Introduction
Hepatocellular carcinoma (HCC) is the fifth most
common malignancy worldwide and the third most common cause of
cancer-related deaths. Although the diagnosis and treatment of HCC
have greatly improved, more than 1.23 million new cases are
diagnosed each year and the 5-year survival rate is still <15%
(1,2). Surgical resection is the important way
to cure the HCC, unfortunately, most patients (approximately 80%)
with HCC are unsuitable for surgery because of early invasion and
extrahepatic metastases, underlying liver disease or comorbidities
(3). Therefore, surgical resection
combined with chemotherapy or chemotherapy alone was used as a hope
to prolong the patient's life. However, abnormal metabolic
environment of HCC cells will restrict the curative effect and
survival, because of resistance to chemotherapy after long-term
drugs used (4,5). For example, Etoposide, 5-Fluorouracil,
Cisplatin, and their combinations have demonstrated
multi-resistance for the treatment of HCC (6). Oxaliplatin, a third-generation platinum
compound, has been widely used in the treatment of a variety of
cancers and achieved excellent results in clinical applications
(7). It is also considered to be a
potential advantage drug of HCC because of the few cytotoxic in
terms of nephrotoxicity and myelosuppression (8). Previous studies showed that oxaliplatin
could increase the autophagy role of HCC cells, which eventually
result in a tumor resistance response in HCC (8). However, the mechanism of
oxaliplatin-resistance in HCC remains undetermined.
Sphingosine kinase is a conserved lipid kinase that
catalyzes the phosphorylation of sphingosine to form
sphingosine-1-phosphate (S1P), which potentially regulated the
biological processes, including cell proliferation, motility,
differentiation, apoptosis, and angiogenesis (9). Two isoforms of mammalian Sphingosine
kinase 1 and 2 (SphK1 and SphK2) have been cloned and characterized
(9). Recently, the role of SphKs in
cancer (SphK1 in particular) has received considerable attention.
Elevated expression of SphK1 has been observed in multiple types of
cancer, and promoted cell survival and growth (10,11).
Furthermore, high SphK1 expression has been associated with poor
prognosis in patients with glioblastoma or breast cancer (12,13).
Previous studies also found that blockade of the SphK1 signaling
pathway may reprogram cellular responsiveness to tamoxifen and
abrogate antiestrogen resistance in human breast cancer cells
(14). SphK1 also involved the
resistance of chemotherapy drugs in colorectal cancer and
pancreatic cancer (15,16), but little is known about the SphK1 in
HCC.
In the present study, we demonstrate that high SphK1
expression levels were correlated with shorter overall survival,
and it is one of the important factors that facilitates resistance
to the oxaliplatin in HCC. Furthermore, knockdown of SphK1 markedly
suppressed the phosphorylations of AKT serine/threonine kinase and
glycogen synthase kinase-3β (Akt and GSK3β) in SK-Hep1 cells, and
suggested that SphK1 promote oxaliplatin resistance of HCC cell via
modulating the Akt/GSK3β pathway. Thus, these findings suggest that
the cellular response to oxaliplatin can be reprogrammed by
manipulation of the SphK1 pathway, and may provide a new way to
overcome oxaliplatin resistance in the treatment of HCC.
Materials and methods
Patients and tissue samples
A total of 21 paired human primary HCC tissues and
adjacent non-tumors tissues (at least 3 cm from the edge of tumor)
were obtained from patients who were recruited into a clinical
trial at Xinchang People's Hospital between 2008 and 2010. The 21
HCC patients were classified into two groups according to the
median level of SphK1 expression: The median level and above
considered as high expression (n=11), and lower than the median
level considered as low (n=10). All patients have no received
irradiation or chemotherapy before surgery and the final diagnosis
was based on the pathological diagnosis after surgery. The present
study was approved by the ethics review board of Xinchang People's
Hospital, and all patients signed informed consent forms.
Cell culture and transfection
Five HCC cell lines (Hep3B, HepG2, HCCLM3 and
SK-Hep1) were obtained from the American Type Culture Collection
(ATCC), and normal liver cell line (HL-7702) was kept at our
laboratory. All cells were maintained in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 2
mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin
(Invitrogen). For transfection, HCCLM3 and SK-Hep1 cells were
seeded in 12-well plates (2×105 cells/well). After 18 h,
the cells were transfected with 2 targeting SphK1 siRNA
oligonucleotides or nonsilencing siRNA oligonucleotides (Qiagen AB,
Sollentuna, Sweden) by using the HiPerFect Transfection Reagent
(Qiagen) according to the manufacturer's protocol. After 72 h of
incubation, cells were collected for western blotting and qRT-PCR
analysis.
Measurement of mRNA expression level
by quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from tissues or cultured
cells using TRIzol reagent (Invitrogen) as the manufacturer's
protocol. For mRNA analyses, 1.0 µg of total RNA was used to
synthesize cDNA using the RevertAid™ First Strand cDNA Synthesis
kit (Fermentas; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Equal amounts of cDNAs were taken for a real-time quantitative PCR
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) using SYBR-Green as a dye (Roche, Mannheim, Germany). The
relative quantity of SphK1 mRNA was normalized using the 18S rRNA
as control. The following primers were used: SphK1 forward primer,
5′-CTGGCAGCTTCCTTGAACCAT-3′, and reverse primer,
5′-TGTGCAGAGACAGCAGGTTCA-3′; SphK2 forward primer,
5′-GCTCAACTGCTCACTGTTGC-3′, and reverse primer,
5′-GCAGGTCAGACACAGAACGA-3′; 18S rRNA forward primer,
5′-AAACGGCTACCACATCCAAG-3′, and reverse primer,
5′-CCTCCAATGGATCCTCGTTA-3′. All reactions were performed in
triplicate.
Sphingosine kinase assay
SphK1 activity was measured by using Sphingosine
Kinase 1 Assay kit (Novus Biologicals, Littleton, CO, USA)
according to the manufacturer's instructions. Firstly, protein
extracts (30 µg) were incubated in reaction buffer, 100 µM
sphingosine and 10 µM ATP for 1 h at 37°C. Then, luminescence
attached ATP detector was then added to stop the kinase reaction.
Kinase activity was measured using BioTek Microplate Readers
(BioTek, Winooski, VT, USA). The results were obtained from 3
independent experiments, each reactions were performed in
triplicate.
MTT survival assay
Oxaliplatin (Sigma-Aldrich, St. Louis, MO, USA) was
dissolved in 100% dimethyl sulfoxide and diluted with DMEM to the
desired concentration. The effects of oxaliplatin on HCC cells
survival were determined by MTT (Sigma-Aldrich) assay. Cells were
seeded into 96-well plates (5×103 cells/well) and
cultured for 48 h with oxaliplatin concentrations between 0, 20, 40
and 60 µg/ml. After treatments, cells were incubated with 20 µl MTT
to each well and then incubated at 37°C for 4 h. Following the
incubation, MTT solution was removed and added 200 µl DMSO into
each well to dissolve the formazan crystals. The plates were shook
for 5 min and subjected to absorbance reading at 570 nm. Percentage
of cell survival was determined as [(Mean OD of test wells-Mean OD
of blank group)/(Mean OD of negative group-Mean OD of blank
group)]x100%. The results were obtained from 3 independent
experiments, each reactions were performed in triplicate.
Western blot analysis
For western blot analysis, cells were lysed with
RIPA lysis buffer supplemented with cOmplete Protease Inhibitor
EASYpacks EDTA-Free (Roche). The lysate were subject to
centrifugation at 12,000 g for 10 min at 4°C and remove the cell
debris. Then, supernatant concentrations were measured by the BCA
Assay kit (Beyotime Biotech, Jiangsu, China) according to the
standard protocol. Sample (50 µg/lane) were separated by 10%
SDS-PAGE. The separated proteins were transferred to PVDF western
blot analysis membrane (Roche) and probed with rabbit anti-SphK1
(1:1,500; Abcam, Cambridge, UK), SphK2 (1:2,000; Abcam), Akt
(1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
Phospho-Akt (Ser473) (1:1,000; Santa Cruz), GSK3β (1:1,000; CST,
Beverly, MA, USA), Phospho-GSK3β (1:1,000; CST) and β-actin
(1:1,000; CST) primary antibodies. Target protein bands visualized
were detected with HRP-linked goat anti-rabbit secondary antibodies
(1:2,000; Abcam).
Clonogenic survival assay
Log-phase cells were plated overnight and treated
with 60 µg/ml oxaliplatin for 48 h. After treatment, cells were
harvested, and 500 cells/well were plated in triplicate into 60 mm
dishes. After two weeks, cells were fixed using 4%
parafor-maldehyde and stained with crystal violet. The number of
colonies per plate was assessed and then expressed as percent
survival relative to untreated cells.
Statistical analysis
Data analyses were performed using SPSS 15.0. All
data are presented as means ± SEM of a minimum of three or more
replicates. P<0.05 was considered to indicate a statistically
significant difference..
Results
Upregulation of SphK1 correlates with
poor prognosis in HCC
To investigate the role of SphK in HCC, we collected
tumor tissues and adjacent non-tumors tissues from 21 HCC patients
(Table I), and the transcriptional
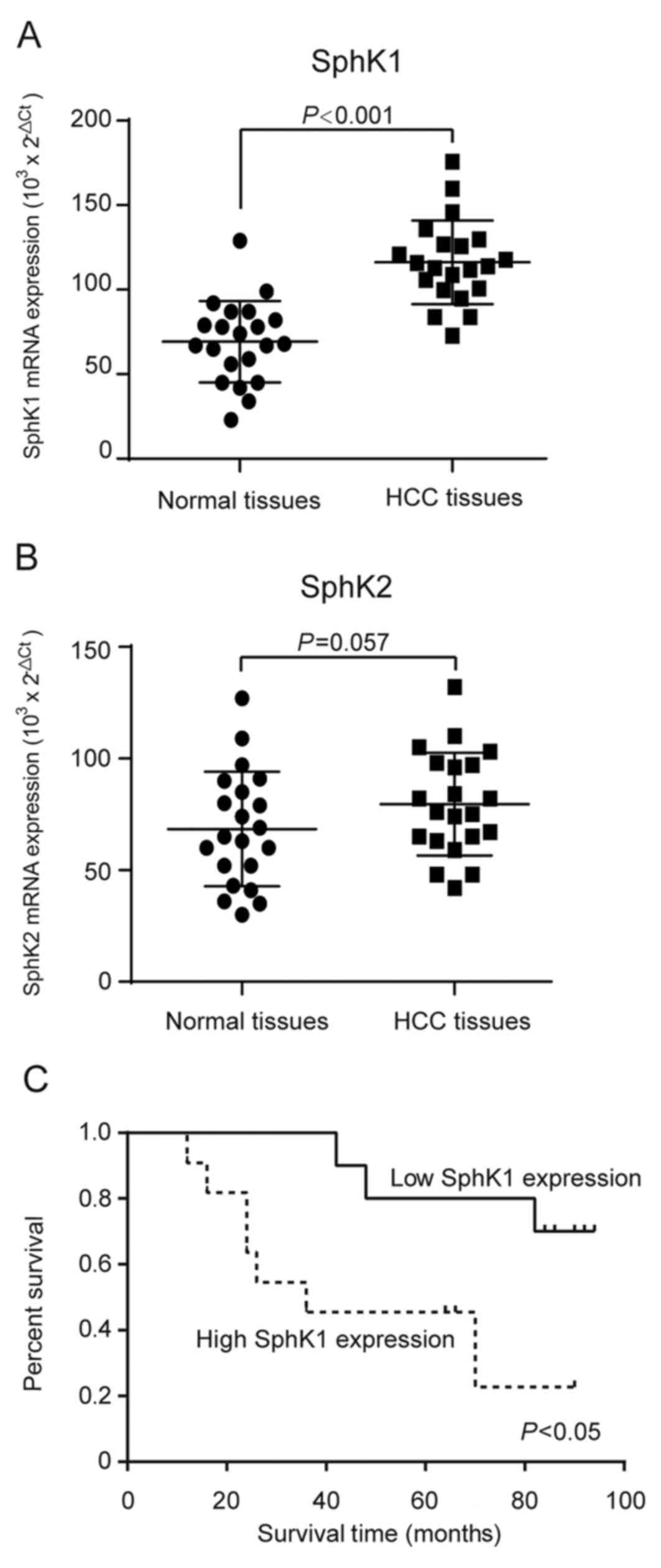
levels of SphK1 and SphK2 were analyzed by qRT-PCR. Our results
demonstrated that the transcriptional levels of SphK1, but not
SphK2, were significantly upregulated in HCC tissues compared with
that in adjacent non-tumors tissues (Fig. 1A-B). Additionally, Kaplan-Meier
survival analysis was used to further evaluate the correlations
between the SphK1 transcriptional levels and prognosis in patients
with HCC. The 21 patients with HCC were classified into a high
expression group (n=11) and a low expression group (n=10). The
result showed that patients with high SphK1 expression had
significantly shorter overall survival than those with low SphK1
expression (Fig. 1C).
 | Table I.Association between Sphk1 expression
and clinicopathological features of 21 hepatocellular carcinoma
patients. |
Table I.
Association between Sphk1 expression
and clinicopathological features of 21 hepatocellular carcinoma
patients.
| Characteristics | Cases (21) | Sphk1 mRNA
expression | P-value |
|---|
| Gender |
|
| 0.612 |
| Male | 11 | 15 |
|
|
Female | 10 | 8 |
|
| Age, years |
|
| 0.463 |
|
<50 | 15 | 14 |
|
| ≥50 | 6 | 9 |
|
| Tumor stage |
|
| 0.048 |
| I/II | 12 | 5 |
|
|
III/IV | 9 | 11 |
|
| Tumor size, cm |
|
| 0.023 |
|
<5 | 25 | 9 |
|
| ≥5 | 22 | 14 |
|
| Lymph node
metastasis |
|
| 0.135 |
|
Negative | 18 | 15 |
|
|
Positive | 3 | 9 |
|
SphK1 is overactivated in the
oxaliplatin-resistant HCC cell lines
On the basis of the suggested correlation between
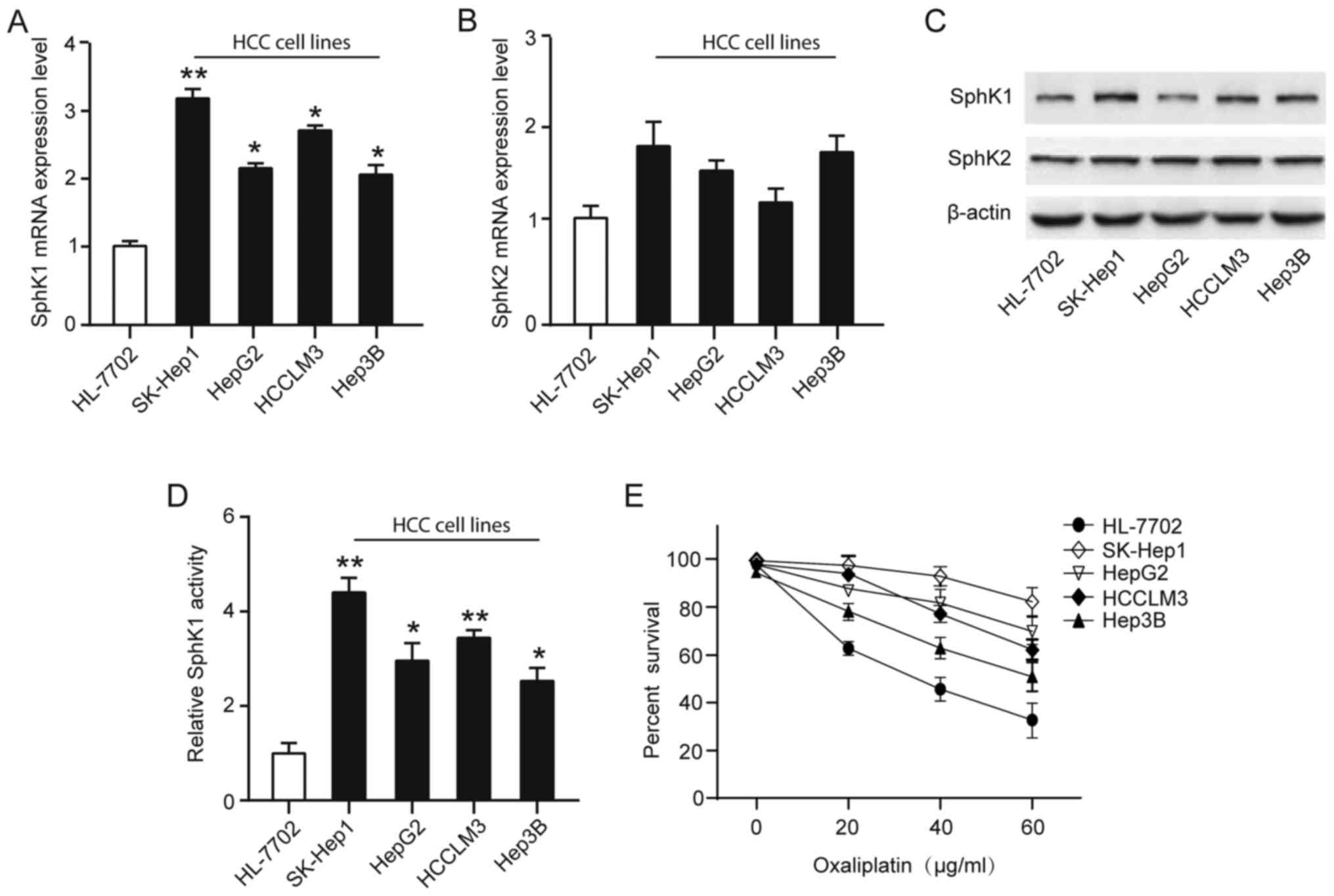
SphK and resistance to anticancer-targeted agents, we analyzed
SphK1 and SphK2 expression and activity in four human HCC cell
lines (Hep3B, HepG2, HCCLM3 and SK-Hep1) and normal liver cell line
(HL-7702). Although both SphK1 and SphK2 isoforms are responsible
for total cellular SphK activity, we found the transcriptional
levels of SphK1, but not SphK2, were significantly upregulated in
HCC cell lines compared with that in normal liver cell line
(Fig. 2A-B). Similarly, SphK1
protein expression levels were significantly upregulated in HCC
cell lines, especially in SK-Hep1, a high invasiveness and
metastasis cell line (Fig. 2C). To
explore whether HCC cell lines resistance toward the oxaliplatin
correlates with a dysregulation of the SphK1, a cell survival assay
using increasing concentrations of oxaliplatin (0–60 µg/ml) for 48
h showed a greater resistance to oxaliplatin of HCC cell lines, and
found the activity of SphK1 were positively associated with
resistance to oxaliplatin (Fig.
2D-E). Taken together, these observations suggest that the
elevated levels in SphK1 expression and activity in HCC cells may
associate with the oxaliplatin-resistant phenotype.
Downregulation of SphK1 decrease
resistant to oxaliplatin in HCC cells
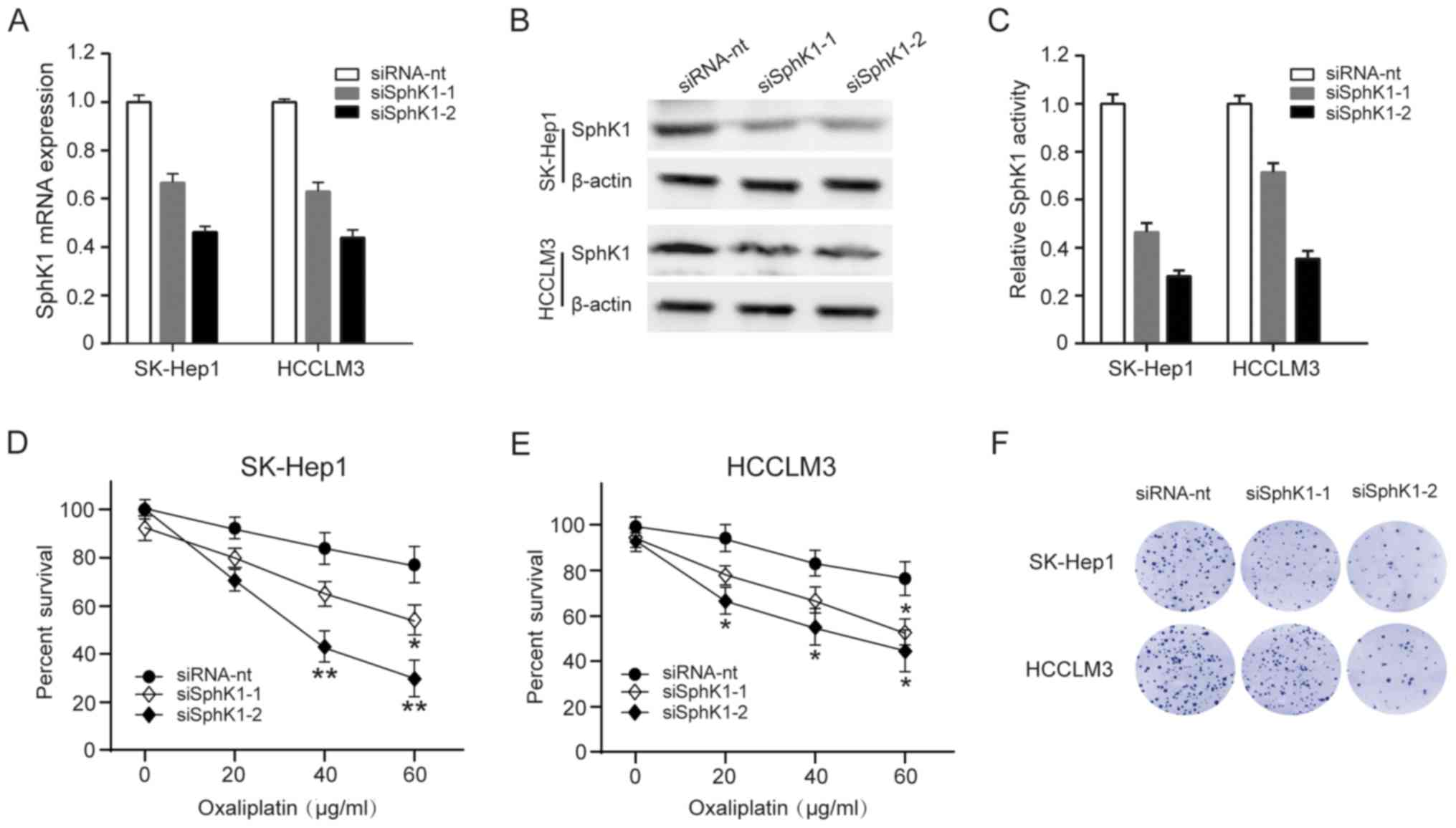
To investigate whether downregulating SphK1 activity
in HCC cells might decrease resistant to oxaliplatin, we used 2
siRNAs to deplete SphK1 in SK-Hep1 and HCCLM3 cells, and the
effectiveness was analyzed by qRT-PCR and western blot analysis
(Fig. 3A-B). At the same time, SphK1
activity was also decrease in SK-Hep1 and HCCLM3 cells (Fig. 3C). We then explored whether
decreasing SphK1 activity affects SK-Hep1 and HCCLM3 cells survival
and sensitivity to oxaliplatin. Compared with mock-transfected
cells, both SK-Hep1 and HCCLM3 cells were less resistant to
oxaliplatin by a cell survival assay analysis (Fig. 3D-E). Similarly, clonogenic survival
of SK-Hep1 and HCCLM3 cells was also significantly compromised in
SphK1-depleting cells (Fig. 3F).
These results indicate that downregulation of SphK1 make HCC cells
sensitive to oxaliplatin-induced cell death.
SphK1 promotes HCC cells survival
through activating Akt/GSK3β signaling pathway
High levels of SphK1 have been related to
chemotherapy drug resistance in certain cancers, which are closely
related with maintaining cancer signaling pathways (17,18).
Previous studies have shown that inhibition of SphK1 contributes to
apoptosis by Akt signaling pathways (19,20).
Therefore, we speculated whether SphK1 affect HCC cells survival in
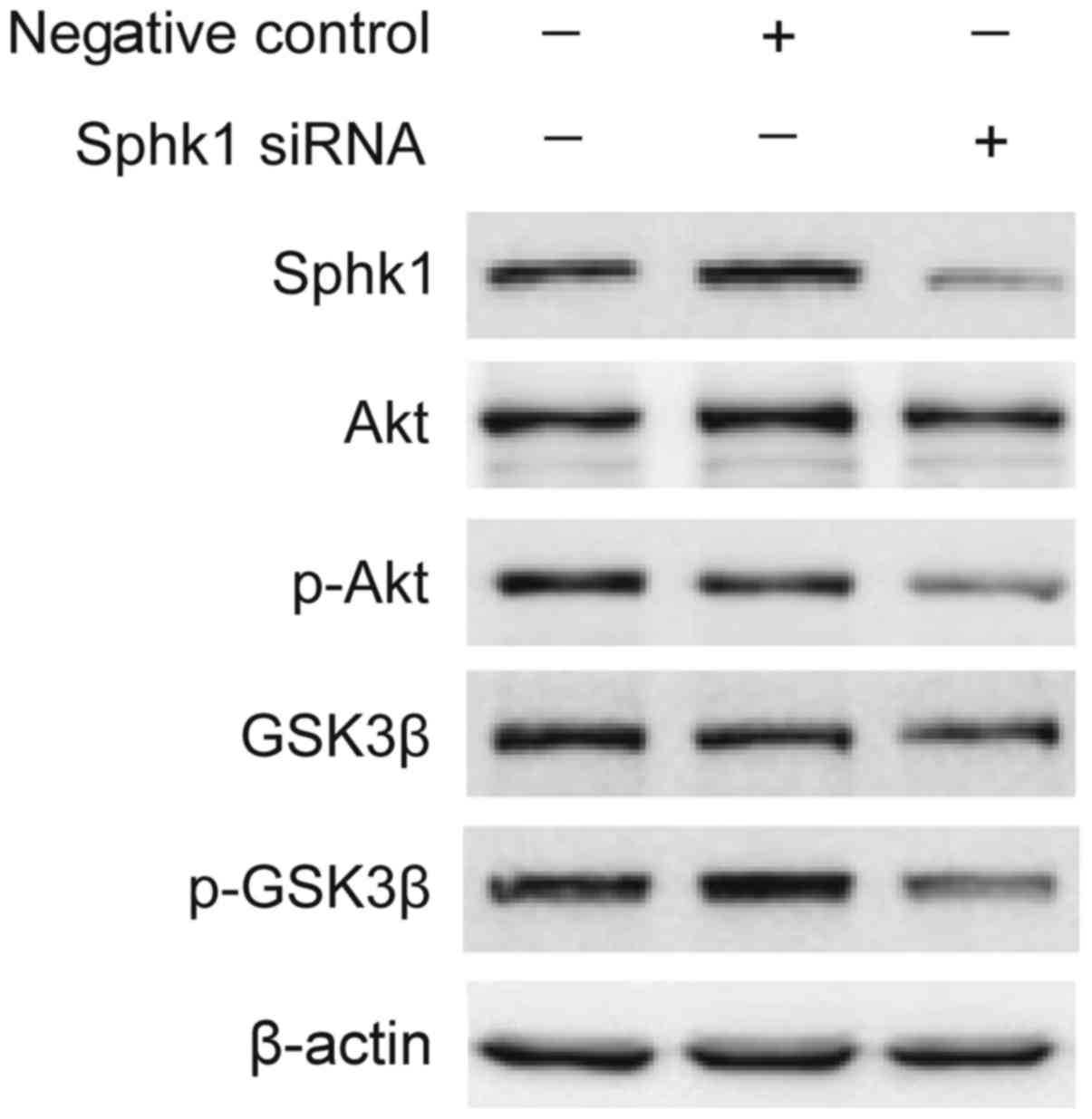
oxaliplatin through Akt signaling pathways. As shown in Fig. 4, knockdown of SphK1 markedly
suppressed the phosphorylations of Akt and GSK3β in SK-Hep1 cells,
while there was no signicantly reduction in the level of Akt and
GSK3β protein. Therefore, we speculate that SphK1 promotes HCC
cells survival in oxaliplatin through activating Akt/GSK3β
signaling pathway.
Discussion
HCC is characterized as a highly chemoresistant
cancer with no effective systemic therapy. Although patients have
been given surgical or locoregional therapies, they often have poor
prognosis because of high tumor recurrence or tumor progression.
Therefore, identifying biological markers associated with the
resistance of HCC is important for providing the scientific
rationale for the development of new therapeutic targets.
Sphingolipid metabolites are ceramide, sphingosine,
ceramide-1-phosphate (C1P), and sphingosine-1-phosphate (S1P)
(9). These lipid mediators involved
in cells life as bioactive signaling molecules, for example,
ceramide and sphingosine can active the apoptosis pathways whereas
S1P and C1P primarily exert mitogenic effects (21,22).
Therefore, the balance between lipid mediators has been considered
as a converter determining the biological processes of cells
(21,22). SphK1, as one of the key enzymes,
regulates the ceramide/S1P conversion that contributes to
determining whether a cell undergoes apoptosis or proliferates
(17,23). Previous studies found that SphK1 is
elevated in various types of cancers, functioning as an oncogene in
tumorigenesis, however, little is known about the role of SphK1 in
HCC progression. In the present study, we found that SphK1 is
upregulated in HCC and that high expression of SphK1 significantly
associated with poorer overall survival in HCC patients.
Overexpression of SphK1 may confer the development
of resistance to chemotherapeutics, whereas disruption of SphK1/S1P
signaling may restore or improve chemotherapy sensitivity (24). For example, raised SphK1 has been
associated with chronic myeloid leukemia resistance to imatinib and
pancreatic cancer cell resistance to gemcitabine (16,25).
Conversely, SphK1 inhibition can sensitize CML and pancreatic
cancer cells to the proapoptotic effects of gemcitabine and
imatinib, respectively, apparently by increasing the ceramide/S1P
ratio and thereby enabling C18-ceramide-dependent apoptosis to
proceed (16,25). Moreover, blockade of the SphK1
signaling pathway may reprogram cellular responsiveness to
tamoxifen and abrogate antiestrogen resistance in human breast
cancer cells (14). Consistent with
previous studies, our results found that knockdown of SphK1 by
siRNA markedly surpressed the ability of resistant oxaliplatin to
in HCC cells.
Although our evidence has highlighted the
oxaliplatin resistence effect of SphK1 in HCC, the molecular
mechanism is still unclear. To explore the mechanisms underlying
the resistence effects of SphK1, we tested the effect of SphK1 on
the AKT/GSK3β pathways, which have been demonstrated to be involved
in tumor progression (24). We have
demonstrated in the present study that p-Akt and p-GSK3β was
downregulated in SphK1-knocked down HCC cells, suggested Akt/GSK3β
activation through SphK1 signaling is associated with oxaliplatin
resistance in HCC cells. However, the molecular mechanism
underlying SphK1-mediated activation of Akt/GSK3β pathways, as well
as its biological outcome, needs to be further delineated. In
addition, several other issues also remain to be addressed. For
example, it would be very meaningful to know whether other pathways
are also involved in mediating the resistant effect of SphK1 in HCC
cells, and what other malignant phenotypes of HCC cells could also
be modulated by upregulated SphK1. These issues are under further
investigation in the laboratory. Nevertheless, understanding the
role of SphK1 in HCC progression will not only advance our
knowledge of the mechanisms underlying HCC survival, but also will
help establish SPHK1 as a potential therapeutic target for the
treatment of HCC.
References
|
1
|
Stewart B and Wild CP: World Health
OrganizationWorld cancer report 2014. IARC Press; Lyon: 2003
|
|
2
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang JH, Wang CC, Hung CH, Chen CL and Lu
SN: Survival comparison between surgical resection and
radiofrequency ablation for patients in BCLC very early/early stage
hepatocellular carcinoma. J Hepatol. 56:412–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Szakács G, Paterson JK, Ludwig JA,
Booth-Genthe C and Gottesman MM: Targeting multidrug resistance in
cancer. Nat Rev Drug Discov. 5:219–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomas MB, O'Beirne JP, Furuse J, Chan AT,
Abou-Alfa G and Johnson P: Systemic therapy for hepatocellular
carcinoma: Cytotoxic chemotherapy, targeted therapy and
immunotherapy. Ann Surg Oncol. 15:1008–1014. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uzma A and Tim M: Are there opportunities
for chemotherapy in the treatment of hepatocellular cancer? J
Hepatol. 56:686–695. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alcindor T and Beauger N: Oxaliplatin: A
review in the era of molecularly targeted therapy. Curr Oncol.
18:18–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du H, Yang W, Chen L, Shi M, Seewoo V,
Wang J, Lin A, Liu Z and Qiu W: Role of autophagy in resistance to
oxaliplatin in hepatocellular carcinoma cells. Oncol Rep.
27:143–150. 2012.PubMed/NCBI
|
|
9
|
Alemany R, van Koppen CJ, Danneberg K, Ter
Braak M, Meyer Zu and Heringdorf D: Regulation and functional roles
of sphingosine kinases. Naunyn Schmiedebergs Arch Pharmacol.
374:413–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pyne NJ and Pyne S: Sphingosine
1-phosphate and cancer. Nat Rev Cancer. 10:489–503. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Heffernan-Stroud LA and Obeid LM:
Sphingosine kinase 1 in cancer. Adv Cancer Res. 117:201–235. 2012.
View Article : Google Scholar
|
|
12
|
Van Brocklyn JR, Jackson CA, Pearl DK,
Kotur MS, Snyder PJ and Prior TW: Sphingosine kinase-1 expression
correlates with poor survival of patients with glioblastoma
multiforme: Roles of sphingosine kinase isoforms in growth of
glioblastoma cell lines. J Neuropathol Exp Neurol. 64:695–705.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruckhäberle E, Rody A, Engels K, Gaetje R,
von Minckwitz G, Schiffmann S, Grösch S, Geisslinger G, Holtrich U,
Karn T and Kaufmann M: Microarray analysis of altered sphingolipid
metabolism reveals prognostic significance of sphingosine kinase 1
in breast cancer. Breast Cancer Res Treat. 112:41–52. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sukocheva O, Wang L, Verrier E, Vadas MA
and Xia P: Restoring endocrine response in breast cancer cells by
inhibition of the sphingosine kinase-1 signaling pathway.
Endocrinology. 150:4484–4492. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rosa R, Marciano R, Malapelle U, Formisano
L, Nappi L, D'Amato C, D'Amato V, Damiano V, Marfè G, Del Vecchio
S, et al: Sphingosine kinase 1 overexpression contributes to
cetuximab resistance in human colorectal cancer models. Clin Cancer
Res. 19:138–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guillermet-Guibert J, Davenne L,
Pchejetski D, Saint-Laurent N, Brizuela L, Guilbeau-Frugier C,
Delisle MB, Cuvillier O, Susini C and Bousquet C: Targeting the
sphingolipid metabolism to defeat pancreatic cancer cell resistance
to the chemotherapeutic gemcitabine drug. Mol Cancer Ther.
8:809–820. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maceyka M, Harikumar KB, Milstien S and
Spiegel S: Sphingosine-1-phosphate signaling and its role in
disease. Trends Cell Biol. 22:50–60. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kunkel GT, Maceyka M, Milstien S and
Spiegel S: Targeting the sphingosine-1-phosphate axis in cancer,
inflammation and beyond. Nat Rev Drug Discov. 12:688–702. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song L, Xiong H, Li J, Liao W, Wang L, Wu
J and Li M: Sphingosine kinase-1 enhances resistance to apoptosis
through activation of PI3K/Akt/NF-κB pathway in human non-small
cell lung cancer. Clin Cancer Res. 17:1839–1849. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji F, Mao L, Liu Y, Cao X, Xie Y, Wang S
and Fei H: K6PC-5, a novel sphingosine kinase 1 (SphK1) activator,
alleviates dexamethasone-induced damages to osteoblasts through
activating SphK1-Akt signaling. Biochem Biophys Res Commun.
458:568–575. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ponnusamy S, Meyers-Needham M, Senkal CE,
Saddoughi SA, Sentelle D, Selvam SP, Salas A and Ogretmen B:
Sphingolipids and cancer: Ceramide and sphingosine-1-phosphate in
the regulation of cell death and drug resistance. Future Oncol.
6:1603–1624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Patwardhan GA and Liu YY: Sphingolipids
and expression regulation of genes in cancer. Prog Lipid Res.
50:104–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rosen H, Gonzalez-Cabrera PJ, Sanna MG and
Brown S: Sphingosine 1-phosphate receptor signaling. Annu Rev
Biochem. 78:743–768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ader I, Malavaud B and Cuvillier O: When
the sphingosine kinase 1/sphingosine 1-phosphate pathway meets
hypoxia signaling: New targets for cancer therapy. Cancer Res.
69:3723–3726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marfe G, Di Stefano C, Gambacurta A,
Ottone T, Martini V, Abruzzese E, Mologni L, Sinibaldi-Salimei P,
de Fabritis P, Gambacorti-Passerini C, et al: Sphingosine kinase 1
overexpression is regulated by signaling through PI3K, AKT2, and
mTOR in imatinib-resistant chronic myeloid leukemia cells. Exp
Hematol. 39:653–665.e6. 2011. View Article : Google Scholar : PubMed/NCBI
|