Introduction
Preeclampsia (PE) is a pregnancy-specific
multisystem disorder that contributes substantially to maternal,
fetal, and neonatal mortality and morbidity rates worldwide
(1). Despite extensive research,
causes of PE remain unclear, and removal of the placenta is the
only effective cure for the disease (2). Researchers have speculated that
placental defects, especially shallow placental implantation, can
induce the clinical disease (3,4). Others
have suggested that long noncoding RNAs (lncRNAs) in human placenta
may play roles in regulating trophoblast cell invasion and inducing
PE (5,6).
LncRNAs comprise a diverse class of RNA transcripts
that exceed 200 nucleotides in length with limited protein-coding
potential, which were believed to have surprisingly complex and
diverse functions (7). Misregulation
of lncRNAs have been identified to be involved in various disorders
such as cancer, cardiovascular disease and PE (8). In addition to their possible role in
the biological functions, circulating lncRNAs have been
characterized as a new class of potential biomarkers in the
diagnosis and prognosis of different types of malignant tumors and
for possibly predicting and monitoring treatment response (9,10).
Recently, two papers described the differentially expressed lncRNAs
in PE placenta using microarray, and revealed that many of these
lncRNAs may be involved in the pathological mechanisms of PE
(11,12). Therefore, We postulated that
placenta-related lncRNAs might be also released into the maternal
circulation in pregnancy and could be utilized to detect and
monitor PE.
Materials and methods
Participant enrollment
All study participants were pregnant and
non-pregnant women of the Department of Obstetrics and Gynecology
of the Affiliated Third Hospital of Guangzhou Medical University
(Guangzhou, China) between February 2015 and February 2017. This
case-control study involved the following sets of cases and
controls: late-onset PE (LOPE) patients (diagnosed after 34 weeks,
n=52) and gestational age (GA)-matched healthy pregnant women
(control group 1, n=52); early-onset PE (EOPE) patients (diagnosed
before 34 weeks, n=58) and GA-matched healthy pregnant women who
underwent routine outpatient antenatal examination and did not
develop hypertension during their pregnancies (control group 2,
n=58). Of these, 21 LOPE patients and all EOPE patients received
anti-hypertensive drugs and/or magnesium sulfate prior to and
following the termination of pregnancy. In addition, 30 healthy
never-pregnant women were also recruited in this study. All
pregnant women had singleton pregnancies and none of the PE
patients had any other maternal complications. PE is defined
according to the criteria of the American College of Obstetricians
and Gynecologists (1). Demographic
and clinical characteristics of all participants are summarized in
Table I. This study was approved by
the ethics committee of the Third Affiliated Hospital of Guangzhou
Medical University. Informed consent was obtained from all
individual participants included in the study.
 | Table I.Demographic and clinical
characteristics of the study groups. |
Table I.
Demographic and clinical
characteristics of the study groups.
| Variable | LOPE (n=52) | EOPE (n=58) | Control group 1
(n=52) | Control group 2
(n=58) | Non-pregnant
(n=30) |
|---|
| Age (years) | 31.1±5.5 |
30.9±6.1 |
30.8±5.7 |
30.4±4.3 |
31.0±2.3 |
| Prepregnancy BMI
(kg/m2) | 21.6±2.5 |
21.7±3.1 |
21.4±2.3 |
21.5±2.9 |
21.5±2.6 |
| Nulliparity n
(%) | 26 (50.0%) | 30 (51.7%) | 25 (48.1%) | 29 (50.0%) | 30 (100.0%) |
| SBP (mmHg) |
162.8±18.3a |
171.1±22.1a | 119.1±7.2 | 118.8±7.7 | 117.5±6.2 |
| DBP (mmHg) |
103.9±13.1a |
108±17.4a |
78.4±6.7 |
77.9±6.5 |
77.8±5.8 |
| 24-h urine protein
(g) |
3.1±2.2 |
4.9±3.9 | ND | ND | ND |
| GA at blood
collection (weeks) | 38.2±2.4 |
31.9±3.0 |
38.8±1.6 |
32.0±1.9 | 0 |
| Birth weight
(g) |
3,090.7±509.1a | 1,575.2±364.7 |
3,456.1±474.4 | ND | 0 |
Sample collection and preparation
Peripheral venous blood samples were collected from
all of the participants, particularly blood samples from women who
had delivered were obtained within 48 h before termination of
pregnancy, without spontaneous or nonspontaneous labor. Up to 2.5
ml of blood was drawn from each nonfasting subject into an
anticoagulation tube containing K2EDTA. Blood was
processed within 30 min of collection. Plasma was collected via a
two-step centrifugation protocol of 1,800 g for 10 min at 4°C and
12,000 g for 10 min at 4°C. This process ensured the thorough
elimination of peripheral blood cells and platelets. Separated
plasma samples were transferred to an RNase/DNase-free tube and
stored at −80°C until total RNA extraction. Blood samples with
hemolysis were excluded from analysis.
Placental tissues were obtained during elective
caesarean sections (C/S) from women who delivered in the absence of
labor. Of the 117 placental tissue samples, 40 were from the LOPE
group, 35 were from control group 1, and 42 were from the EOPE
group. Placental tissue that was about 2 cm away from the
attachment site of the umbilical cord in the placenta was randomly
dissected within 5 min of delivery, including a full-thickness of
placenta. Harvested samples were washed extensively in cold saline
immediately after resection and stored in sufficient quantities of
RNAlater (AM7020; Ambion, Thermo Fisher Scientific, Inc., Waltham,
MA, USA) to protect RNA from degradation. Liquid was removed after
24 h of storage, and placental samples were stored at −80°C until
use.
LncRNA microarray analysis
For microarray analysis, 8 randomly and blindly
selected placental samples from LOPE patients and matched controls
(4 samples per group) were used to extract total RNA. The
expression profiles of the placental lncRNAs were detected using
the Agilent Human lncRNA Microarray V4.0 (OE Biotech, Shanghai,
China). The threshold for a dysregulated lncRNA was set as a
fold-change (FC) value of 2.0 or greater. Gene Ontology (GO;
ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz,
October 2015) and Kyoto Encyclopedia of Genes and Genomes (KEGG;
http://www.genome.jp/kegg/, Release
76.0) analyses were applied to explore the roles of the
differentially expressed lncRNAs that exhibited a significance
value of P<0.05.
RNA extraction from placenta and
plasma
Total RNA was extracted from placenta tissue using
Trizol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The RNA integrity was
evaluated using agarose gel electrophoresis stained with GelRed.
For plasma, total RNA was extracted from 800 µl of each sample
using the mirVana PARIS kit (1556; Ambion, Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
average recovery volume was 45 µl, and the RNA yield calculated
using spectrophotometry was approximately 11.3 ng/µl.
Quantitative polymerase chain reaction
(qPCR)
A total of 1 µg or 7 µl of RNA from each placental
tissue or plasma was reverse transcribed to cDNA using the
PrimeScript™ RT reagent kit with gDNA Eraser (RR047A; Takara
Biotechnology Co., Ltd., Dalian, China) in a 20-µl reaction volume.
All primers used in this study were designed and synthesized by
Shanghai Generay Biotech Co., Ltd. (Shanghai, China), a ccording
ddto the sequences obtained from the National Center for
Biotechnology Information Database. Then, qPCR was performed on a
StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, Inc.)
using SYBR® Premix EX Tag™ II (RR820A; Takara
Biotechnology Co., Ltd.) in a 20-µl reaction volume, and each
sample was analyzed in triplicate and the specificity of each PCR
reaction was confirmed by melt curve analysis. The qPCR reaction
conditions were as follows: A denaturation step at 95°C for 30 sec,
followed by 40 PCR cycles at 95°C for 5 sec and 60°C for 30 sec.
Relative expression levels of lncRNAs were quantified based on the
Cquantification cycle values and normalized to the
internal control housekeeping gene glyceraldehyde 3-phosphate
dehydrogenase (GAPDH), which has been shown to be the
optimal endogenous control for lncRNA expression in tissues and
plasma (10,13). Expression levels were calculated by
using the ΔCq method. A smaller ΔCq value
indicates higher expression, and relative lncRNA expression was
calculated by the 2−ΔΔCq method normalized to
GAPDH.
Statistical analyses
All statistical analyses were performed with the
Statistical Package for Social Sciences software version 22.0
(SPSS, Inc., Chicago, IL, USA). Comparisons between two subgroups
were estimated by using the independent samples t-test or
Mann-Whitney U-test for continuous data and Fisher's exact test for
categorical data. Comparisons between multiple subgroups were
performed by one-way analysis of variance (ANOVA). Spearman's rank
analysis was used for correlation analysis. All reported P-values
are two-sided, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Microarray analysis and functional
prediction
We identified 163 lncRNAs that were differentially
expressed between LOPE placentas and healthy controls, with 38 up-
and 125 downregulated lncRNAs. NONHSAT084322
(CUST_4347_PI429545410) was the most upregulated (FC=9.202518) and
NONHSAT028367 (CUST_33686_PI429545402) was the most
downregulated lncRNA (FC=16.553854). The lncRNA microarray
expression results have been uploaded to NCBI Gene Expression
Omnibus (GEO) and are accessible with the GEO series accession
number GSE97898 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97898).
Using the same criteria as the lncRNAs, we found 14
upregulated and 67 downregulated mRNA transcripts. The most
upregulated and downregulated mRNA transcripts were GSTT1
(A_23_P254944) and KRT6A (A_33_P3292886), with FCs of 9.89536 and
22.835077, respectively. Hierarchical clustering of the expression
of those lncRNAs and mRNAs based on centered Pearson correlation
clearly separated LOPE from normal tissues (Fig. 1). Differentially expressed lncRNAs
were clustered into hundreds of GO and KEGG pathway annotations.
According to enrichment counting, the significantly enriched GO
terms corresponded to biological processes, involving
musculoskeletal movement, immune responses, and metabolic
processes, and the most enriched KEGG pathways included oxidative
phosphorylation and metabolic pathways (Fig. 2).
Confirmation of lncRNAs by qPCR
For practical purposes, to validate the microarray
results and to increase the detection probability of plasma
lncRNAs, we first selected differentially expressed lncRNAs that
have high normalized probe signal values on the array platform, and
then subjected to qPCR validation in placental tissues from the
LOPE patients (n=40) and controls (n=35) delivered by caesarean
section. Demographic and clinical characteristics of all of the
pregnant women are summarized in Table
II. Finally, 9 highest expressed lncRNAs in placental tissues
were selected as candidate targets for subsequent circulating
lncRNA assay, and their primers used are shown in Table III. The qPCR results showed the
same tendencies (upregulation or downregulation) as the microarray
data (Fig. 3).
 | Table II.Demographic and clinical
characteristics of LOPE patients and controls delivered by
caesarean section. |
Table II.
Demographic and clinical
characteristics of LOPE patients and controls delivered by
caesarean section.
| Variable | LOPE (n=40) | Control (n=35) | P-value |
|---|
| Age (years) | 31.0±5.8 |
30.9±5.7 | 0.757 |
| Prepregnancy BMI
(kg/m2) | 21.5±2.9 |
21.3±2.4 | 0.533 |
| Nulliparity n
(%) | 21 (52.5%) | 18 (51.4%) | 0.425 |
| SBP (mmHg) | 163.2±18.4 | 120.1±7.8 | <0.001 |
| DBP (mmHg) | 103.8±13.7 |
79.3±6.5 | <0.001 |
| 24-h urine protein
(g) |
3.3±2.4 | ND | <0.001 |
| Gestational age at
delivery (weeks) | 38.3±2.3 |
38.9±1.3 | 0.086 |
| Birth weight
(g) |
3,068±625.7 |
3,422±437.6 | <0.001 |
 | Table III.The nine lncRNAs and their primers
used in this study. |
Table III.
The nine lncRNAs and their primers
used in this study.
| Target ID | FC(abs) | Primer sequnce (5′
to 3′) | Product (bp) |
|---|
|
NONHSAT145880 | 2.5↑ | Forward
TGTCAAGAAATACCCTGAGCC | 165 |
|
|
| Reverse
AGCCTGGGCGACAAAGTGA |
|
|
ENST00000587240 | 2.2↑ | Forward
TTTACCCCTCAACATACACC | 144 |
|
|
| Reverse
TACGGCTTCCAACAATGA |
|
|
NONHSAT116812 | 2.6↓ | Forward
GAGGCGGATGGCTGTGAC | 87 |
|
|
| Reverse
CCCTTGGGGCCTGAGTAG |
|
|
NONHSAT104536 | 2.6↓ | Forward
CTTGGGTTCAAGGCTTTGTT | 143 |
|
|
| Reverse
TCTCCTCTGCACTGGCTGTT |
|
|
FR339600 | 2.5↓ | Forward
GCAGAGCCAACATACACC | 59 |
|
|
| Reverse
AGGCAAGGCACATACAAA |
|
|
NONHSAG018907 | 2.1↓ | Forward
ACCTGAAAAGTGGAAATGATAG | 96 |
|
|
| Reverse
AGTAAGAGCAGCAACACACA |
|
|
TCONS_l2_00014782 | 2.1↓ | Forward
CCTCCTGACAGCCCCATTA | 184 |
|
|
| Reverse
GAGGCATCACCCAGTTGTTT |
|
|
NONHSAT134432 | 2.0↓ | Forward
AGTGGAGAGGGTGAGGAG | 146 |
|
|
| Reverse
GGAGGAAGGAACAGAGGA |
|
|
NONHSAG024318 | 2.0↓ | Forward
GGGTGCAGTTAGGTCACCCT | 113 |
|
|
| Reverse
CAGCACAAATTCCTCCGTCT |
|
| GAPDH | ND | Forward
AAGAAGGTGGTGAAGCAGG | 144 |
|
|
| Reverse
GTCAAAGGTGGAGGAGTGG |
|
Presence of placental lncRNAs in human
plasma
To explore whether the nine placenta-related lncRNAs
were present at detectable levels in maternal plasma, we used qPCR
to examine lncRNA expression levels in plasma samples from the LOPE
group and control group 1 (52 samples per group, 104 samples total)
and from the EOPE group and control group 2 (58 samples per group,
116 samples total). As shown in Fig.
4, the expression level of NONHSAT116812 and
NONHSAT145880 was significantly lower or higher in PE
patients than in healthy controls with a 100% detection rate,
respectively. The other seven lncRNAs had detection rates below 30%
in both PE plasma samples and healthy controls and, therefore, were
excluded from subsequent analyses. To verify that these two lncRNAs
were present in plasma samples which were obtained from PE patients
and healthy controls, the qPCR products (165 bp of
NONHSAT145880 and 87 bp of NONHSAT116812) were
followed by Sanger sequencing with the primers used for qPCR, using
BigDye™ Terminator v3.1 Cycle Sequencing kit (Applied Biosystems:
4337455) by Guangzhou Ige Biotech Co., Ltd. (Guangzhou, China). As
expected, their sequences were identical to those derived from
NONHSAT116812 (http://noncode.org/show_rna.php?id=NONHSAT116812)
and NONHSAT145880 (http://noncode.org/show_rna.php?id=NONHSAT145880)
(Fig. 5). However, these two lncRNAs
were also detected in plasma samples from non-pregnant individuals
at a rate of 100%, whereas other lncRNAs were not detectable.
Therefore, we compared expression levels of NONHSAT116812
and NONHSAT145880 in plasma samples from pregnant women
(n=52 from the LOPE group, n=52 from control group 1, n=58 from the
EOPE group, and n=58 from control group 2) and non-pregnant women
(n=30) by measuring the ΔCt value in the plasma. As expected, we
found significantly larger ΔCt values of these two lncRNAs in
never-pregnant women than in pregnant women, and significantly
smaller ΔCt values in PE patients than in healthy pregnant women
(Table IV).
 | Table IV.Comparison of plasma ΔCq value in
each group. |
Table IV.
Comparison of plasma ΔCq value in
each group.
| LncRNA | LOPE (n=52) | EOPE (n=58) | Control group 1
(n=52) | Control group 2
(n=58) | Non-pregnant
(n=30) |
|---|
|
NONHSAT116812 |
5.308±1.509a |
5.419±1.736a | 4.295±1.210 | 4.336±1.427 |
7.319±1.385b |
|
NONHSAT145880 |
5.131±1.308a |
5.071±1.565a | 5.993±1.450 | 6.018±1.603 | 7.208
±1.267b |
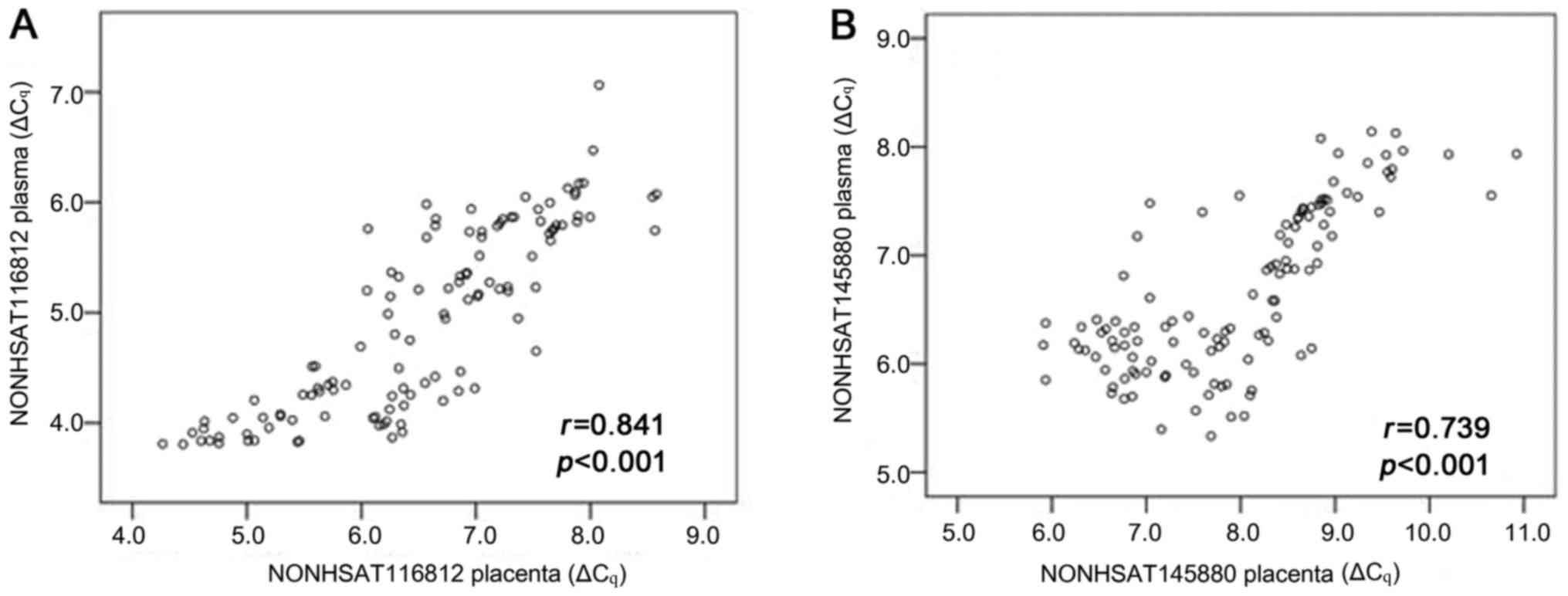
Correlation of lncRNA levels between
plasma and placenta
To determine whether expression levels of lncRNAs in
plasma and placentas were related, we compared expression levels of
NONHSAT116812 and NONHSAT145880 in paired plasma and
placental samples from 117 pregnant women delivered by C/S (n=42
from EOPE group, n=40 from LOPE group, and n=35 from control group
1). The data showed that plasma and placental measurements were
strongly correlated for NONHSAT116812 (r=0.841, P<0.001)
and NONHSAT145880 (r=0.739, P<0.001) (Fig. 6).
Evaluation of plasma lncRNAs as
biomarkers for detecting PE
We evaluated the ability of these two
placenta-related lncRNAs to be used as plasma biomarkers for PE.
Receiver operating characteristic (ROC) curves and areas under the
ROC curves (AUCs) were obtained from the data of all pregnant
women. Strong separation was found between ROC curves from the LOPE
patients (n=52) and the healthy controls (n=52).
NONHSAT116812 showed an AUC of 0.828 (95% CI: 0.750–0.907,
P<0.001), sensitivity of 86.5%, and specificity of 63.5%.
NONHSAT145880 showed an AUC of 0.815 (95% CI: 0.734–0.896,
P<0.001), sensitivity of 82.7%, and specificity of 67.3%. ROC
curves also discriminated the EOPE patients (n=58) from the healthy
controls (n=58), with an AUC of 0.864 (95% CI: 0.797–0.930,
P<0.001), 72.4% sensitivity, and 91.4% specificity for
NONHSAT116812, and with an AUC of 0.859 (95% CI:
0.792–0.925, P<0.001), 79.3% sensitivity, and 75.9% specificity
for NONHSAT145880 (Fig.
7).
 | Figure 7.Evaluation of plasma lncRNAs for
detection of PE. ROC curves were drawn with the data of plasma
lncRNAs from 52 patients with late-onset PE and 52 healthy
controls, and from 58 patients with early-onset PE and 58 healthy
controls, respectively. (A) ROC-AUC for late-onset PE
(NONHSAT116812, 0.828, P<0.001; NONHSAT145880,
0.815, P<0.001); (B) ROC-AUC for early-onset PE
(NONHSAT116812, 0.864, P<0.001; NONHSAT145880,
0.859, P<0.001). lncRNAs, long noncoding RNAs; PE, preeclampsia;
ROC, receiver operating characteristic. |
Discussion
PE is a serious disease that cannot be cured except
removal of the placenta. Early identification of women at high risk
of developing PE would enable surveillance and intervention, with
the potential for drastically improving pregnancy outcomes. To
date, although numerous protein biomarkers of placental origin have
been strongly associated with PE, these lack adequate accuracy to
be used clinically (14,15). The circulating placental RNAs
biomarkers have obvious advantage over protein biomarkers, changes
of which in gene expression might be detectable before changes in
protein levels occur and reflect alterations in placental function
(16). However, little is known
about the value of circulating placenta-related lncRNAs as
potential biomarkers for PE.
In this study, we first selected LOPE placental
tissues and healthy controls for lncRNA microarray analysis and a
total of 163 differentially expressed lncRNAs (FC ≥2.0) were
identified. The results are consistent with a previous study that
described the expression patterns of 738 differentially expressed
lncRNAs (FC ≥1.5) in placentas from patients with LOPE (11). However, a recent study evaluated the
differential expression of lncRNAs in placental samples from EOPE
patients and preterm controls and identified 28,824 dysregulated
lncRNAs (FC ≥2.0) (12). The
distinct expression profiles of lncRNAs may be because EOPE is more
serious than LOPE and placental defects are more prevalent
(17,18). However, these studies have shown that
aberrantly expressed lncRNAs in placenta may function in the
pathogenesis and development of PE.
Further, 9 highest expressed lncRNAs in placental
tissues were evaluated by qPCR to measure their expression levels
in plasma. We found that the expression levels of
NONHSAT116812 and NONHSAT145880 were significantly
lower or higher in plasma from LOPE patients compared to healthy
controls with a 100% detection rate, and the trends were consistent
with the results in placental samples. Interestingly, the
expression levels of the two lncRNAs in plasma from EOPE patients
have the same trends with those of LOPE patients. However, our
results are inconsistent with a previous study that revealed higher
expression levels of circulating placental RNAs in EOPE compared
with LOPE (19). This
inconsistencies may be due to the existence of a wide range of
heterogeneity between the studied populations, with samples
collected across various gestations and processing procedures
(20).
In addition, the expression levels of these two
lncRNAs in placenta and plasma from the same individual correlated
strongly. While other lncRNAs were also detectable in plasma, the
detection rates were less than 30% in plasma samples from PE
patients and corresponding controls. Moreover, to explore whether
the nine lncRNAs were specific to pregnancy, we measured their
expression levels in plasma samples from non-pregnant women and the
results showed thatNONHSAT116812 and NONHSAT145880
were detectable with a detection rate of 100%, but their expression
levels were significantly lower than those in pregnant women, and
the other lncRNAs were not detectable in non-pregnant women.
Therefore, We hypothesized that these two lncRNAs are expressed not
only in the placenta but also in other tissues of the body and, for
the first time, NONHSAT116812 was detected in the placenta
(http://noncode.org/show_rna.php?id=NONHSAT145880;
http://noncode.org/show_rna.php?id=NONHSAT116812).
To futher verify that these two lncRNAs were present in plasma, the
qPCR products were validated by a traditional Sanger-based method.
As expected, their sequences were identical to those derived from
NONHSAT145880 and NONHSAT116812. Taken together,
these data provided strong evidence that placenta-related lncRNAs
could be released into the circulation and their different
expression patterns in plasma could be utilized as biomarkers for
detecting and monitoring PE. Recently, several studies have
suggested that cell-free lncRNAs are detectable in human plasma and
serum and may be utilized as minimally invasive biomarkers for
disease prediction, diagnosis, and prognosis (21–23).
Although the precise mechanism of lncRNA release into the
extracellular environment is not completely understood, recent
studies have shown that circulating lncRNAs are packaged into
microparticles or other secondary structures that are protected
from endogenous RNases (24,25).
Finally, the diagnostic power of the two circulating
placenta-related lncRNAs was evaluated for PE. ROC curves
illustrated a strong separation between PE patients and
corresponding controls indicating that NONHSAT116812 and
NONHSAT145880 have high diagnostic power for the detection
of LOPE and EOPE. Thus, these two lncRNAs could be worthy of
further research when seeking novel biomarkers for predicting and
monitoring onset of PE. However, our study has some limitations,
such as a modest sample size, use of a qPCR method with relatively
low-sensitivity, and lack of an in-depth functional investigation
of placenta-related lncRNAs. Therefore, prospective cohort studies
are required to determine whether these findings are the
consequence or cause of PE, and to determine the clinical
applicability of using these molecules as the early markers of
PE.
In conclusion, aberrantly expressed placenta-related
lncRNAs might play a key or partial role in the pathogenesis of PE.
More importantly, differentially expressed lncRNAs can be released
into the maternal circulation, particularly NONHSAT116812
and NONHSAT145880 had high diagnostic power for PE. Further
research is required to measure circulating placenta-related
lncRNAs as potential tools for PE prediction and management.
Acknowledgements
We gratefully thank Professor Bolan Yu and Dr Hao
Yan for providing statistical support and conducting the
experiments. We appreciate all of the nurses who collected blood
samples. We thank the staff in the medical records room for
supplying case information and the biobank of the Third Affiliated
Hospital of Guangzhou Medical University for sample support. This
study was supported by the National Natural Science Foundation (no.
81671533) and National Key R&D Program of China (no.
2017YFC1001402).
References
|
1
|
American College of Obstetricians and
Gynecologists; Task Force on Hypertension in Pregnancy:
Hypertension in pregnancy. Report of the American college of
obstetricians and Gynecologists' task force on hypertension in
pregnancy. Obstet Gynecol. 122:1122–1131. 2013.PubMed/NCBI
|
|
2
|
Baumann MU, Bersinger NA, Mohaupt MG, Raio
L, Gerber S and Surbek DV: First-trimester serum levels of soluble
endoglin and soluble fms-like tyrosine kinase-1 as first-trimester
markers for late-onset preeclampsia. Am J Obstet Gynecol.
199:266.e1–6. 2008. View Article : Google Scholar
|
|
3
|
Fu G, Ye G, Nadeem L, Ji L, Manchanda T,
Wang Y, Zhao Y, Qiao J, Wang YL, Lye S, et al: MicroRNA-376c
impairs transforming growth factor-β and nodal signaling to promote
trophoblast cell proliferation and invasion. Hypertension.
61:864–872. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He G, Xu W, Chen Y, Liu X and Xi M:
Abhealthy apoptosis of trophoblastic cells is related to the
up-regulation of CYP11A gene in placenta of preeclampsia patients.
PLoS One. 8:e596092013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Zou Y, Wang W, Zuo Q, Jiang Z,
Sun M, De W and Sun L: Down-regulated long non-coding RNA MEG3 and
its effect on promoting apoptosis and suppressing migration of
trophoblast cells. J Cell Biochem. 116:542–550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen H, Meng T, Liu X, Sun M, Tong C, Liu
J, Wang H and Du J: Long non-coding RNA MALAT-1 is downregulated in
preeclampsia and regulates proliferation, apoptosis, migration and
invasion of JEG-3 trophoblast cells. Int J Clin Exp Pathol.
8:12718–12727. 2015.PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
10
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker for
diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He X, He Y, Xi B, Zheng J, Zeng X, Cai Q,
Ouyang Y, Wang C, Zhou X, Huang H, et al: LncRNAs expression in
preeclampsia placenta reveals the potential role of LncRNAs
contributing to preeclampsia pathogenesis. PLoS One. 8:e814372013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Long W, Rui C, Song X, Dai X, Xue X, Lu Y,
Shen R, Li J, Li J and Ding H: Distinct expression profiles of
lncRNAs between early-onset preeclampsia and preterm controls. Clin
Chim Acta. 463:193–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Z, Guo X, Li G, Shi Y and Li L: Long
noncoding RNAs as potential biomarkers in gastric cancer:
Opportunities and challenges. Cancer Lett. 371:62–70. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu P, van den Berg C, Alfirevic Z, O'Brien
S, Röthlisberger M, Baker PN, Kenny LC, Kublickiene K and Duvekot
JJ: Early pregnancy biomarkers in Pre-eclampsia: A systematic
review and meta-analysis. Int J Mol Sci. 16:23035–23056. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kleinrouweler CE, Cheong-See FM, Collins
GS, Kwee A, Thangaratinam S, Khan KS, Mol BW, Pajkrt E, Moons KG
and Schuit E: Prognostic models in obstetrics: Available, but far
from applicable. Am J Obstet Gynecol. 214(79–90): e362016.
|
|
16
|
Tsui NB, Jiang P, Wong YF, Leung TY, Chan
KC, Chiu RW, Sun H and Lo YM: Maternal plasma RNA sequencing for
genome-wide transcriptomic profiling and identification of
pregnancy-associated transcripts. Clin Chem. 60:954–962. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ogge G, Chaiworapongsa T, Romero R,
Hussein Y, Kusanovic JP, Yeo L, Kim CJ and Hassan SS: Placental
lesions associated with maternal underperfusion are more frequent
in early-onset than in late-onset preeclampsia. J Perinat Med.
39:641–652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Raymond D and Peterson E: A critical
review of early-onset and late-onset preeclampsia. Obstet Gynecol
Surv. 66:497–506. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kodama M, Miyoshi H, Fujito N, Samura O
and Kudo Y: Plasma mRNA concentrations of placenta-specific 1
(PLAC1) and pregnancy associated plasma protein A (PAPP-A) are
higher in early-onset than late-onset pre-eclampsia. J Obstet
Gynaecol Res. 37:313–318. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Farina A: The Role of RNAs and microRNAs
in non-invasive prenatal diagnosis. J Clin Med. 3:440–452. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Amorim M, Salta S, Henrique R and Jerónimo
C: Decoding the usefulness of non-coding RNAs as breast cancer
markers. J Transl Med. 14:2652016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Sun J, Wang J, Song Y, Gao P, Shi
J, Chen P and Wang Z: Long nonconding RNAs in gastric cancer:
Functions and clinical applications. Onco Targets Ther. 9:681–697.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chauhan R and Lahiri N: Tissue- and
serum-associated biomarkers of hepatocellular carcinoma. Biomark
Cancer. 8 Suppl 1:S37–S55. 2016.
|
|
24
|
Redman CW, Tannetta DS, Dragovic RA,
Gardiner C, Southcombe JH, Collett GP and Sargent IL: Review: Does
size matter? Placental debris and the pathophysiology of
pre-eclampsia. Placenta. 33 Suppl:S48–S54. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar : PubMed/NCBI
|