Introduction
Ischemic stroke (IS) occurs due to a lack of blood
flow to the brain, which is induced by various risk factors,
including high blood pressure, tobacco smoking, alcohol drinking,
obesity, high blood cholesterol and diabetes mellitus, and results
in cell death and improper functioning of part of the brain
(1,2). IS patients always have symptoms
including inability to move or inconvenience on one side of the
body (3). Techniques, such as a
neurological examination, CT scans, MRI scans and Doppler
ultrasound, have been used for diagnosis and determining the type
and causes of IS (4,5). Current studies found that blood tests
and gene detection may help to find the potential causes of IS.
Many genes are closely associated with IS disease.
For instance, phosphodiesterase 4D (PDE4D) participated in
atherosclerosis, which is a primary pathological process for IS
disease (6). In addition, transfer
of the Kallikrein gene was confirmed to inhibit apoptosis and
promote glial cell migration, and then protect against IS (7). Liu et al (8) confirmed that polymorphisms of heat
shock protein 70 may increase the risk of IS in smoking patients.
In Caucasians and northern Han Chinese, the single-nucleotide
polymorphisms of interleukin-1 (IL-1) and PDE4D were also confirmed
to increase the IS risk (9).
Especially in young women and adults, IS risk and
cigarette smoking exhibited a strong dose-response relationship
(10). In addition, heavy and
light-moderate drinking exerted different effects on IS (11). Although cigarette smoking and alcohol
drinking had a significant influence on IS, to the best of our
knowledge, very little research has been done in the field of
related molecule mechanisms. The biomarkers and related pathways of
IS disease have been investigated through bioinformatics analysis
(12). Based on this foundation, the
aim of the present study was to focus on the special influence of
cigarette smoking and alcohol drinking on gene expression in IS
samples. Blood genomic expression profile was used to screen
differentially expressed genes of IS specially induced by smoking
and drinking history, and related functions and pathways were
investigated.
Materials and methods
DEG screening of smoking or drinking
induced-IS
The expression profile of E-GEOD-22255 was obtained
from ArrayExpress archive (http://www.ebi.ac.uk/arrayexpress/), which was
deposited by Krug et al (13). This profile contained 40 samples,
including 20 IS samples (7 IS samples without smoking or drinking
history and 13 IS samples with smoking or drinking history) and 20
controls (9 normal controls and 11 controls with smoking or
drinking history). The platform of this chip was GPL570
(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 array. The
expression value of probe sets was calculated by algorithm of
robust multi-chip average (RMA) in three steps: Background
correction, standardization and summarizing (14). According to the information of the
Affymetrix official website, these processed probe sets were
annotated and included with the threshold of >0.8 median NUSE
<1.2 and >-0.25 median RLE <0.25. Thus, NUSE was the value
of normalized unscaled standard errors, while RLE was the value of
relative log expression.
Significance analysis of microarray (SAM) was used
to construct a d score for each gene to measure the correlation
degree between of gene expression and grouping (15). In this process, exchangeable factors
were obtained by calculating the mean absolute deviation. The
statistical value of each gene was calculated using the formula:
di = ri/(si +
s0).
Thus, ri reflected the difference
of genes in average level, and si reflected
overall change of samples.
Then, >1,000 permutations were applied to
simulate of the distribution of d score. In this study, this method
was used on samples to screen the two groups of DEGs: IS samples
without smoking or drinking history compared with normal controls,
IS samples with smoking or drinking history compared with controls
with smoking or drinking history. Multiple test was used to
calculate P-value of each gene.
To screen IS-related DEGs induced by smoking and
drinking history, DEGs of samples with smoking or drinking history
were deducted by DEGs of samples without smoking or drinking
history based on the same gene symbols.
Functional and pathway enrichment
analysis
Gene ontology (GO) analysis is a method used to
select the significant functions of gene groups based on the GO
database (16), and the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database is a strong tool
for biological metabolic analysis and metabolic network research
(17). In this study, these
databases were used for functional and pathway enrichment analysis
of smoking- or drinking-induced DEGs. To define the results more
precisely, Fisher's exact test and multiple comparative test were
used to calculate the P-value and FDR value, respectively.
Construction of pathway relationship
network
The KEGG database was also used to construct a
pathway relationship network. This network was able to show the
signal transduction relationship between significant pathways, and
importantly, upstream and downstream signal pathways were also
identified.
Construction of gene co-expression
network
Key DEGs induced by smoking and drinking history
were obtained by inserting DEGs in significant functions and
pathways. Based on the expression value of genes, the gene
co-expression network was constructed and analyzed. Genes with mean
connectedness of >1 were involved.
Results
DEGs screening
Compared with normal controls, 128 DEGs were
screened in IS samples without smoking or drinking history. At the
same time, 465 DEGs induced by smoking and drinking and other
factors were obtained. DEGs specially induced by smoking and
drinking were screened based on the same gene symbol, and 319
special DEGs were obtained.
Functional and pathway enrichment
analysis
These special DEGs were enriched in different
functions, including inflammatory response (FDR=2.39E-17), immune
response (FDR=1.71E-12), apoptotic process (FDR=7.42E-12) and
negative regulation of apoptotic process (FDR=3.91E-09) (Table I). Simultaneously, these DEGs also
participated in various pathways, such as nuclear factor-κB (NF-κB)
signaling pathway (FDR=1.77E-12), influenza A (FDR=1.90E-11),
legionellosis (FDR=5.96E-11) and NOD-like receptor signaling
pathway (FDR=6.53E-11) (Table
II).
 | Table I.Top 10 functions enriched by special
DEGs. |
Table I.
Top 10 functions enriched by special
DEGs.
| GO ID | GO name | Diff gene counts in
GO | Enrichment score | P-value | FDR |
|---|
| GO:0006954 | Inflammatory
response | 24 | 14.50814 | 1.83E-20 | 2.39E-17 |
| GO:0006955 | Immune response | 21 | 10.66927 | 2.62E-15 | 1.71E-12 |
| GO:0006915 | Apoptotic
process | 26 | 7.089543 | 1.70E-14 | 7.42E-12 |
| GO:0043066 | Negative regulation
of apoptotic process | 20 | 7.368978 | 1.20E-11 | 3.91E-09 |
| GO:0008285 | Negative regulation
of cell proliferation | 17 | 8.46815 | 5.63E-11 | 1.47E-08 |
| GO:0045429 | Positive regulation
of nitric oxide biosynthetic process | 7 | 41.61016 | 5.84E-10 | 1.27E-07 |
| GO:0045087 | Innate immune
response | 19 | 6.115986 | 9.50E-10 | 1.77E-07 |
| GO:0002237 | Response to
molecule of bacterial origin | 5 | 99.07182 | 1.32E-09 | 2.15E-07 |
| GO:0045944 | Positive regulation
of transcription from RNA polymerase II promoter | 21 | 5.289427 | 1.54E-09 | 2.24E-07 |
| GO:0006935 | Chemotaxis | 10 | 15.37321 | 2.48E-09 | 3.04E-07 |
 | Table II.Top 10 pathways enriched by special
DEGs. |
Table II.
Top 10 pathways enriched by special
DEGs.
| Pathway ID | Pathway name | Different gene
counts in pathway | Gene amount in
pathway | Enrichment
score | P-value | FDR |
|---|
| 4064 | NF-κB signaling
pathway | 13 | 92 | 25.1987 | 1.19E-14 | 1.77E-12 |
| 5164 | Influenza A | 15 | 179 | 14.94379 | 2.55E-13 | 1.90E-11 |
| 5134 | Legionellosis | 10 | 55 | 32.4235 | 1.20E-12 | 5.96E-11 |
| 4621 | NOD-like receptor
signaling pathway | 10 | 57 | 31.28584 | 1.75E-12 | 6.53E-11 |
| 5132 | Salmonella
infection | 11 | 88 | 22.29116 | 6.03E-12 | 1.68E-10 |
| 4060 | Cytokine-cytokine
receptor interaction | 16 | 267 | 10.6864 | 6.78E-12 | 1.68E-10 |
| 4380 | Osteoclast
differentiation | 12 | 135 | 15.85149 | 3.72E-11 | 7.91E-10 |
| 4062 | Chemokine signaling
pathway | 13 | 192 | 12.07438 | 1.66E-10 | 3.09E-09 |
| 5323 | Rheumatoid
arthritis | 10 | 94 | 18.9712 | 3.06E-10 | 5.07E-09 |
| 5142 | Chagas disease
(American trypanosomiasis) | 10 | 105 | 16.98374 | 9.26E-10 | 1.29E-08 |
Construction of pathway relationship
network
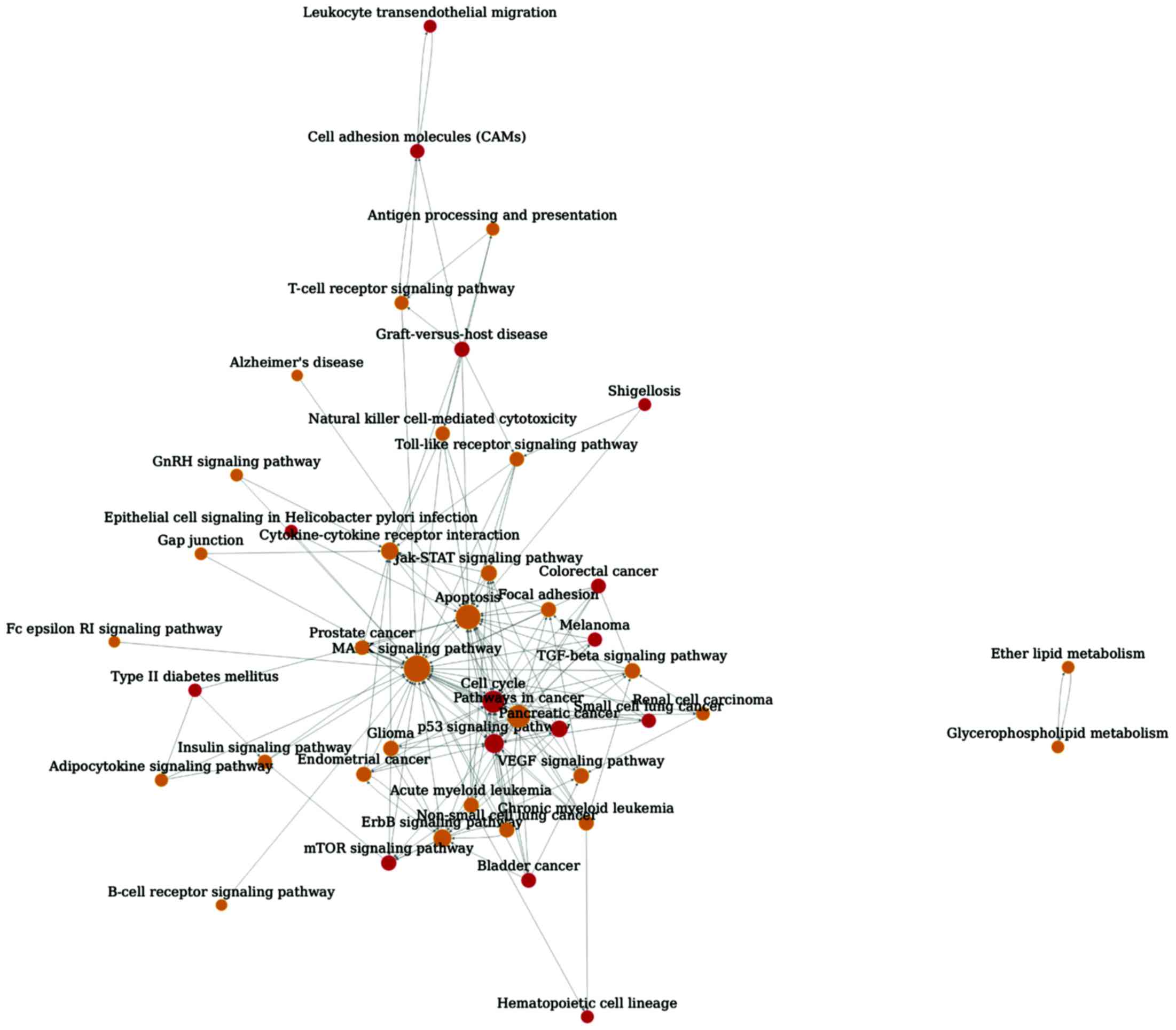
The pathway relationship network with 44 nodes and
161 edges were constructed (Fig. 1).
In this network, the hub nodes were MAPK signaling pathway
(degree=32), apoptosis (degree=27) and pathways in cancer
(degree=23). Of note, pathways in cancer (out-degree=23,
in-degree=0) and cytokine-cytokine receptor interaction
(out-degree=0, in-degree=12) were the upstream and downstream
pathway, respectively.
Construction of gene co-expression
network
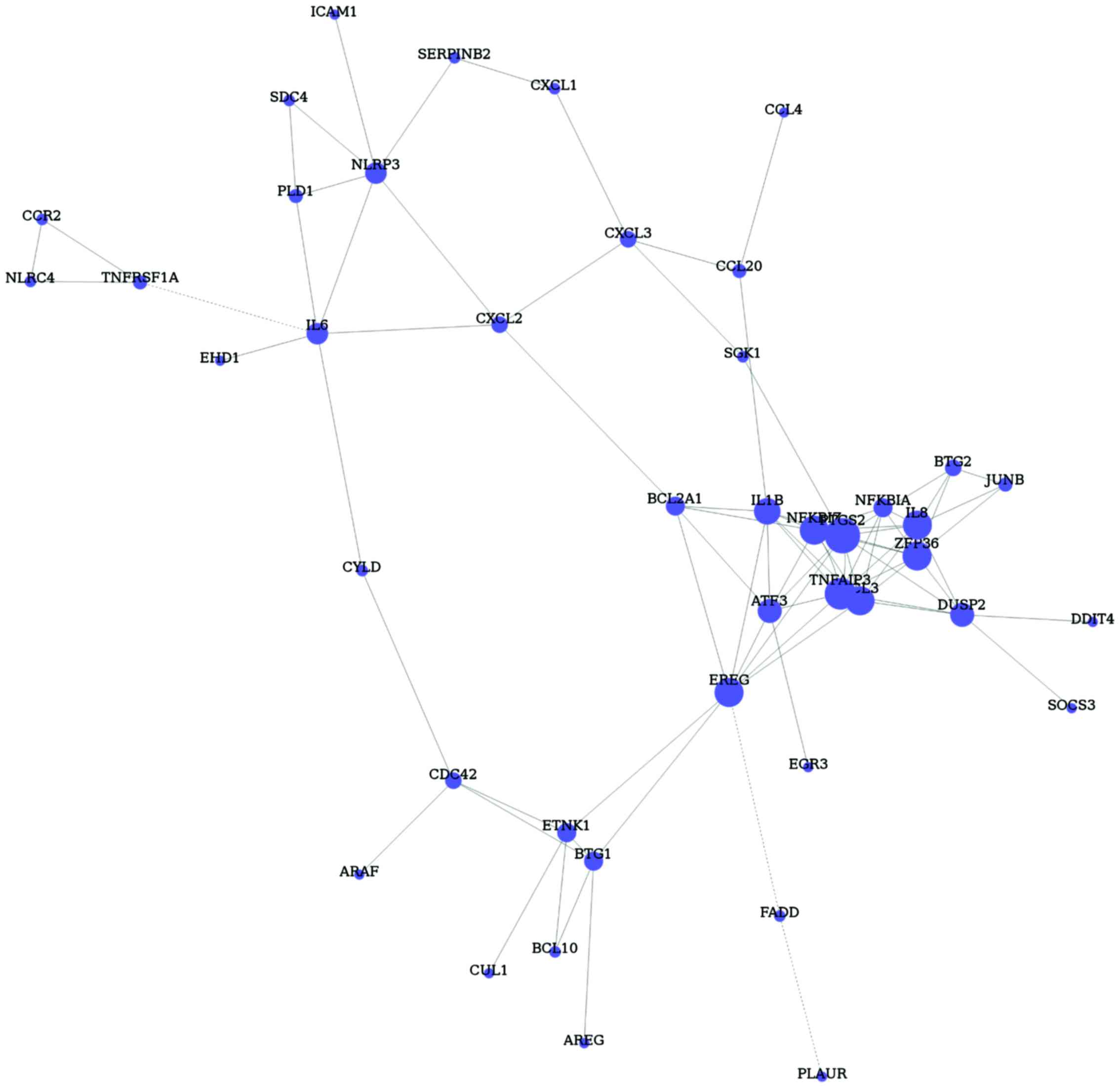
The DEGs in GO terms and pathways were inserted and
87 key genes were obtained (Fig. 2).
According to the mean connectedness of these genes, the gene
co-expression network was constructed with 43 nodes and 88 edges.
The hub nodes were prostaglandin-endoperoxide synthase 2 (PTGS2,
degree=11), TNF-α-induced protein 3 (TNFAIP3, degree=10), ZFP36
ring finger protein (ZFP36, degree=9) and NFKB inhibitor ζ (NFKBIZ,
degree=9). Importantly, these four nodes were found to have
positive relationships.
Discussion
Smoking and alcohol drinking were confirmed to be
risk factors for IS. However, the causes of smoking and alcohol
drinking induced IS are still unknown (11,18). In
this study, various factors of IS except smoking and alcohol
drinking were excluded. Importantly, several key DEGs were
screened, including PTGS2, TNFAIP3, ZFP36 and
NFKBIZ.
PTGS2, an isozyme of
prostaglandin-endoperoxide synthase (PTGS), was a critical enzyme
in prostaglandin biosynthesis (19).
Kunze et al (5) confirmed
that in the environment with smoking, ΔNp63 could bind to PTGS2
promoter, and further influence the process of inflammatory
response. Moreover, a case control study showed that smoking and
alcohol drinking induced substantial differences between esophageal
squamous cell carcinoma samples and controls (20). The overexpression of PTGS2 also
played an important role in the pathogenesis of IS. In
African-Americans with the variant of G-765C allele of PTGS2, IS
was found with a significantly higher incidence rate (21). As shown in this study, PTGS2 was
enriched in various functions and pathways, including inflammatory
response, negative regulation of cell proliferation, as well as the
NF-κB and VEGF signaling pathways. Moreover, the NF-κB signaling
pathway was confirmed to activate the synthesis of inducible PTGS2
in the brain (22). Liu et al
(23) found that electroacupuncture
could target the NF-κB signaling pathway and inhibit inflammatory
injury for IS. Thus, we inferred that PTGS2 is a potential
biomarker for smoking- and drinking-induced IS disease by
participating in inflammatory response and the NF-κB signaling
pathway.
Furthermore, TNFAIP3 and PTGS2 were
positively associated in this study. The expression of TNFAIPS was
always induced by tumor necrosis factor. In 2010, Lodolce et
al (24) confirmed that TNFAIP3
was a ubiquitin-modifying enzyme, and genetic polymorphisms of
TNFAIP2 were also found to affect the autoimmunity regulation.
Under inflammatory conditions, TNFAIP3 was a negative-feedback
regulator of NF-κB activation (25).
Furthermore, TNFAIP was confirmed to be a negative regulatory of
Toll-like receptor signaling pathway, which could lead to
inflammatory effects (26).
Similarly in this study, TNFAIP3 was involved in inflammatory,
innate immune response, NF-κB signaling pathway and NOD-like
receptor signaling pathway.
ZFP36 was also screened with a higher degree
in the gene co-expression network, and enriched in negative
regulation of transcription from RNA polymerase II, response to
stress and vasculogenesis. In a previous study (27) ZFP36 was found to be a critical gene
for obesity-related metabolic complications by detecting the
cholesterol level and omental adipose tissue ZFP36 mRNA levels.
Furthermore, decreased regulation of the metabolic syndrome may
significantly affect the prevalence of stroke and related
disability (28). As shown in a
study by Kurl et al who conducted an exercise stress test,
an increase in systolic blood pressure was independently associated
with the risk of IS and other types of stroke (29). In addition, the stress of acute
ischemic cerebral insults may increase secretion of stress
hyperglycemia, and further induce hyperglycemia following stroke
(30). Nicotine in cigarette was
also confirmed to promote vasculogenesis by affecting endothelial
progenitor cells (31). Cerebral
ischemic stroke could also be treated by stem cell transplantation
and angiogenesis (32).
NFKBIZ, another important key DEG, is a
member of the ankyrin-repeat family, induced by lipopolysaccharide
(33). It is known to participate in
inflammatory responses to lipopolysaccharide by interacting with
NF-B proteins through ankyrin-repeat domains (34). Similarly with the results in the
present study, NFKBIZ was involved in inflammatory response,
transcription, DNA-dependent, and transcriptional misregulation in
cancer. As known, early inflammatory response may potentiate
ischemic injury, whereas late response may induce contrary function
in stroke. In acute experimental stroke,
phosphatidylinositol-3-kinase inhibitors were found to suppress the
activation of DNA-dependent protein kinases, which played a
critical role in blood-brain barrier dysfunction (35). Thus, NFKBIZ was inferred to be
a key IS-related gene.
In conclusion, the screened DEGs, such as PTGS2,
TNFAIP3, ZFP36 and NFKBIZ, are potential biomarkers of
smoking- and drinking-induced IS disease, participating in various
functions, such as inflammatory response. Thus, giving up alcohol
and tobacco, and detecting the abovementioned biomarkers is of
significance for IS prevention and treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZM was reponsible for DEG screening. MG, SZ and XS
helped with construction of pathway relationship network. FW and HL
participated in data analysis. ZM and ZC contributed to
construction of gene co-expression network. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Collins P: Risk factors for cardiovascular
disease and hormone therapy in women. Heart. 92 Suppl
3:iii24–iii28. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Urban PP, Wicht S, Vukurevic G, Fitzek C,
Fitzek S, Stoeter P, Massinger C and Hopf HC: Dysarthria in acute
ischemic stroke: lesion topography, clinicoradiologic correlation,
and etiology. Neurology. 56:1021–1027. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen WC, Cheng HC and Datsen GW:
Pharmaceutical composition for preventing/treating brain injury and
enhancing recovery of sequelae and manufacture thereof. US Patent
US 20150290272 A1. Filed April 9, 2015; issued October 15.
2015.
|
|
4
|
Wong CB and Wong JC: A novel method to
quantify carotid artery stenosis by Doppler ultrasound: Using the
continuity principle. Int J Angiol. 19:e86–e90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kunze E, Pham M, Raslan F, Stetter C, Lee
JY, Solymosi L, Ernestus RI, Vince GH and Westermaier T: Value of
perfusion CT, transcranial doppler sonography, and neurological
examination to detect delayed vasospasm after aneurysmal
subarachnoid hemorrhage. Radiol Res Pract.
2012:2312062012.PubMed/NCBI
|
|
6
|
Gretarsdottir S, Thorleifsson G,
Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T,
Gudmundsdottir T, Bjarnadottir SM, Einarsson OB, Gudjonsdottir HM,
et al: The gene encoding phosphodiesterase 4D confers risk of
ischemic stroke. Nat Genet. 35:131–138. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia CF, Yin H, Borlongan CV, Chao L and
Chao J: Kallikrein gene transfer protects against ischemic stroke
by promoting glial cell migration and inhibiting apoptosis.
Hypertension. 43:452–459. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu J, Cheng J, Peng J, Han S, Yu L and
Nie S: Effects of polymorphisms of heat shock protein 70 gene on
ischemic stroke, and interaction with smoking in China. Clin Chim
Acta. 384:64–68. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li N, He Z, Xu J, Liu F, Deng S and Zhang
H: Association of PDE4D and IL-1 gene polymorphism with ischemic
stroke in a Han Chinese population. Brain Res Bull. 81:38–42. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhat VM, Cole JW, Sorkin JD, Wozniak MA,
Malarcher AM, Giles WH, Stern BJ and Kittner SJ: Dose-response
relationship between cigarette smoking and risk of ischemic stroke
in young women. Stroke. 39:2439–2443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Klatsky AL, Armstrong MA, Friedman GD and
Sidney S: Alcohol drinking and risk of hospitalization for ischemic
stroke. Am J Cardiol. 88:703–706. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barr TL, Conley Y, Ding J, Dillman A,
Warach S, Singleton A and Matarin M: Genomic biomarkers and
cellular pathways of ischemic stroke by RNA gene expression
profiling. Neurology. 75:1009–1014. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krug T, Gabriel JP, Taipa R, Fonseca BV,
Domingues-Montanari S, Fernandez-Cadenas I, Manso H, Gouveia LO,
Sobral J, Albergaria I, et al: TTC7B emerges as a novel risk factor
for ischemic stroke through the convergence of several genome-wide
approaches. J Cereb Blood Flow Metab. 32:1061–1072. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smita S, Katiyar A, Pandey DM, Chinnusamy
V, Archak S and Bansal KC: Identification of conserved drought
stress responsive gene-network across tissues and developmental
stages in rice. Bioinformation. 9:72–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kutalik Z, Inwald J, Gordon SV, Hewinson
RG, Butcher P, Hinds J, Cho KH and Wolkenhauer O: Advanced
significance analysis of microarray data based on weighted
resampling: A comparative study and application to gene deletions
in Mycobacterium bovis. Bioinformatics. 20:357–363. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blake JA and Harris MA: The Gene Ontology
(GO) project: Structured vocabularies for molecular biology and
their application to genome and expression analysis. Curr Protoc
Bioinformatics: Chapter 7. Unit 7.2. 2008. View Article : Google Scholar
|
|
17
|
Jing LS, Shah FF, Mohamad MS, Hamran NL,
Salleh AH, Deris S and Alashwal H: Database and tools for metabolic
network analysis. Biotechnol Bioprocess Eng. 19:568–585. 2014.
View Article : Google Scholar
|
|
18
|
Shah RS and Cole JW: Smoking and stroke:
The more you smoke the more you stroke. Expert Rev Cardiovasc Ther.
8:917–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosaka T, Miyata A, Ihara H, Hara S,
Sugimoto T, Takeda O, Takahashi E and Tanabe T: Characterization of
the human gene (PTGS2) encoding prostaglandin-endoperoxide synthase
2. Eur J Biochem. 221:889–897. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu HM, Kuo CH, Lee CH, Wu IC, Lee KW, Lee
JM, Goan YG, Chou SH, Kao EL, Wu MT, et al: Polymorphism in COX-2
modifies the inverse association between Helicobacter pylori
seropositivity and esophageal squamous cell carcinoma risk in
Taiwan: A case control study. BMC Gastroenterol. 9:372009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeldin D: The G-765C promoter polymorphism
in cyclooxygenase-2 (PTGS2), aspirin utilization and cardiovascular
disease risk: The Atherosclerosis Risk in Communities (ARIC) study.
Pharmacotherapy. 26:e872006.
|
|
22
|
Nadjar A, Tridon V, May MJ, Ghosh S,
Dantzer R, Amédée T and Parnet P: NFkappaB activates in vivo the
synthesis of inducible Cox-2 in the brain. J Cereb Blood Flow
Metab. 25:1047–1059. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu W, Wang X, Zheng Y, Shang G, Huang J,
Tao J and Chen L: Electroacupuncture inhibits inflammatory injury
by targeting the miR-9-mediated NFκB signaling pathway following
ischemic stroke. Mol Med Rep. 13:1618–1626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lodolce JP, Kolodziej LE, Rhee L, Kariuki
SN, Franek BS, McGreal NM, Logsdon MF, Bartulis SJ, Perera MA,
Ellis NA, et al: African-derived genetic polymorphisms in TNFAIP3
mediate risk for autoimmunity. J Immunol. 184:7001–7009. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vereecke L, Beyaert R and van Loo G:
Genetic relationships between A20/TNFAIP3, chronic inflammation and
autoimmune disease. Biochem Soc Trans. 39:1086–1091. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hung YY, Lin CC, Kang HY and Huang TL:
TNFAIP3, a negative regulator of the TLR signaling pathway, is a
potential predictive biomarker of response to antidepressant
treatment in major depressive disorder. Brain Behav Immun.
59:265–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bouchard L, Tchernof A, Deshaies Y,
Marceau S, Lescelleur O, Biron S and Vohl MC: ZFP36: A promising
candidate gene for obesity-related metabolic complications
identified by converging genomics. Obes Surg. 17:372–382. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seet RC, Zhang Y, Wijdicks EF and
Rabinstein AA: Impact of obesity and metabolic syndrome in stroke
patients undergoing intravenous thrombolysis. Stroke.
43:A1562012.
|
|
29
|
Kurl S, Laukkanen JA, Rauramaa R, Lakka
TA, Sivenius J and Salonen JT: Systolic blood pressure response to
exercise stress test and risk of stroke. Stroke. 32:2036–2041.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
O'Connell JE and Gray CS: The stress
response to acute stroke. Stress Health. 7:239–243. 1991.
|
|
31
|
Heeschen C, Chang E, Aicher A and Cooke
JP: Endothelial progenitor cells participate in nicotine-mediated
angiogenesis. J Am Coll Cardiol. 48:2553–2560. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wei L, Keogh CL, Whitaker VR, Theus MH and
Yu SP: Angiogenesis and stem cell transplantation as potential
treatments of cerebral ischemic stroke. Pathophysiology. 12:47–62.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oonuma T, Morimatsu M, Ochiai K, Iwanaga T
and Hashizume K: Role of NF-kappaB in constitutive expression of
MAIL in epidermal keratinocytes. J Vet Med Sci. 69:279–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ishiguro-Oonuma T, Ochiai K, Hashizume K,
Iwanaga T and Morimatsu M: NFKBIZ regulates the proliferation and
differentiation of keratinocytes. Jpn J Vet Res. 63:107–114.
2015.PubMed/NCBI
|
|
35
|
Jin R, Song Z, Yu S, Piazza A, Nanda A,
Penninger JM, Granger DN and Li G: Phosphatidylinositol-3-kinase
gamma plays a central role in blood-brain barrier dysfunction in
acute experimental stroke. Stroke. 42:2033–2044. 2011. View Article : Google Scholar : PubMed/NCBI
|