Introduction
A number of long non-coding RNAs (lncRNAs) have been
identified in the past decade, and previous results link specific
lncRNAs to many physiological processes and to various diseases,
including cancer and chronic pain (1–3).
Investigation into the tissue and subcellular localization of
lncRNAs is necessary to determine their function and underlying
mechanisms. Metastasis-associated lung adenocarcinoma transcript 1
(MALAT1) is an abundant, ubiquitously expressed lncRNA (4). It has previously been reported that
MALAT1 is expressed in the nervous system and regulates lung cancer
and glioma (4–6).
In situ hybridization (ISH) is a useful tool
for the quantification and localization of specific RNAs within
cultured cells or tissue sections. In ISH, an oligonucleotide probe
is used to detect the RNA of interest through complementary base
pairing (7). Historically, ISH was
performed with radioactive probes; however, the handling of
radioactive materials has many risks, and the method of image
capture was time consuming with this technique (7). These disadvantages were overcome with
the advent of fluorescence in situ hybridization (FISH),
which uses fluorescently labeled probes. The utility of FISH is
increased when it is combined with other techniques; for example,
immunofluorescence in situ hybridization (immuno-FISH) is a
combination of FISH and immunohistochemistry that enables the
detection of RNAs and proteins in the same samples (8). Variations of the immuno-FISH method
have previously been documented. Nehmé et al (9) reported that treatment with proteinase K
(PK) increased the sensitivity of FISH, but decreased the signal of
immunofluorescence staining in a study of 65-kDa glutamic acid
decarboxylase mRNA and three proteins [neuronal nuclei (NeuN), FBJ
murine osteosarcoma viral oncogene homolog B and tyrosine
hydroxylase] in frozen brain sections. Although the author provided
a method to correct this problem (9), the method was complicated and its
application in studies of noncoding RNA has not been validated. de
Planell-Saguer et al (10)
reported an immuno-FISH method for detecting non-coding RNAs in
paraffin-embedded tissues and cultured cells; however, they did not
report its application in frozen tissue sections.
In the present study, a modified immuno-FISH
protocol was used to investigate the expression and distribution of
lncRNA MALAT1 and its association with the protein markers of
neurons, microglia and astrocytes in 10-µm frozen spinal cord
slices from rats. The modified protocol was also compared with
other reported protocols.
Materials and methods
Animals
Adult male Sprague Dawley rats (n=6, 200–250 g, 6–7
weeks old; Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai,
China) were housed under a 12-h light/dark cycle, at 23–25°C and
45–50% humidity and provided with free access to food and water.
All surgical and experimental procedures were approved by the
Animal Ethics Committee of Fudan University (Shanghai, China).
Reagents
To prepare a 1% sodium pentobarbital solution, 5 g
sodium pentobarbital (cat. no. 69020181; Sinopharm Chemical Reagent
Co., Ltd., Shanghai, China) was dissolved in 500 ml distilled
(d)H2O, and the solution was stored at 4°C in the dark.
To prepare 1 l of 4% paraformaldehyde, 40 g paraformaldehyde was
added to 1 l of 1X phosphate-buffered saline (PBS) and heated
gradually to 60°C with continuous stirring to dissolve the
paraformaldehyde. The pH was subsequently adjusted to 7.4 with
NaOH. To prepare a 10 or 30% sucrose solution, 10 or 30 g sucrose
(cat. no. 10021418; Sinopharm Chemical Reagent Co., Ltd.) was added
to 100 ml dH2O. To prepare 1 l of antigen unmasking
buffer (10 mM sodium citrate), 2.94 g sodium citrate tribasic salt
dihydrate
(C6H5Na3O7·2H2O,
cat. no. 10019418; Sinopharm Chemical Reagent Co., Ltd.) was added
to 1 l dH2O. The pH was adjusted to 6.0 and the solution
was subsequently filtered (pore diameter, 75 µm). To prepare 1 l of
20X saline-sodium citrate (SSC), 175.2 g NaCl and 88.2 g sodium
citrate tribasic salt dihydrate were dissolved in 800 ml
dH2O. The pH was adjusted to 7.0, and dH2O
was added to bring the volume to 1 l. To prepare 100 ml of
prehybridization buffer, 3 g bovine serum albumin (BSA; cat. no.
69003433; Sinopharm Chemical Reagent Co., Ltd.) was added to 100 ml
of 4X SSC. The resulting solution was used to prepare 0.1, 1, 2 and
4X SSC. To prepare 10 ml of hybridization buffer, 1 g dextran
sulfate (cat. no. S14047; Shanghai Yuanye Biotechnology Co., Ltd.,
Shanghai, China) and 1 ml deionized formamide (cat. no. 30091218;
Sinopharm Chemical Reagent Co., Ltd.) were added to 9 ml of 4X SSC.
To prepare 100 ml of blocking buffer, 1 g BSA and 0.1 ml Tween-20
were added to 100 ml of 1X PBS. To prepare 50 ml of
4′,6-diamidino-2-phenylindole (DAPI) staining solution, 0.5 µl DAPI
stock solution was diluted in 50 ml of 1X PBS.
Riboprobes and antibodies
All DNA oligonucleotides, including three probes for
rat lncRNA malat1 (Malat1-a, -b and -c) and one probe for GAPDH
(positive control; Table I), were
designed and synthesized and labeled on the 5′-end with Alexa Fluor
633 by Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Probes were stored at −20°C in powder form. To prepare a 250
nM working solution, probes were diluted in hybridization buffer.
Antibodies against NeuN, Iba1 and GFAP were used as biomarkers of
neurons, microglia and astrocytes, respectively (11). The primary and Alexa Fluor
488-conjugated secondary antibodies used for immunofluorescence
were purchased from Abcam (Cambridge, MA, USA; Table II) and diluted with blocking buffer
to 1:500 and 1:400, respectively.
 | Table I.Riboprobes used in the present
study. |
Table I.
Riboprobes used in the present
study.
| Name | Tm,
°C | Sequence, 5′-3′ | Length, bp | GC, % |
|---|
| Malat1-a | 66.1 |
GGGCCGTTATAAGAGTCGACTGTCGCATGTACGAAGGCATGAG | 43 | 53.5 |
| Malat1-b | 66.1 |
GCGGTTCGTTGGAGGAAGCTAGGAAGAAGGAGCCGAAATGATG | 43 | 53.5 |
| Malat1-c | 62.4 |
GGCTGGTAGTTTATTCTTTTCCCCCTCCCTTAACAAGACTTG | 42 | 45.2 |
| GAPDH | 64.0 |
CTTGTGACAAAGTGGACATTGTTGCCATCAACGACCCCTTCATTG | 45 | 46.7 |
 | Table II.Antibodies used in the present
study. |
Table II.
Antibodies used in the present
study.
| Name |
Primary/secondary | Company | Cat. no. | Conjugated | Host | Dilution |
|---|
| Anti-NeuN | Primary | Abcam | Ab177487 | No | Rabbit | 1:500 |
| Anti-Iba-1 | Primary | Abcam | Ab5076 | No | Goat | 1:500 |
| Anti-GFAP | Primary | Abcam | Ab7260 | No | Rabbit | 1:500 |
| Anti-rabbit IgG | Secondary | Abcam | Ab150129 | Alexa fluor 488 | Donkey | 1:400 |
| Anti-goat IgG | Secondary | Abcam | Ab150073 | Alexa fluor 488 | Donkey | 1:400 |
Tissue preparation
Rats were anesthetized with an intraperitoneal
injection of 1% sodium pentobarbital solution (40 mg/kg) and
transcardially perfused with 1X PBS and 500 ml of 4%
paraformaldehyde in phosphate buffer (0.1 M, pH 7.4). The spinal
cord was harvested at the cervical level and specimens were
immersed in 4% paraformaldehyde for 4 h at 4°C. The tissues were
then sequentially immersed in 10 and 30% sucrose until the spinal
cords fell to the bottom. Spinal cords were embedded in optimal
cutting temperature compound (cat. no. 4583; Sakura Finetek USA,
Inc., Torrance, CA, USA) and mounted on a microtome stage.
Horizontal sections (10 µm) were mounted onto pre-warmed
poly-l-lysine-coated slides (cat. no. P4981; Thermo Fisher
Scientific, Inc.).
Antigen retrieval and dehydration
Following desiccation for 20 min at room
temperature, slides were rinsed twice with 1X SSC (5 min/wash). The
slides were placed into a Coplin jar filled with antigen unmasking
buffer, which was placed into an autoclave and heated to 100°C for
5 min and subsequently cooled to room temperature. Sections were
then washed three times with 1X SSC (5 min/wash). The tissue
sections were dehydrated in a graded series of ethanol (50, 70, 90
and 100%; 3 min each) and air-dried for 10 min.
For comparison, some slides were treated with PK
instead of treatment with antigen unmasking buffer and autoclaving,
according to the method described by Nehmé et al (9). For this method, the slides were
immersed in a buffer containing 0.1 M Tris-HCl (pH 8), 50 mM EDTA
(pH 8) and 10 µg/ml PK (Roche Diagnostics, Basel, Switzerland) for
25 min at 37°C.
Prehybridization
The prehybridization buffer was warmed to ~47°C
[usually 22–25°C below the melting temperature (Tm) of
probes] and 200 µl warmed prehybridization buffer was pipetted onto
each slide and prehybridized for 20 min in a humid chamber (47°C).
The probes (Malat1-a, -b and -c and GAPDH) were diluted with
hybridization buffer and denatured at 65°C for 10 min, then stored
on ice.
Hybridization
The prehybridization buffer was removed and 100 µl
aliquots of each probe (or same volume of hybridization buffer
without probes as negative control) were added to separate slides
and hybridized overnight (4–16 h) at the same temperature used in
the prehybridization step. From this step on, all slides were kept
in the dark.
Post-hybridization
Following hybridization, the slides were rinsed with
the following solutions: 4X SSC (twice at 5 min/wash), 2X SSC (5
min), 1X SSC (5 min), and 0.1X SSC (5 min) at the same temperature
as in the hybridization step. The slides were then rinsed with 1X
PBS for 5 min at room temperature.
Immunofluorescence staining
Following in situ hybridization, the sections
were blocked in blocking buffer for 30 min at 37°C. From this step
on, sections were kept in a humid chamber to protect the tissue
sections from drying out. The blocking buffer was removed and 200
µl primary antibody diluted in blocking buffer was added (Table II), and the sections were incubated
overnight at 4°C in a humid chamber. Sections were rinsed three
times with 1X PBS (5 min/wash), then incubated with corresponding
secondary antibody (200 µl diluted in blocking buffer) for 2 h at
37°C. Slides were subsequently rinsed three times with 1X PBS (5
min/wash). At this point, all FISH and immunofluorescence staining
steps were complete. However, if the nucleus needed to be observed,
nuclear staining with DAPI was performed. Finally, a few drops of
antifade (cat. no. P36965; Thermo Fisher Scientific, Inc.) were
added to the slides and a coverslip was placed. The slides were
sealed with nail polish.
Image acquisition and analysis
Images were captured with a confocal laser-scanning
microscope (FluoView™ FV1000; Olympus Corporation, Tokyo, Japan)
equipped with a digital camera and image analysis system (FV10-ASW
4.0; Olympus Corporation). All images were analyzed using Image J
1.44 (National Institutes of Health, Bethesda, MD, USA). Brightness
and contrast were adjusted. Positive cells and the background
surrounding positive cells were selected as regions of interest.
The ratio of integral optical density (IOD) between the positive
cells and background signal was calculated using Image J 1.44. All
statistical analyses using Student's t-test and histograms were
completed with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla,
CA, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
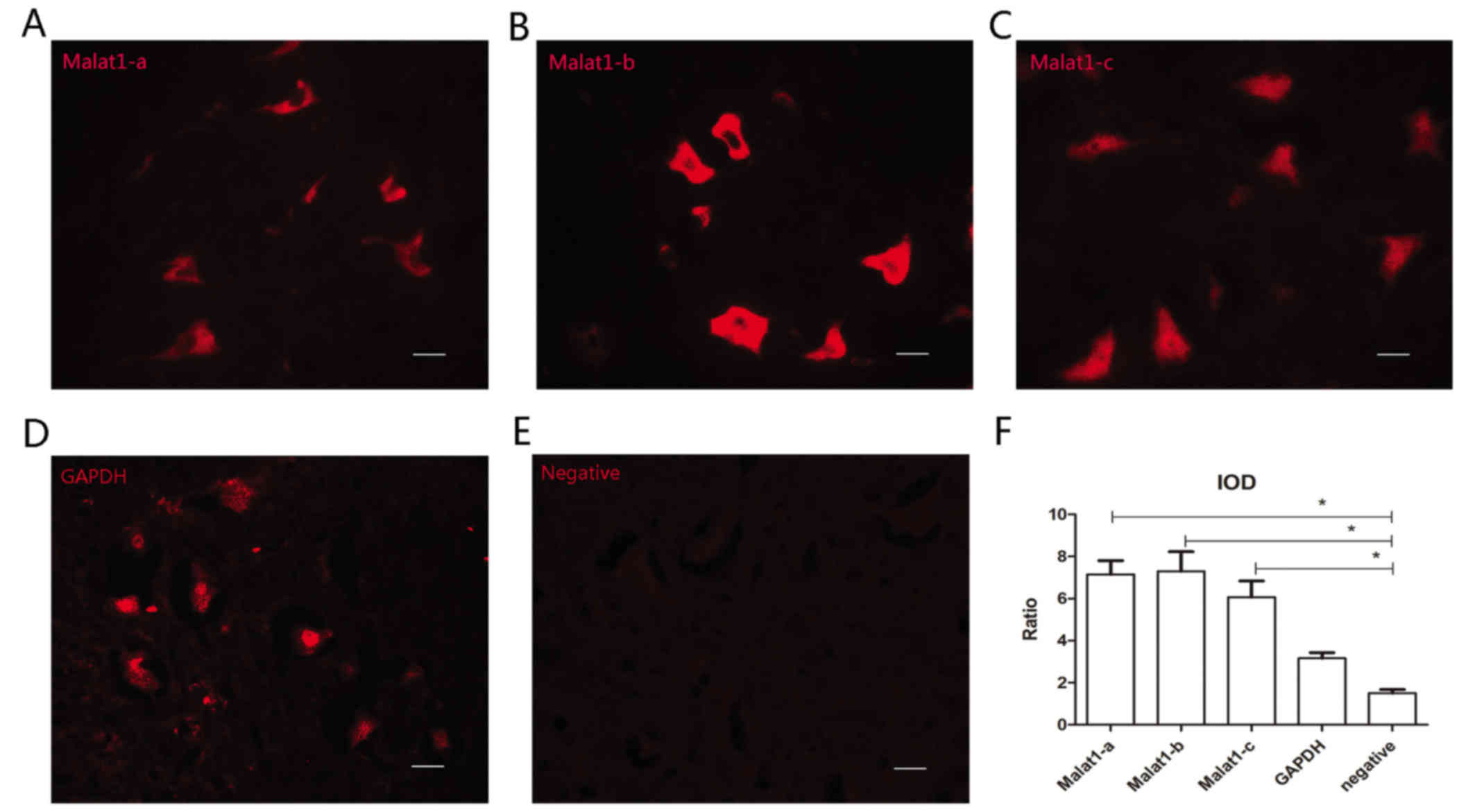
All three probes are effective for
immuno-FISH
Satisfactory fluorescent signals were obtained from
all three malat1 probes (Malat1-a, -b and -c; Fig. 1A-C). During FISH, GAPDH served as a
positive control and sections without probes (hybridization buffer
only) served as negative controls (Fig.
1D and E). The IOD ratio of Malat1-a, -b, -c group was
respectively compared with negative control using Student's t-test.
All three Malat1 probes were demonstrated to be effective (n=6,
P<0.05); however, the Malat1-b probe exhibited the highest
signal amplitude (Fig. 1F).
Therefore, Malat1-b was used for subsequent experiments.
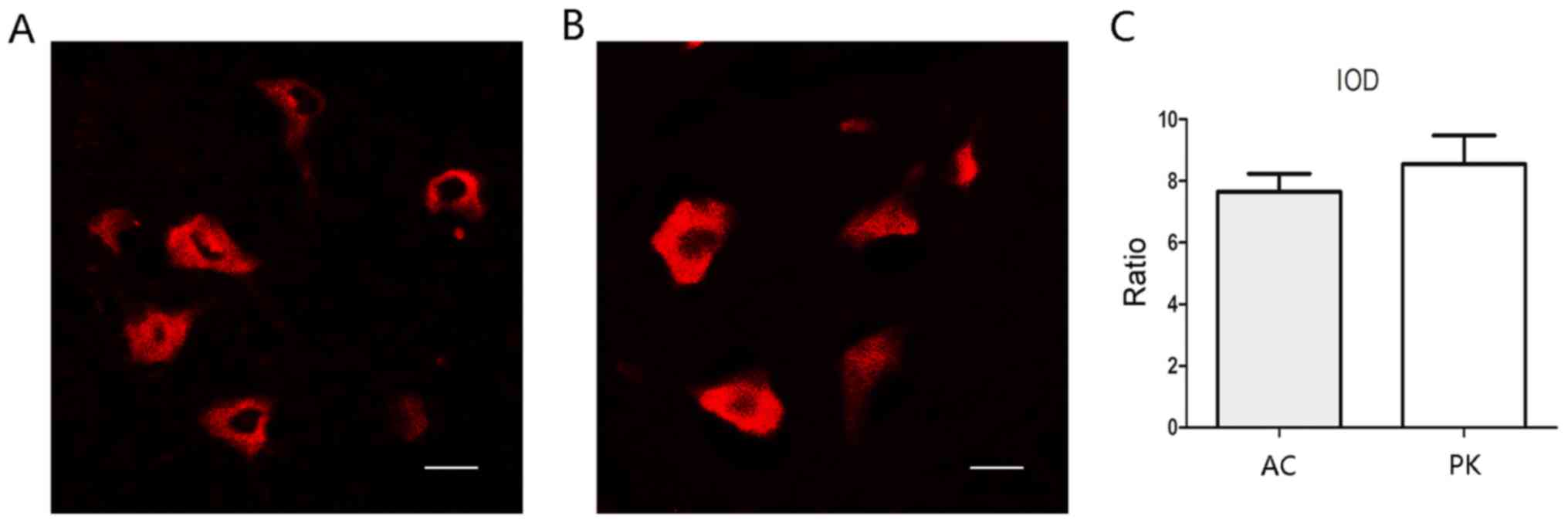
Antigen retrieval by autoclaving and
PK treatment has similar effects on FISH
Two antigen retrieval methods, autoclaving and PK
treatment, were compared. Using the same imaging parameters, both
methods achieved notable FISH signals (Fig. 2). The IOD ratio of positive cells to
background signal was calculated for each group, and did not differ
significantly between the two groups (Fig. 2C).
Modified immuno-FISH is effective
The Malat1-b probe was used to detect the expression
of malat1 (Fig. 3; labeled red).
Using immuno-FISH, malat1 and three different proteins were
successfully co-detected. NeuN-positive cells were double labeled
with malat1 (green-red merge; Fig.
3A), while cells positive for glial fibrillary acidic protein
and ionized calcium binding adaptor molecule 1 (Iba-1) were a
single color (green; Fig. 3B and C).
These results indicated that Malat1 is expressed in neurons but not
in microglia or astrocytes.
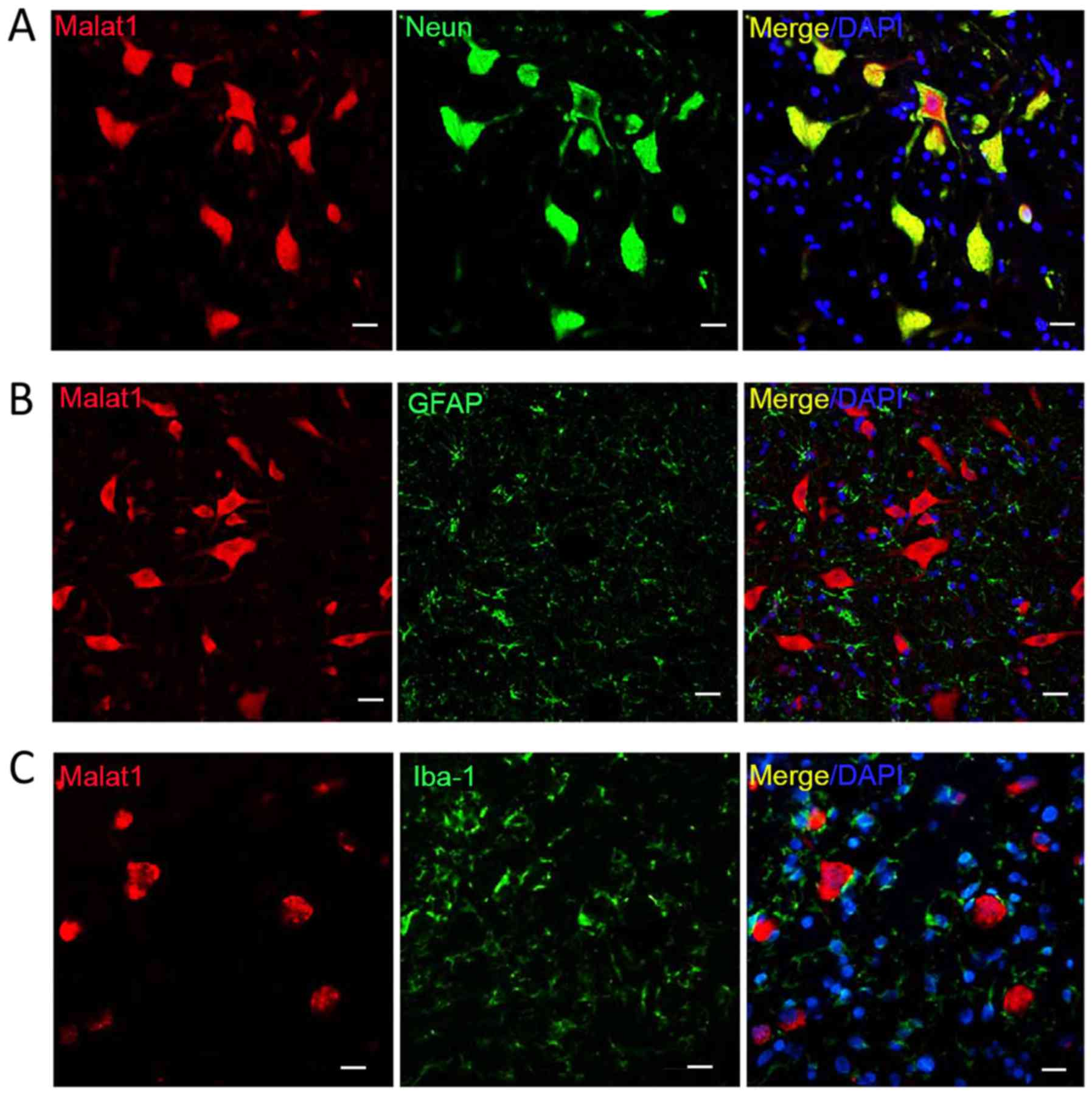 | Figure 3.Double labeling for Malat1 and protein
markers. Co-detection of Malat1 with (A) NeuN, (B) GFAP and (C)
Iba-1. Only NeuN-positive cells were double labeled. Scale bar, 20
µm, magnification, ×100. Malat, metastasis-associated lung
adenocarcinoma transcript; NeuN, neuronal nuclei; GFAP, glial
fibrillary acidic protein; Iba-1, ionized calcium binding adaptor
molecule 1; DAPI, 4′,6-diamidino-2-phenylindole. |
Discussion
Immuno-FISH is a reliable method for the double
staining of RNA and cellular proteins (10). It is typically used to detect the
localization and abundance of target RNA expression at the
histological level (9). LncRNAs are
among the most abundant classes of non-coding RNAs (12). Many lncRNAs have been implicated in
the functioning of the nervous system and development of disease
(13). In the present study, a
modified protocol of immuno-FISH was developed. Using this
protocol, high-quality fluorescent signals and histological data
were obtained that provided information about the expression and
distribution of lncRNA malat1 and three different proteins in the
spinal cord.
Pretreatment of tissue sections is a critical step
for obtaining satisfactory fluorescence signals, and this step is
modified in different protocols. The purpose of pretreatment is to
ensure signal specificity and minimize non-specific background
signals (14,15). Acetylation, which negatively charges
sections to reduce the adsorption of negatively-charged probes, is
widely used as a pretreatment step to reduce background staining
(10,16,17).
However, in some protocols, acetylation has been reported as a
dispensable step (10,18). As such, in the present study,
sections were not treated with acetylation, which simplified the
protocol and saved time.
In traditional RNA FISH procedures, PK treatment may
be used to improve sensitivity, as it denatures many proteins in
the membrane and within RNA-protein complexes, making it easier for
the probe to cross membranes and bind to target RNA (9). However, previous results have indicated
that PK may also denature proteins of interest, which makes it
incompatible with immunofluorescence (9). In the present study, sodium
citrate-based antigen unmasking and autoclaving were used for
antigen retrieval, and high-quality FISH and immunofluorescence
signals were obtained. High temperatures may also denature membrane
proteins and RNA-protein complexes, unmasking the antigenic sites
of target proteins and thus improving antibody-antigen reactions
(19). In addition, high pressure
prevents solutions from boiling, which otherwise causes tissue
sections to peel away from slides.
The reagents used in the current protocol were
readily available, relatively non-toxic and inexpensive.
Researchers may prepare the antigen unmasking, prehybridization,
and hybridization buffers using common reagents following the
simple formulas listed above. Other reported hybridization buffer
solutions include lauroyl sarcosine (18), Denhardt's solution, transfer RNA or
dithiothreitol (9), though these may
be difficult to obtain and/or store.
Another advantage of the current protocol was that
it was time-efficient. All of the steps undertaken in the present
study were performed within 24–32 h. The variation in the time
required to complete the protocol was mainly due to the antibodies
used for immunofluorescence, as different antigen-antibody
reactions required different incubation periods. For instance, it
was possible to label NeuN in 2 h, whereas Iba-1 required >6 h
at 4°C. Thus, an overnight reaction at 4°C is preferred in the
majority of immunohistochemistry procedures (3,9,20). In instances where the protocol is
applied using other antibodies and the antibody-antigen reaction is
not satisfactory, longer reaction times at 4°C should be
considered.
Malat1 is an abundant lncRNA that has been
demonstrated to regulate the progression of many diseases (4). In the present study, the distribution
of malat1 in the spinal cord of rats was assessed using a simple
immuno-FISH protocol, and it was demonstrated that malat1 was
expressed in neurons but not in microglia or astrocytes. Despite
the lack of a signal amplification method, satisfactory
fluorescence signals were obtained. However, the abundance of some
lncRNAs is low, for example BACE1-AS and lnc-DC (21,22), and
in these cases signal amplification treatments, including tyramide
amplification and specially designed riboprobes, may be necessary
to obtain detectable fluorescence.
In conclusion, the present study performed a simple
immuno-FISH protocol for the detection of malat1 lncRNA and protein
markers of neurons, microglia and astrocytes in frozen spinal cord
sections of rats. Advantages of the modified immune-FISH protocol
include the ready availability of reagents and general speed of the
method. This protocol may be adapted for other tissues and RNAs,
and may also be used for FISH or immunofluorescence alone.
Acknowledgements
The present study was supported by the Ministry of
Science and Technology of China (973 Program; grant no.
2014CB542204 to Yun Wang) and the Science and Technology Commission
of Shanghai Municipality (grant no. 16ZR1404200 to Yuzhou Liu).
References
|
1
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao X, Tang Z, Zhang H, Atianjoh FE, Zhao
JY, Liang L, Wang W, Guan X, Kao SC, Tiwari V, et al: A long
noncoding RNA contributes to neuropathic pain by silencing Kcna2 in
primary afferent neurons. Nat Neurosci. 16:1024–1031. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gutschner T, Hammerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kryger R, Fan L, Wilce PA and Jaquet V:
MALAT-1, a non protein-coding RNA is upregulated in the cerebellum,
hippocampus and brain stem of human alcoholics. Alcohol.
46:629–634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han Y, Wu Z, Wu T, Huang Y, Cheng Z, Li X,
Sun T, Xie X, Zhou Y and Du Z: Tumor-suppressive function of long
noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and
inactivation of ERK/MAPK signaling. Cell Death Dis. 7:e21232016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pardue ML and Gall JG: Molecular
hybridization of radioactive DNA to the DNA of cytological
preparations. Proc Natl Acad Sci USA. 64:600–604. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fuller CE and Perry A: Fluorescence in
situ hybridization (FISH) in diagnostic and investigative
neuropathology. Brain Pathol. 12:67–86. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nehmé B, Henry M and Mouginot D: Combined
fluorescent in situ hybridization and immunofluorescence: Limiting
factors and a substitution strategy for slide-mounted tissue
sections. J Neurosci Methods. 196:281–288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Planell-Saguer M, Rodicio MC and
Mourelatos Z: Rapid in situ codetection of noncoding RNAs and
proteins in cells and formalin-fixed paraffin-embedded tissue
sections without protease treatment. Nat Protoc. 5:1061–1073. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Meng X, Yang F, Ouyang T, Liu B, Wu C and
Jiang W: Specific gene expression in mouse cortical astrocytes is
mediated by a 1740bp-GFAP promoter-driven combined adeno-associated
virus 2/5/7/8/9. Neurosci Lett. 593:45–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clark MB and Mattick JS: Long noncoding
RNAs in cell biology. Semin Cell Dev Biol. 22:366–376. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qureshi IA, Mattick JS and Mehler MF: Long
non-coding RNAs in nervous system function and disease. Brain Res.
1338:20–35. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Namekawa SH and Lee JT: Detection of
nascent RNA, single-copy DNA and protein localization by immunoFISH
in mouse germ cells and preimplantation embryos. Nat Protoc.
6:270–284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sui QQ, Zhu J, Li X, Knight GE, He C,
Burnstock G, Yuan H and Xiang Z: A modified protocol for the
detection of three different mRNAs with a new-generation in situ
hybridization chain reaction on frozen sections. J Mol Histol.
47:511–529. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Just L, Timmer M, Tinius J, Stahl F,
Deiwick A, Nikkhah G and Bader A: Identification of human cells in
brain xenografts and in neural co-cultures of rat by in situ
hybridisation with Alu probe. J Neurosci Methods. 126:69–77. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Silahtaroglu AN, Nolting D, Dyrskjøt L,
Berezikov E, Møller M, Tommerup N and Kauppinen S: Detection of
microRNAs in frozen tissue sections by fluorescence in situ
hybridization using locked nucleic acid probes and tyramide signal
amplification. Nat Protoc. 2:2520–2528. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Urbanek MO and Krzyzosiak WJ: RNA FISH for
detecting expanded repeats in human diseases. Methods. 98:115–123.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
White AK, Hansen-Lardy L, Brodersen BW,
Kelling CL, Hesse RA and Duhamel GE: Enhanced immunohistochemical
detection of infectious agents in formalin-fixed, paraffin-embedded
tissues following heat-mediated antigen retrieval. J Vet Diagn
Invest. 10:214–217. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vicuña L, Strochlic DE, Latremoliere A,
Bali KK, Simonetti M, Husainie D, Prokosch S, Riva P, Griffin RS,
Njoo C, et al: The serine protease inhibitor SerpinA3N attenuates
neuropathic pain by inhibiting T cell-derived leukocyte elastase.
Nat Med. 21:518–523. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|