
meta-analysis.
Study characteristics
In total, 8,503 participants (4,842 OA patients and
3,661 controls) were eligible for inclusion, involving three
different regions, each of which encompassed a distinct human race,
and four types of OA pathogenesis research. Cohort studies included
137 patients and 234 controls while case-control studies comprised
4,705 patients and 3,427 controls. A total of 4 articles focused on
Asians, 4 on Europeans and 2 on Americans. Among them, 7 studies
examined KOA and 4 studies focused on hand or hip OA. Participants
included in both case and control groups were all OA patients and
healthy controls confirmed by standard diagnostic criteria (K/L
grading system5, Criteria of American Rheumatology
College, clinical and radiologic diagnosis), respectively.
Polymorphisms of the ASPN gene were detected using multiplex
polymerase chain reaction (PCR) and single nucleotide polymorphism
(SNP) analyses, which were performed using the GenomeLab SNPstream
Genotyping System (Beckman Coulter, Fullerton, CA, USA). The
characteristics of all included studies are presented in Table II.
 | Table II.Baseline characteristics of the 10
articles (including 11 studies) included in the meta-analysis. |
Table II.
Baseline characteristics of the 10
articles (including 11 studies) included in the meta-analysis.
|
|
|
|
|
|
|
|
| OA | Control |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Author year | Design | Na (OA/Control) | Ageb (OA/Control) | Diagnostic
criteriac | Country | Ethnicity | OA site | D12 | D13 | D14 | D15 | D16 | D17 | D18 | Others | D12 | D13 | D14 | D15 | D16 | D17 | D18 | Others | Score | HWE | (Refs.) |
|---|
| Mustafa 2005 | Case-control | 1,247/748 | 65.0/69.0 | KL grade
system | UK | Caucasians | Hip/Knee | 130 | 1,183 | 352 | 498 | 216 | 60 | 34 | 5 | 76 | 752 | 190 | 289 | 124 | 26 | 22 | 7 | 9 | No | (18) |
| Kizawa 2005 | Case-control | 986/374 | 65.4/28.8 | KL grade
system | Japan | Honshu | Knee/Hip | – | 1,190 | 154 | – | – | – | – | 628 | – | 479 | 36 | – | – | – | – | 233 | 12 | Yes | (27) |
| Kizawa 2005 | Cohort | 137/234 | 75.3/73.6 | KL grade
system | Japan | Honshu | Knee | – | 163 | 30 | – | – | – | – | 81 | – | 314 | 22 | – | – | – | – | 132 | 12 | Yes | (27) |
| Jiang 2006 | Case-control | 218/454 | 58.1/56.3 | KL grade
system | China | Han | Knee | 61 | 300 | 41 | 11 | 15 | 5 | 0 | 3 | 186 | 604 | 44 | 29 | 39 | 3 | 3 | – | 11 | Yes | (28) |
| Kaliakatsos
2006 | Case-control | 155/190 | 70.3/69.1 | KL grade
system | Greece | Thessaly | Knee | 20 | 118 | 47 | 84 | 20 | 11 | 6 | 4 | 17 | 189 | 53 | 74 | 30 | 16 | 0 | 1 | 7 | No | (29) |
| Duval 2006 | Case-control | 723/294 | 65/> 55 | ACR criteria | Spanish | Caucasians | Hip/Knee/Hand | 52 | 627 | 172 | 362 | 144 | 40 | 28 | 23 | 31 | 248 | 74 | 150 | 55 | 12 | 10 | 8 | 6 | No | (26) |
| Atif 2008 | Case-control | 775/551 | 77.8/69.0 | KL grade
system | USA | Caucasian | Hand/Knee | – | 750 | 206 | 169 | – | – | – | – | – | 268 | 77 | 114 | – | – | – | – | 10 | No | (19) |
| Song 2008 | Case-control | 190/376 | 60/47.7 | Radiology and
pathology | Korean | – | Knee | 60 | 265 | 22 | 13 | 15 | 5 | – | – | 118 | 483 | 65 | 28 | 51 | 7 | – | – | 9 | No | (30) |
| Arellano 2013 | Case-control | 218/222 | 57.99/52.67 | KL grade
system | Mexico | Torreon
Coahuila | Knee | 12 | 205 | 91 | 69 | 38 | 19 | 1 | 1 | 32 | 204 | 107 | 66 | 22 | 8 | 2 | 3 | 10 | Yes | (31) |
| Jazayeri 2013 | Case-control | 100/100 | 62.5/63 | Clinical and
radiologic diagnosis | Iranians | Fars | Knee | 4 | 82 | 32 | 52 | 22 | 7 | 0 | 1 | 4 | 91 | 40 | 45 | 12 | 6 | 1 | 1 | 7 | No | (3) |
| Juchtmans 2015 | Case-control | 93/118 | 56.4/51.8 | Clinical and
radiologic diagnosis | Mexico | Mexican
mestizo | Knee | – | 7 | 123 | 25 | 21 | 9 | – | 1 | – | 6 | 134 | 47 | 32 | 16 | – | 1 | 12 | Yes | (4) |
Methodological quality
Studies were generally of good quality with a mean
score of 9.55 (Table II), of which
eight studies (66.67%) had a score equal to or greater than 8,
while the three other studies (33.33%) received a score lower than
8. Two-thirds of the studies included in this meta-analysis were
thus classified as of high quality.
Results of overall meta-analysis
The association between ASPN polymorphisms and
susceptibility to OA is shown in Table
III. The results of the analyses of heterogeneity showed that
there was heterogeneity on D12 in seven studies (P=0.02,
I2=62%), on D13 (P=0.01,
I2=55%) and D14 (P=0.0001,
I2=71%) in eleven studies, and on D15 (P=0.005,
I2=63%) in nine studies. Therefore,
random-effects models were used for the meta-analysis of D12-D15.
There was no significant heterogeneity on either D16 (P=0.07,
I2=47%) or D17 (P=0.37, I2=8%)
in eight studies, or on D18 (P=0.38, I2=6%) in
six studies. Consequently, fixed-effects models were used to
calculate the pooled ORs of D16, D17 and D18. The pooled OR and its
95% CI for the D17 allele vs. others combined indicated that D17
alleles were a statistically significant risk factor for OA
development (OR=1.33, 95% CI: 1.02–1.73). No statistically
significant association was found between OA and ASPN D12, D13,
D14, D15, D16 or D18 alleles.
 | Table III.Summary of ORs and 95% CIs of the
ASPN polymorphisms and OA susceptibility. |
Table III.
Summary of ORs and 95% CIs of the
ASPN polymorphisms and OA susceptibility.
|
|
| Sample size | Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|---|
| Comparisons | No. of studies | OA | Control | OR | 95% CI | P-value | Modela | P-value |
I2 (%) |
|---|
| D12 vs. all the
others | 7 | 9,684 | 7,322 | 0.81 | 0.61–1.07 | 0.14 | R | 0.02 | 62 |
| D13 vs. all the
others | 11 | 9,684 | 7,322 | 0.94 | 0.84–1.05 | 0.25 | R | 0.01 | 55 |
| D14 vs. all the
others | 11 | 9,684 | 7,322 | 1.15 | 0.94–1.40 | 0.17 | R |
0.0001 | 71 |
| D15 vs. all the
others | 9 | 9,684 | 7,322 | 0.97 | 0.70–1.15 | 0.71 | R | 0.005 | 63 |
| D16 vs. all the
others | 8 | 9,684 | 7,322 | 1.03 | 0.89–1.19 | 0.72 | F | 0.07 | 47 |
| D17 vs. all the
others | 8 | 9,684 | 7,322 | 1.33 |
1.02–1.73 | 0.04 | F | 0.37 | 8 |
| D18 vs. all the
others | 6 | 9,684 | 7,322 | 1.07 | 0.72–1.59 | 0.75 | F | 0.38 | 6 |
Test of heterogeneity
Because the included studies showed significant
heterogeneity for D12, D13, D14 and D15, univariate and
multivariate meta-regression analyses were performed to explore the
source of this heterogeneity. For the reason that the number of
studies focusing on D12 was only 7 and the frequency of D12 alleles
reported in the original studies was small, meta-regression
analyses were only conducted on D13, D14 and D15. Meta-regression
analyses were conducted according to a random-effects model.
Univariate analyses were performed separately for the following
variables: sample size, continent of origin and OA site. Therefore
results showed that there was no statistically significant
association between independent variables (sample size, continent
of origin and OA site) and the dependent variable (LogOR) for D13,
D14, and D15 (Figs. 2–4). However, judging from adjusted R-squared
values, the sample size, continent of origin and OA site could all
impact the development of OA to a certain degree (Figs. 2–4).
Multivariate analysis results showed that sample size played a role
in the association between D13 and OA risk (P=0.039; Table IV).
 | Table IV.Multivariate meta-regression analysis
of the association between D13, D14, D15 and the risk of OA. |
Table IV.
Multivariate meta-regression analysis
of the association between D13, D14, D15 and the risk of OA.
| Genotype | Dependent
variablesa | OR | SE | t | P-value | 95% CI |
|---|
| D13 | Continent of
origin | 0.8417473 | 0.1296135 | −1.12 | 0.296 | 0.5901624,
1.200582 |
|
| OA site | 0.7689406 | 0.1452108 | −1.39 | 0.202 | 0.4974702,
1.188553 |
|
| N | 1.576952 | 0.290733 |
2.47 | 0.039 |
1.030815, 2.412438 |
| D14 | Continent of
origin | 1.366192 | 0.4471786 |
0.95 | 0.368 | 0.6422559,
2.906132 |
|
| OA site | 1.054285 | 0.4480117 |
0.12 | 0.904 | 0.3957158,
2.808876 |
|
| N | 0.9812867 | 0.3917458 | −0.05 | 0.963 | 0.3908288,
2.463799 |
| D15 | Continent of
origin | 1.147952 | 0.5037435 |
0.31 | 0.764 | 0.3922823,
3.359297 |
|
| OA site | 1.145223 | 0.6270224 |
0.25 | 0.813 | 0.2999584,
4.372395 |
|
| N | 0.6839221 | 0.3202035 | −0.81 | 0.448 | 0.2175071,
2.150502 |
Subgroup analysis
After meta-regression analysis, we conducted a
subgroup analysis according to study sample size. Indeed, when
sample size was greater than 1,000 the heterogeneity of the D13
gene decreased and became not statistically significant (both
Ps=0.08). The results indicated that when the sample size
>1,000, D17 alleles increased the risk for the development of OA
(OR=1.46, 95% CI: 1.03–2.07, P<0.05; Table V). There was no significant
association between the other ASPN polymorphisms (D12, D13, D14,
D15, D16, D18) and OA in the other subgroup analyses (Tables VI and VII). According to the score of the
included studies, we performed a subgroup analysis with score ≥8
and score <8 and the results are presented in Table VIII, which indicated that when
study design and quality score >8, D17 alleles increased the
risk for OA (OR=1.45, 95% CI: 1.04–2.02, P<0.05).
 | Table V.Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to sample
size. |
Table V.
Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to sample
size.
|
|
|
| Sample size | Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Comparison | Sample size | No. of studies | OA | Control | OR | 95% CI | P-value | Modela | P-value | I2
(%) |
|---|
| D12 vs. the
others | <1,000 | 3 | 1,460 | 1,728 | 0.79 | 0.29–2.17 | 0.65 | R | 0.01 | 76 |
|
| >1,000 | 4 | 8,278 | 5,594 | 0.83 | 0.64–1.08 | 0.16 | R | 0.07 | 57 |
| D13 vs. the
others | <1,000 | 5 | 1,460 | 1,728 | 0.81 | 0.64–1.03 | 0.09 | R | 0.08 | 51 |
|
| >1,000 | 6 | 8,278 | 5,594 | 0.99 | 0.89–1.10 | 0.87 | F | 0.08 | 49 |
| D14 vs. the
others | <1,000 | 5 | 1,460 | 1,728 | 1.18 | 0.81–1.72 | 0.39 | R | 0.005 | 73 |
|
| >1,000 | 6 | 8,278 | 5,594 | 1.14 | 0.89–1.45 | 0.32 | R | 0.001 | 75 |
| D15 vs. the
others | <1,000 | 4 | 1,460 | 1,728 | 1.09 | 0.78–1.54 | 0.60 | R | 0.05 | 62 |
|
| >1,000 | 5 | 8,278 | 5,594 | 0.89 | 0.73–1.09 | 0.25 | R | 0.04 | 61 |
| D16 vs. the
others | <1,000 | 4 | 1,460 | 1,728 | 1.21 | 0.90–1.62 | 0.21 | R | 0.06 | 60 |
|
| >1,000 | 4 | 8,278 | 5,594 | 0.97 | 0.82–1.15 | 0.75 | F | 0.22 | 33 |
| D17 vs. the
others | <1,000 | 4 | 1,460 | 1,728 | 1.15 | 0.30–1.62 | 0.52 | F | 0.16 | 43 |
|
| >1,000 | 4 | 8,278 | 5,594 | 1.46 |
1.03–2.07 | 0.03 | F | 0.68 | 0 |
| D18 vs. the
others | <1,000 | 2 | 1,460 | 1,728 | 2.21 | 0.68–7.20 | 0.19 | R | 0.10 | 57 |
|
| >1,000 | 2 | 8,278 | 5,594 | 0.96 | 0.72–1.47 | 0.86 | F | 0.66 |
|
 | Table VI.Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to OA site. |
Table VI.
Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to OA site.
|
|
|
| Sample size | Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Comparison | OA site | No. of studies | OA | Control | OR | 95% CI | P-value | Modela | P-value |
I2 (%) |
|---|
| D12 vs. all the
others | Knee | 5 | 2,222 | 3,388 | 0.79 | 0.51–1.20 | 0.27 | R | 0.01 | 68 |
|
| Knee/Hand/Hip | 2 | 7,462 | 3,934 | 0.86 | 0.57–1.30 | 0.48 | R | 0.12 | 58 |
|
| Overall | 7 | 9,684 | 7,322 | 0.81 | 0.61–1.07 | 0.14 | R | 0.02 | 62 |
| D13 vs. all the
others | Knee | 7 | 2,222 | 3,388 | 0.93 | 0.75–1.16 | 0.54 | R | 0.005 | 68 |
|
| Knee/Hand/Hip | 4 | 7,462 | 3,934 | 0.93 | 0.86–1.02 | 0.12 | F | 0.33 | 12 |
|
| Overall | 11 | 9,684 | 7,322 | 0.94 | 0.84–1.05 | 0.25 | R | 0.01 | 55 |
| D14 vs. all the
others | Knee | 7 | 2,222 | 3,388 | 1.18 | 0.83–1.68 | 0.35 | R | 0.35 | 77 |
|
| Knee/Hand/Hip | 4 | 7,462 | 3,934 | 1.10 | 0.90–1.36 | 0.35 | R | 0.05 | 62 |
|
| Overall | 11 | 9,684 | 7,322 | 1.15 | 0.94–1.40 | 0.17 | R |
0.0001 | 71 |
| D15 vs. all the
others | Knee | 6 | 2,222 | 3,388 | 1.04 | 0.80–1.36 | 0.76 | F | 0.10 | 45 |
|
| Knee/Hand/Hip | 3 | 7,462 | 3,934 | 0.89 | 0.70–1.15 | 0.37 | R |
0.007 | 80 |
|
| Overall | 9 | 9,684 | 7,322 | 0.97 | 0.70–1.15 | 0.71 | R |
0.005 | 63 |
| D16 vs. all the
others | Knee | 6 | 2,222 | 3,388 | 0.98 | 0.77–1.25 | 0.88 | R | 0.02 | 62 |
|
| Knee/Hand/Hip | 2 | 7,462 | 3,934 | 1.06 | 0.88–1.28 | 0.57 | F | 0.92 | 0 |
|
| Overall | 8 | 9,684 | 7,322 | 1.03 | 0.89–1.19 | 0.72 | F | 0.07 | 47 |
| D17 vs. all the
others | Knee | 6 | 2,222 | 3,388 | 1.27 | 0.87–1.85 | 0.22 | F | 0.19 | 33 |
|
| Knee/Hand/Hip | 2 | 7,462 | 3,934 | 1.38 | 0.95–2.02 | 0.09 | F | 0.96 | 0 |
|
| Overall | 8 | 9,684 | 7,322 | 1.33 |
1.02–1.73 | 0.04 | F | 0.37 | 8 |
| D18 vs. all the
others | Knee | 4 | 2,222 | 3,388 | 1.51 | 0.55–4.11 | 0.42 | F | 0.14 | 45 |
|
| Knee/Hand/Hip | 2 | 7,462 | 3,934 | 1.00 | 0.65–1.54 | 1.00 | F | 0.65 | 0 |
|
| Overall | 6 | 9,684 | 7,322 | 1.07 | 0.72–1.59 | 0.75 | F | 0.38 | 6 |
 | Table VII.Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to continent of
origin. |
Table VII.
Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to continent of
origin.
|
|
|
| Sample size | Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Comparison | Continent of
origin | No. of studies | OA | Control | OR | 95% CI | P-value | Modela | P-value |
I2 (%) |
|---|
| D12 vs. all the
others | Asia | 3 | 3,262 | 3,076 | 0.81 | 0.55–1.18 | 0.26 | R | 0.13 | 50 |
|
| Europe | 3 | 5,800 | 3,566 | 0.96 | 0.66–1.40 | 0.85 | R | 0.12 | 52 |
|
| America | 1 |
622 |
680 | 0.36 | 0.19–0.72 | 0.003 | – | – | – |
|
| Overall | 7 | 9,684 | 7,322 | 0.81 | 0.61–1.07 | 0.14 | R | 0.02 | 62 |
| D13 vs. all the
others | Asia | 5 | 3,262 | 3,076 | 0.95 | 0.78–1.16 | 0.61 | R | 0.02 | 65 |
|
| Europe | 4 | 5,800 | 3,566 | 0.90 | 0.77–1.06 | 0.21 | R | 0.03 | 67 |
|
| America | 2 |
622 |
680 | 1.06 | 0.82–1.38 | 0.63 | F | 0.53 | 0 |
|
| Overall | 11 | 9,684 | 7,322 | 0.94 | 0.84–1.05 | 0.25 | R | 0.01 | 55 |
| D14 vs. all the
others | Asia | 5 | 3,262 | 3,076 | 1.34 | 0.39–1.07 | 0.25 | R | 0.0002 | 82 |
|
| Europe | 4 | 5,800 | 3,566 | 1.03 | 0.91–1.17 | 0.62 | F | 0.57 | 0 |
|
| America | 2 |
622 |
680 | 1.10 | 0.62–1.94 | 0.75 | R | 0.03 | 80 |
|
| Overall | 11 | 9,684 | 7,322 | 1.15 | 0.94–1.40 | 0.17 | R | 0.0001 | 71 |
| D15 vs. all the
others | Asia | 3 | 3,262 | 3,076 | 1.03 | 0.73–1.43 | 0.88 | F | 0.56 | 0 |
|
| Europe | 4 | 5,800 | 3,566 | 0.99 | 0.76–1.30 | 0.97 | R | 0.0005 | 83 |
|
| America | 2 |
622 |
680 | 0.85 | 0.50–1.44 | 0.54 | R | 0.10 | 64 |
|
| Overall | 9 | 9,684 | 7,322 | 0.97 | 0.81–1.16 | 0.71 | R | 0.005 | 63 |
| D16 vs. all the
others | Asia | 3 | 3,262 | 3,076 | 0.86 | 0.61–1.23 | 0.41 | R | 0.03 | 70 |
|
| Europe | 3 | 5,800 | 3,566 | 1.03 | 0.86–1.23 | 0.75 | F | 0.68 | 0 |
|
| America | 2 |
622 |
680 | 1.26 | 0.86–1.87 | 0.24 | R | 0.05 | 75 |
|
| Overall | 8 | 9,684 | 7,322 | 1.03 | 0.89–1.19 | 0.72 | F | 0.07 | 47 |
| D17 vs. all the
others | Asia | 3 | 3,262 | 3,076 | 1.63 | 0.81–3.28 | 0.17 | F | 0.48 | 0 |
|
| Europe | 3 | 5,800 | 3,566 | 1.26 | 0.89–1.77 | 0.19 | F | 0.52 | 0 |
|
| America | 2 |
622 |
680 | 1.32 | 0.38–4.57 | 0.66 | R | 0.04 | 77 |
|
| Overall | 8 | 9,684 | 7,322 | 1.32 | 0.99–1.77 | 0.06 | F | 0.37 | 8 |
| D18 vs. all the
others | Asia | 2 | 3,262 | 3,076 | 0.31 | 0.03–2.75 | 0.29 | F | 0.96 | 0 |
|
| Europe | 3 | 5,800 | 3,566 | 1.16 | 0.76–1.77 | 0.49 | F | 0.14 | 49 |
|
| America | 1 |
622 |
680 | 0.51 | 0.05–5.62 | 0.58 | – | – | – |
|
| Overall | 6 | 9,684 | 7,322 | 1.07 | 0.72–1.59 | 0.75 | F | 0.38 | 6 |
 | Table VIII.Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to study design
and quality scores. |
Table VIII.
Subgroup analysis of pooled ORs and
95% CIs of ASPN polymorphisms and OA risk according to study design
and quality scores.
|
|
|
| Sample size | Test of
association | Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Comparison | Scores | No. of studies | OA | Control | ORa | OR 95%
CIb | P-value | Modelc | P-value |
I2 (%) |
|---|
| D12 vs. the
others | ≥8 | 4 | 7,728 | 6,154 | 0.75 | 0.52–1.09 | 0.14 | R | 0.008 | 75 |
|
| <8 | 3 | 1,956 | 1,168 | 0.94 | 0.53–1.67 | 0.84 | F | 0.02 | 46 |
| D13 vs. the
others | ≥8 | 8 | 7,728 | 6,154 | 0.97 | 0.87–1.08 | 0.55 | F | 0.06 | 47 |
|
| <8 | 3 | 1,956 | 1,168 | 0.83 | 0.59–1.16 | 0.28 | R | 0.02 | 76 |
| D14 vs. the
others | ≥8 | 8 | 7,728 | 6,154 | 1.24 | 0.97–1.60 | 0.09 | R | <0.0001 | 77 |
|
| <8 | 3 | 1,956 | 1,168 | 0.94 | 0.76–1.17 | 0.59 | F | 0.55 | 0 |
| D15 vs. the
others | ≥8 | 6 | 7,728 | 6,154 | 0.86 | 0.69–1.07 | 0.18 | R | 0.03 | 60 |
|
| <8 | 3 | 1,956 | 1,168 | 1.19 | 0.88–1.59 | 0.26 | R | 0.10 | 57 |
| D16 vs. the
others | ≥8 | 5 | 7,728 | 6,154 | 1.00 | 0.84–1.19 | 0.98 | R | 0.05 | 58 |
|
| <8 | 3 | 1,956 | 1,168 | 1.10 | 0.84–1.43 | 0.50 | F | 0.18 | 47 |
| D17 vs. the
others | ≥8 | 5 | 7,728 | 6,154 | 1.45 |
1.04–2.02 | 0.03 | F | 0.20 | 33 |
|
| <8 | 3 | 1,956 | 1,168 | 1.13 | 0.72–1.77 | 0.59 | F | 0.64 | 0 |
| D18 vs. the
others | ≥8 | 3 | 7,728 | 6,154 | 0.85 | 0.51–1.42 | 0.55 | F | 0.69 | 0 |
|
| <8 | 3 | 1,956 | 1,168 | 1.48 | 0.77–1.59 | 0.24 | R | 0.14 | 50 |
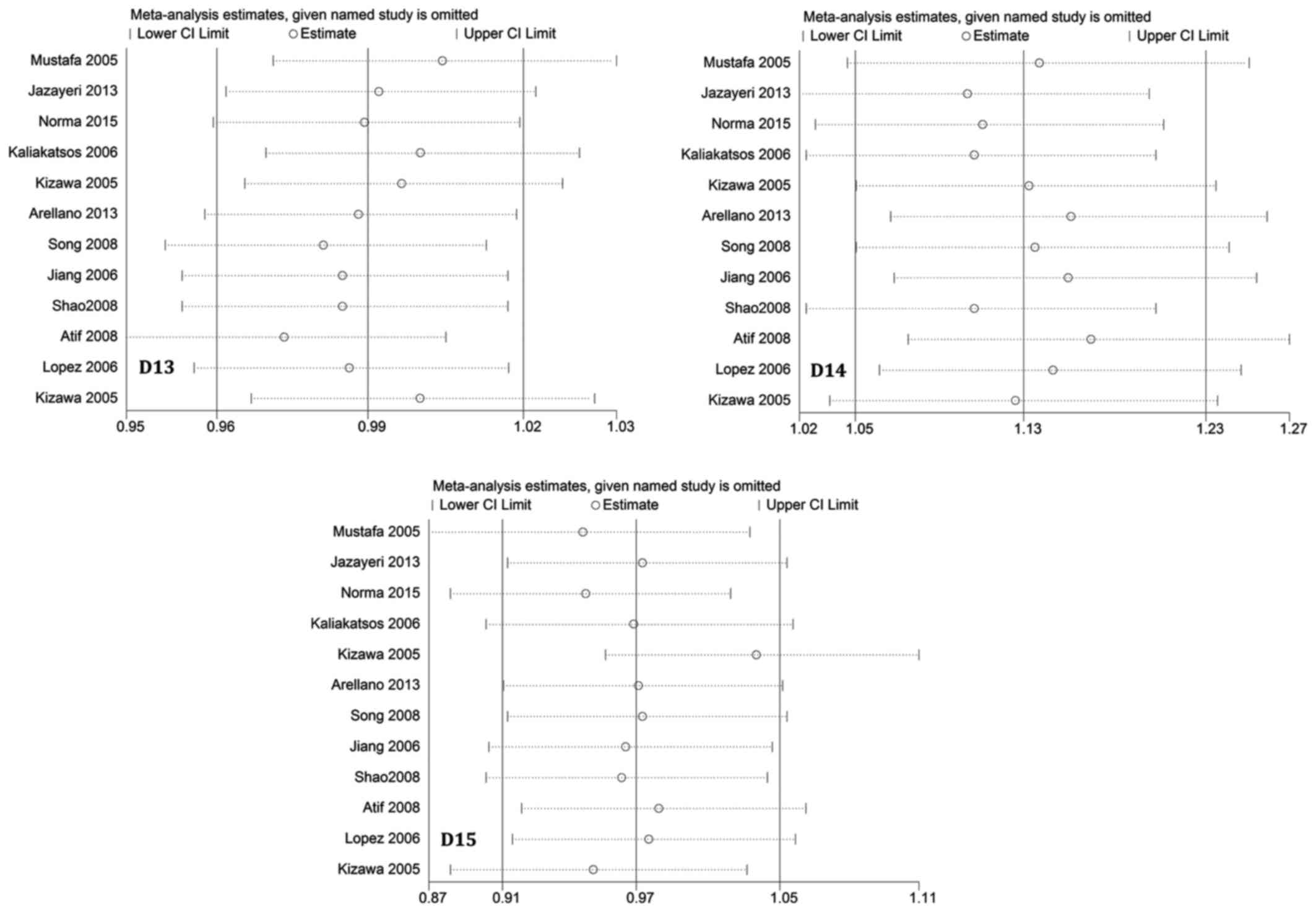
Sensitivity analysis
To assess the influence of each individual study on
the pooled ORs, a sensitivity analysis of sequential removal of
individual studies was performed. Results shown in Fig. 5 suggest that no individual study
significantly affected the pooled ORs, indicating that our
meta-analysis results were stable and reliable. Another sensitivity
analysis performed by excluding HWE-violating studies did not
perturb the overall results.
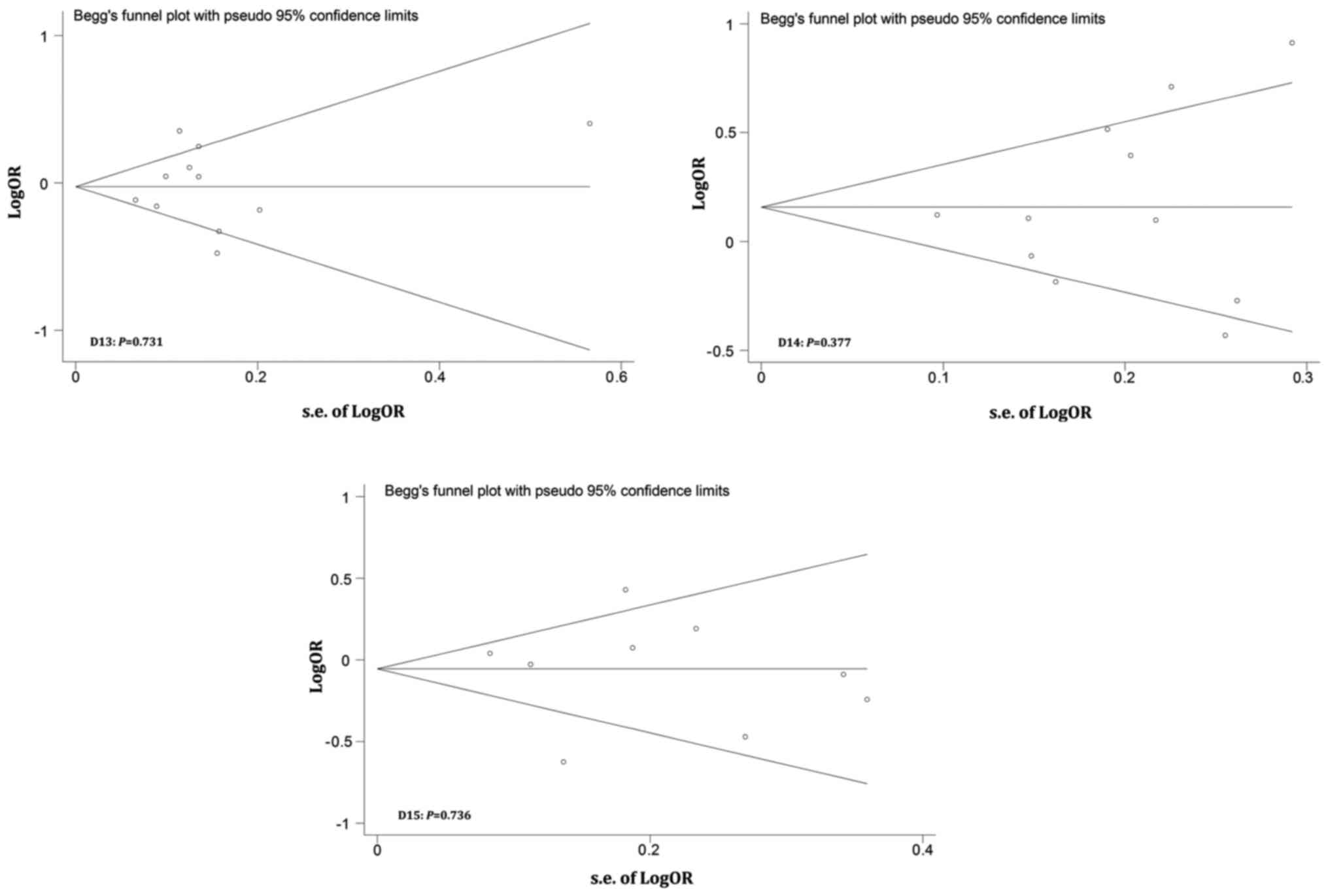
Publication bias
Publication bias was evaluated by performing Begg's
funnel plots and Egger's tests. Fig.
6 shows that the shapes of the Funnel plots were basically
symmetrical and did not reveal obvious evidence of asymmetry. All
P-values from Egger's and Begg's tests were greater than 0.05.
Therefore, our results suggest no evidence of any publication bias
in this meta-analysis.
Discussion
In the last few years, a greater number of
researchers have paid attention to the link between the ASPN gene
and OA. The ASPN gene encodes for a protein that is part of the 95%
CI family (23). This ASPN gene is
abundantly expressed in human articular cartilage with OA. In the
present meta-analysis, we included ten articles (eleven studies)
after reviewing the literature for the association between ASPN
D-repeat polymorphisms and OA susceptibility (6). In the present study, we aimed to
provide more convincing evidence regarding the role of ASPN
D-repeat polymorphisms in the incidence of OA.
According to a widely accepted viewpoint, ASPN is a
newly identified extracellular matrix protein that contains an
N-terminal unique aspartic acid repeat that ranges in length from 8
to 19 residues. The human ASPN gene has 8 exons. It spans 26
kilobases and is located in chromosome 9q31.1–32, which belongs to
the SLRP family. Previous studies have indicated that TGF-β1 plays
an important role as a regulator of cellular differentiation,
proliferation, apoptosis and migration in bone tissue. ASPN can
directly bind to the TGF-β1 receptor and thus inhibit the
expression of cartilage matrix Agc1 and Col2a1 genes, which are
mediated by TGF-β1 (24). The
interaction between ASPN and TGF-β1 can repress the expression of
cartilage matrix genes (25). Based
on the above findings, many researchers believe that ASPN may play
an important role in the occurrence of joint disease (26), including OA.
Many studies have focused on the association between
ASPN polymorphisms and OA, but a consensus has not yet been
reached. Mustafa et al (18)
found that ASPN polymorphism was not a major influence on OA
etiology in Caucasians. However, Kizawa et al (27) conducted a cohort study and a
case-control study and reported a significant association between
the Japanese population's D-repeat polymorphism and OA. Jiang et
al (28) studied 218 patients
and 454 age-matched controls from the Han Chinese population,
revealing for the first time that the OA susceptibility gene was
definitely replicated among different ethnic groups. The frequency
of the D14 allele in the Greek population showed no significant
difference compared with the other alleles, but the frequency of
the D13 allele was significantly lower in KOA patients (29). In contrast, the frequency of the D13
allele was found to be significantly different between female KOA
patients and controls compared with other alleles (30). In addition, Arellano et al
(31) suggested that polymorphisms
of the ASPN gene could influence KOA susceptibility, but they
indicated that this association also required more extensive
research. By investigating the allelic association of the D-repeat
polymorphism with OA, Jazayeri et al (3) suggested that the D15 allele could be
considered a risk allele only for the female Iranian population.
González-Huerta et al (1)
reported that the D14 allele of the ASPN polymorphism had a certain
impact on the etiology of primary OA of the knee. Therefore, a more
objective and accurate conclusion about the link between ASPN
polymorphisms and OA risk is required to provide a robust framework
for future studies in the field.
From an epidemiological point of view, a
case-control study is retrospective and is a causal study allowing
exploration of the etiology of diseases. However, a cohort study is
more effective than a case-control study for investigation of
disease etiology. We conducted an updated comprehensive analysis
based on case-control and cohort studies which focused on ASPN
polymorphisms and OA risk. The present meta-analysis included 7
studies (58.33%) on KOA, 2 studies (16.67%) on hand OA, and 3
(25.00%) on hip OA. In these 12 studies, 5 (41.67%) were conducted
in Asian populations, four (33.33%) in European populations and 2
(16.67%) in American populations. Results of this meta-analysis
showed that the D17 allele was a risk factor for OA and may
increase the incidence of OA (OR=1.33, 95% CI: 1.02–1.73,
P<0.05), particularly in studies with a large sample size
(N>1,000). Results also showed that the other types of alleles
did not play an important role in the development of OA. We
addressed the association between OA susceptibility and ASPN
D-repeat polymorphisms in populations originating from different
continents, and the results remained the same. We did not find any
significant association between ASPN polymorphisms and OA
susceptibility in the subgroup analyses performed according to the
OA site and the continent of origin.
We performed a comprehensive review of the
literature and designed strict inclusion and exclusion criteria to
obtain valid results. Compared with previous studies, our research
was more detailed and in addition we conducted different subgroup
and meta-regression analyses. There were, however, several
limitations in this meta-analysis (10–15,32): i)
Because there were inevitably mixed factors in the study, our group
analysis could not completely encompass them; ii) the results of
this meta-analysis may be misinterpreted due to the limited number
of research studies, the shortage of uniform inclusion criteria and
the existence of heterogeneity; iii) there is a lack of clinical
variables in the literature such as classification of OA severity,
and these variables are likely to be related to ASPN polymorphisms;
iv) in the subgroup analyses, the results may lack efficiency due
to the limited number of studies; and v) since the inclusion of the
literature may have been biased, even after using a regression
test, the inherent bias may have affected the overall analysis.
Moreover, as a complex multifactorial disease, development of OA
may have been affected by genetic or genetic-environmental
interactions between races. Therefore, further research is needed
to elucidate this question.
In summary, this meta-analysis study did not show
any association between ASPN polymorphisms and OA susceptibility in
Asians, Europeans or Americans. Thus, our results do not support
the common viewpoint that the ASPN polymorphisms (except for D17)
constitute risk factors for OA susceptibility in Asians, Europeans
or Americans. We observed that the D17 alleles of ASPN increased
the risk for development of OA if the considered sample size is
greater than 1,000. Because of large heterogeneity in the studies
included in this meta-analysis, we consider that the effects of
ASPN polymorphisms on OA development still require further
confirmation by larger studies performed in homogeneous
populations.
Acknowledgements
The authors thank the researchers of the original
studies included in this meta-analysis.
Funding
The present study was supported by the youth
research project of Shaanxi University of Chinese Medicine
(2015QN05).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author contributions
RZ designed the study. JW, AY, JZ and RZ screened
the literature. NS, XiangwenL, XinghuiL, QL, JL and XR extracted
the data from the literature. JW, RZ and ZK conducted the
meta-analysis and wrote the manuscript. RZ and JW submitted the
study.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
González-Huerta NC, Borgonio-Cuadra VM,
Zenteno JC, Cortés-González S, Duarte-Salazar C and Miranda-Duarte
A: D14 repeat polymorphism of the asporin gene is associated with
primary osteoarthritis of the knee in a Mexican Mestizo population.
Int J Rheum Dis. 20:1935–1941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song GG, Kim JH and Lee YH: A
meta-analysis of the relationship between aspartic acid (D)-repeat
polymorphisms in asporin and osteoarthritis susceptibility.
Rheumatol Int. 34:785–792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jazayeri R, Qoreishi M, Hoseinzadeh H,
Babanejad M, Bakhshi E, Najmabadi H and Jazayeri SM: Investigation
of the asporin gene polymorphism as a risk factor for knee
osteoarthritis in Iran. Am J Orthop (Belle Mead NJ). 42:313–316.
2013.PubMed/NCBI
|
|
4
|
Juchtmans N, Dhollander AA, Coudenys J,
Audenaert EA, Pattyn C, Lambrecht S and Elewaut D: Distinct
dysregulation of the small leucine-rich repeat protein family in
osteoarthritic acetabular labrum compared to articular. cartilage.
67:1–441. 2015.
|
|
5
|
Michael JW, Schlüter-Brust KU and Eysel P:
The epidemiology, etiology, diagnosis, and treatment of
osteoarthritis of the knee. Dtsch Arztebl Int. 107:152–162.
2010.PubMed/NCBI
|
|
6
|
Xing D, Ma XL, Ma JX, Xu WG, Wang J, Yang
Y, Chen Y, Ma BY and Zhu SW: Association between aspartic acid
repeat polymorphism of the asporin gene and susceptibility to knee
osteoarthritis: A genetic meta-analysis. Osteoarthritis Cartilage.
21:1700–1706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woolf AD and Pfleger B: Burden of major
musculoskeletal conditions. Bull World Health Organ. 81:646–656.
2003.PubMed/NCBI
|
|
8
|
Zhang Y and Jordan JM: Epidemiology of
osteoarthritis. Clin Geriatr Med. 26:355–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Haq SA and Davatchi F: Osteoarthritis of
the knees in the COPCORD world. Int J Rheum Dis. 14:122–129. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Minafra L, Bravatà V, Saporito M,
Cammarata FP, Forte GI, Caldarella S, D'Arienzo M, Gilardi MC,
Messa C and Boniforti F: Genetic, clinical and radiographic signs
in knee osteoarthritis susceptibility. Arthritis Res Ther.
16:R912014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fernandes MT, Fernandes KB, Marquez AS,
Cólus IM, Souza MF, Santos JP and Poli-Frederico RC: Association of
interleukin-6 gene polymorphism (rs1800796) with severity and
functional status of osteoarthritis in elderly individuals.
Cytokine. 75:316–320. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rushton MD, Reynard LN, Young DA, Shepherd
C, Aubourg G, Gee F, Darlay R, Deehan D, Cordell HJ and Loughlin J:
Methylation quantitative trait locus analysis of osteoarthritis
links epigenetics with genetic risk. Hum Mol Genet. 24:7432–7444.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bijsterbosch J, Kloppenburg M, Reijnierse
M, Rosendaal FR, Huizinga TW, Slagboom PE and Meulenbelt I:
Association study of candidate genes for the progression of hand
osteoarthritis. Osteoarthritis Cartilage. 21:565–569. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakamura T, Shi D, Tzetis M,
Rodriguez-Lopez J, Miyamoto Y, Tsezou A, Gonzalez A, Jiang Q,
Kamatani N, Loughlin J and Ikegawa S: Meta-analysis of association
between the ASPN D-repeat and osteoarthritis. Hum Mol Genet.
16:1676–1681. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Valdes AM, Loughlin J, Oene MV, Chapman K,
Surdulescu GL, Doherty M and Spector TD: Sex and ethnic differences
in the association of ASPN, CALM1, COL2A1, COMP, and FRZB with
genetic susceptibility to osteoarthritis of the knee. Arthritis
Rheum. 56:137–146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kou I, Nakajima M and Ikegawa S:
Expression and regulation of the osteoarthritis-associated protein
asporin. J Biol Chem. 282:32193–32199. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Neptune ER, Frischmeyer PA, Arking DE,
Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY and Dietz HC:
Dysregulation of TGF-beta activation contributes to pathogenesis in
Marfan syndrome. Nat Genet. 33:407–411. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mustafa Z, Dowling B, Chapman K,
Sinsheimer JS, Carr A and Loughlin J: Investigating the aspartic
acid (D) repeat of asporin as a risk factor for osteoarthritis in a
UK Caucasian population. Arthritis Rheum. 52:3502–3506. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Atif U, Philip A, Aponte J, Woldu EM,
Brady S, Kraus VB, Jordan JM, Doherty M, Wilson AG, Moskowitz RW,
et al: Absence of association of asporin polymorphisms and
osteoarthritis susceptibility in US Caucasians. Osteoarthritis
Cartilage. 16:1174–1177. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Min SK, Nakazato K, Ishigami H and
Hiranuma K: Cartilage intermediate layer protein and Asporin
Polymorphisms are independent risk factors of lumbar disc
degeneration in male collegiate athletes. Cartilage. 5:37–42. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thakkinstian A, McEvoy M, Minelli C,
Gibson P, Hancox B, Duffy D, Thompson J, Hall I, Kaufman J, Leung
TF, et al: Systematic review and meta-analysis of the association
between {beta}2-adrenoceptor polymorphisms and asthma: A HuGE
review. Am J Epidemiol. 162:201–11. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin X, Peng Q, Tang W, Lao X, Chen Z, Lai
H, Deng Y, Mo C, Sui J, Wu J, et al: An updated meta-analysis on
the association of MDM2 SNP309 polymorphism with colorectal cancer
risk. PloS One. 8:e760312013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arellano-Pérez-Vertti RD, Argüello-Astorga
JR, Cortéz-López ME, Zamarripa-Mottú JI, García-Salcedo JJ and
Luna-Ceniceros DA: D-repeat polymorphism in the ASPN gene in knee
osteoarthritis in females in Torreón, Coahuila. Acta Ortop Mex.
28:363–368. 2014.PubMed/NCBI
|
|
24
|
Maris P, Blomme A, Palacios AP, Costanza
B, Bellahcène A, Bianchi E, Gofflot S, Drion P, Trombino GE, Di
Valentin E, et al: Asporin is a fibroblast-derived TGF-β1 inhibitor
and a tumor suppressor associated with good prognosis in breast
cancer. PLoS Med. 12:e10018712015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi D, Dai J, Zhu P, Qin J, Zhu L, Zhu H,
Zhao B, Qiu X, Xu Z, Chen D, et al: Association of the D repeat
polymorphism in the ASPN gene with developmental dysplasia of the
hip: A case-control study in Han Chinese. Arthritis Res Ther.
13:R272011. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Duval E, Bigot N, Hervieu M, Kou I,
Leclercq S, Galéra P, Boumediene K and Baugé C: Asporin expression
is highly regulated in human chondrocytes. Mol Med. 17:816–823.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kizawa H, Kou I, Iida A, Sudo A, Miyamoto
Y, Fukuda A, Mabuchi A, Kotani A, Kawakami A, Yamamoto S, et al: An
aspartic acid repeat polymorphism in asporin inhibits
chondrogenesis and increases susceptibility to osteoarthritis. Nat
Genet. 37:138–144. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang Q, Shi D, Yi L, Ikegawa S, Wang Y,
Nakamura T, Qiao D, Liu C and Dai J: Replication of the association
of the aspartic acid repeat polymorphism in the asporin gene with
knee-osteoarthritis susceptibility in Han Chinese. J Hum Genet.
51:1068–1072. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaliakatsos M, Tzetis M, Kanavakis E,
Fytili P, Chouliaras G, Karachalios T, Malizos K and Tsezou A:
Asporin and knee osteoarthritis in patients of Greek origin.
Osteoarthritis Cartilage. 14:609–611. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Song JH, Lee HS, Kim CJ, Cho YG, Park YG,
Nam SW, Lee JY and Park WS: Aspartic acid repeat polymorphism of
the asporin gene with susceptibility to osteoarthritis of the knee
in a Korean population. Knee. 15:191–195. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arellano RD, Hernández F, García-Sepúlveda
CA, Velasco VM, Loera CR and Arguello JR: The D-repeat polymorphism
in the ASPN gene and primary knee osteoarthritis in a Mexican
mestizo population: A case-control study. J Orthop Sci. 18:826–831.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rodriguez-Lopez J, Pombo-Suarez M, Liz M,
Gomez-Reino JJ and Gonzalez A: Lack of association of a variable
number of aspartic acid residues in the asporin gene with
osteoarthritis susceptibility: Case-control studies in Spanish
Caucasians. Arthritis Res Ther. 8:R552006. View Article : Google Scholar : PubMed/NCBI
|