Introduction
Pancreatic ductal adenocarcinoma (PDAC) accounts for
>90% of all pancreatic cancer cases (1). It represents a severe health risk and
is the fourth leading cause of cancer-associated mortality in
developed countries (2). PDAC is a
highly aggressive malignancy with a poor prognosis; the five-year
survival rate is <5% (3). As PDAC
is often diagnosed at an advanced stage with metastasis, it is
often too late for patients to undergo curative surgery and
traditional chemotherapy is not an effective treatment strategy
(4). Therefore, it is critical to
understand the molecular mechanisms underlying the development and
progression of PDAC to enable the development of novel strategies
to inhibit tumor development, impede tumor growth and reduce the
recurrence rate of the disease.
In mammals, Notch is a highly conserved gene family
that includes the Notch1-4 receptors and their ligands: δ-like
ligand protein (DLL)1, DLL3, DLL4, Jagged1 and Jagged2 (5). The Notch signaling pathway consists of
Notch receptors, their ligands and C-promoter binding factor 1,
suppressor of hairless, Lag-1 (CSL), DNA binding proteins and
downstream target genes that are involved in regulating cell
functions, including proliferation, differentiation and apoptosis
(6,7). Binding of the Notch receptor to its
ligand in adjacent cells activates the Notch signaling pathway
(8). Notch receptor proteins are
sheared by proteolytic enzymes, releasing the C-terminal
intracellular domain (NICD), which then translocates into the
nucleus. In the nucleus, the NICD binds to CSL, changing CSL from a
transcriptional repressor to an activator (9). This leads to the activation of
downstream Notch target genes, including Hes and Hey family genes
(5). The Notch signaling pathway
regulates pancreatic cell differentiation in the developing
pancreas (6) and participates in the
development and progression of PDAC (10–13).
Although previous studies have described the activation of Notch
signaling components in PDAC (14–17), the
link between elevated Notch expression and tumorigenesis in PDAC is
controversial as contradictory results have been reported by
different studies. Mazur et al (11) demonstrated that Notch signaling has a
tumor promoting effect, whereas Hanlon et al (18) demonstrated that it had an inhibitory
tumor effect (11,18). In addition, the expression pattern of
the Notch receptors and ligands in PDAC remains unclear.
The high expression of a potential oncogene means
that it serves a significant role in cancer (17). Therefore, to elucidate the role of
Notch signaling in PDAC, in the current study, immunohistochemical
staining was performed on samples collected from 24 patients with
the necessary associated clinical data. Immunofluorescence staining
and western blot analysis were also performed to detect the
expression of Notch receptors and their ligands in the pancreatic
cancer human pancreatic adenocarcinoma (HPAC) and PANC-1 cell
lines.
Materials and methods
Cell lines
The PDAC cell lines HPAC and PANC-1 and the 293 T
cells and HeLa cell lines were all purchased from the cell bank of
the Chinese Academy of Sciences (Beijing, China). PANC-1 and 293
cells were maintained in Dulbecco's modified Eagle's medium
(DMEM)-high glucose (Hyclone™; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; Zhejiang Tianhang Biotechnology Co., Ltd., Huzhou, China), 1%
penicillin and 1% streptomycin (Beyotime Institute of
Biotechnology, Haimen, China). HPAC cells and HeLa cells were
maintained in RPMI-1640 (GE Healthcare Life Sciences) supplemented
with 10% FBS, 1% penicillin and 1% streptomycin. All cells were
maintained at 37°C in 5% CO2.
A total of 24 PDAC tissues were collected from
patients who underwent surgery for pancreatic cancer at the
Affiliated Center Hospital of Xinxiang Medical University
(Xinxiang, China) from May 2010 to July 2015. PDAC tissues were
then formalin-fixed (10% formalin for 24 h at room temperature) and
paraffin-embedded. The study protocol adhered to The Code of Ethics
of the World Medical Association (Declaration of Helsinki). The
present study was approved by the Ethics Committee of the
Affiliated Center Hospital of Xinxiang Medical University. Written
informed consent was obtained from all patients prior to the
procedure. Patient information is listed in Table I. PDAC tissues were confirmed using
histopathological analysis.
 | Table I.Patient information. |
Table I.
Patient information.
| No. | Age | Sex | TNM | Grade | Stage |
|---|
| 1 | 77 | F | T2N0M0 | 1 | I |
| 2 | 46 | F | T3N0M0 | 1 | II |
| 3 | 56 | F | T2N0M0 | 1 | I |
| 4 | 47 | F | T3N0M0 | 1 | II |
| 5 | 64 | F | T2N0M0 | 1 | I |
| 6 | 77 | M | T2N0M0 | 1 | I |
| 7 | 67 | F | T3N0M0 | 1 | III |
| 8 | 50 | M | T3N0M0 | 1 | II |
| 9 | 48 | F | T3N0M0 | 1 | II |
| 10 | 77 | M | T3N0M0 | 1 | II |
| 11 | 65 | M | T2N0M0 | 1 | I |
| 12 | 47 | M | T3N0M0 | 1 | II |
| 13 | 61 | M | T2N0M0 | 1 | I |
| 14 | 65 | M | T3N0M0 | 2 | II |
| 15 | 57 | M | T3N0M0 | 2 | II |
| 16 | 38 | M | T3N0M1 | 2 | IV |
| 17 | 39 | M | T3N0M0 | 2 | II |
| 18 | 31 | M | T3N0M0 | 2 | II |
| 19 | 42 | M | T3N0M0 | 1 | II |
| 20 | 44 | M | T3N0M0 | 2 | II |
| 21 | 57 | M | T3N0M0 | 2 | II |
| 22 | 59 | M | T3N0M0 | 2 | II |
| 23 | 75 | F | T3N0M1 | 2 | IV |
| 24 | 52 | M | T1N0M0 | 2 | I |
Immunohistochemistry
For histological assessment, immunohistochemical
analysis was performed using 5-µm-thick PDAC tissue sections.
Xylene and graded alcohols were used for dewaxing and rehydration.
Subsequently, sections were treated with citrate salt buffer (pH
6.0) in a microwave for 15 min for antigen retrieval (100°C),
followed by incubation with 3% hydrogen peroxide for 15 min to
block endogenous peroxidase activity at room temperature. The
samples were blocked with 5% donkey blood serum (Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) in PBS for
1 h at room temperature. The primary antibodies used in the
experiments are listed in Table II.
Samples were incubated with the primary antibodies at 4°C
overnight, followed by incubation with secondary horseradish
peroxidase (HRP)-conjugated antibodies (cat. no. SP-9001; OriGene
Technologies, Inc., Beijing, China) for 1 h at room temperature.
Diaminobenzidine and hematoxylin were used for staining (20 sec)
and counterstaining (10 sec), respectively at room temperature.
Following dehydration with graded alcohols and xylene, slides were
sealed with coverslips and neutral gum. The negative control group
was incubated with PBS instead of the primary antibody. Staining
intensities were quantified by two pathologists blinded to the
sample group. The Video Pro32 color image analysis system was used,
using the Grey value and optical density value to analyze the
immunohistochemical positive expression strength. The intensity of
Notch receptors and ligands staining was scored using the following
scoring system: 0 (no appreciable staining; negative), 1 (barely
detectable staining; weak positive), 2 (readily identifiable brown
staining; positive) and 3 (dark brown staining; strong positive).
The total score was calculated by multiplying the percentage of
positive cells and the intensity score. A tumor sample was
considered positive if the score was ≥4 and negative otherwise.
 | Table II.Antibodies used within the study. |
Table II.
Antibodies used within the study.
|
|
| Dilution |
|
|
|---|
|
|
|
|
|
|
|---|
| Antigen | Host species | IHC | WB | Supplier | Cat. no. |
|---|
| Notch1 | Rabbit | 1:50 | 1:500 | SC | sc-6014R |
| Notch2 | Rabbit | 1:500 | 1:2,000 | LS | LS-B399 |
| Notch3 | Rabbit | 1:50 | 1:500 | SC | sc-5593 |
| Notch4 | Rabbit | 1:50 | 1:500 | SC | sc-5594 |
| Jagged1 | Rabbit | 1:50 | 1:500 | SC | sc-8303 |
| Jagged2 | Rabbit | 1:50 | 1:500 | SC | sc-5604 |
| DLL1 | Rabbit | 1:50 | 1:500 | Ab | ab76655 |
| DLL3 | Rabbit | 1:100 | 1:1,000 | CS | 2483s |
| DLL4 | Rabbit | 1:50 | 1:1,000 | BR | HP1274 |
Immunofluorescence
Cell lines from adherent cultures were digested
using 0.25% trypsin with EDTA at 37°C for 8 min and centrifuged at
180 × g for 4 min at room temperature. The cell pellet was
resuspended in complete DMEM-high glucose (10% FBS, 1% penicillin
and 1% streptomycin). Following the preparation of 6-well plates
with coverslips, cell suspensions were added to each well
(3×105/well). Cells were cultured at 37°C in 5%
CO2 for 48 h, washed with PBS and fixed with 4%
paraformaldehyde for 15 min at room temperature. The cells were
subsequently washed with 1% PBS with Triton-100 to penetrate the
cell membrane. Following incubation with 10% donkey serum (cat. no.
017-000-121; Jackson ImmunoResearch Laboratories, Inc.) at room
temperature for 1 h, cells were incubated with primary antibodies
against Notch1, Notch2, Notch3, and Notch4 and their ligands
Jagged1, Jagged2, DLL1, DLL3 and DLL4 (Table II) at 4°C overnight. The signals
were generated following incubation with Alexa Fluor 594-conjugated
donkey anti-rabbit immunoglobulin G (IgG) secondary antibodies,
(dilution, 1:1,000; cat. no. R37119; Invitrogen™; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at room temperature for
1 h. Nuclear staining was performed with DAPI (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) for 5 min at room temperature. Stained
coverslips were visualized using a laser scanning confocal
microscope (Olympus Soft Imaging Solutions GmbH, Münster, Germany)
at magnification ×40 and ×100. The negative control group was
incubated with PBS instead of the primary antibodies.
Western blot analysis
The PDAC cell lines HPAC and PANC-1 were cultured in
culture flasks and collected when they became confluent. Cells were
subsequently homogenized in a radioimmunoprecipitation buffer for
protein extraction [50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton
X-100, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate, 10
µl/ml protease inhibitor cocktail and 1 mM phenylmethylsulfonyl
fluoride; cat. no. P0013B; Beyotime Institute of Biotechnology].
The protein samples were separated by either 8 or 10% SDS-PAGE and
subsequently transferred onto nitrocellulose membranes (Merck
KGaA). Following 3 washes for 10 min/wash with 20 mM Tris-Cl (pH
7.5), 0.15 M NaCl and 0.05% Tween-20 (TBST), cells were blocked
with 5% skimmed milk in TBST for 1 h at room temperature. Membranes
were then incubated overnight with primary antibodies at 4°C
(Table II). The membranes were
subsequently incubated with secondary goat anti-rabbit IgG
HRP-conjugated antibodies (1:1,000; cat. no. 111-625-144; LI-COR
Biosciences, Lincoln, NE, USA) for 1 h at room temperature. Protein
bands were visualized using a chemiluminescence detection system
(Odyssey® two-color infrared fluorescence imaging
system; LI-COR Biosciences). Protein levels were normalized to
GAPDH (1:10,000; cat. no. G9545; Sigma-Aldrich; Merck KGaA) levels
and quantified using ImageJ software version 1.43b (National
Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. Statistical analysis was performed using SPSS software,
version 19.0 (IBM Corp., Armonk, NY, USA). Pearson's correlation
co-efficient was used to identify whether there were correlations
between the expression of Notch receptors and their ligands.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Positive rate and intensity of Notch
receptor expression
Rabbit polyclonal anti-Notch1-4 antibodies were used
to detect the expression of Notch1-4 in human PDAC tissues
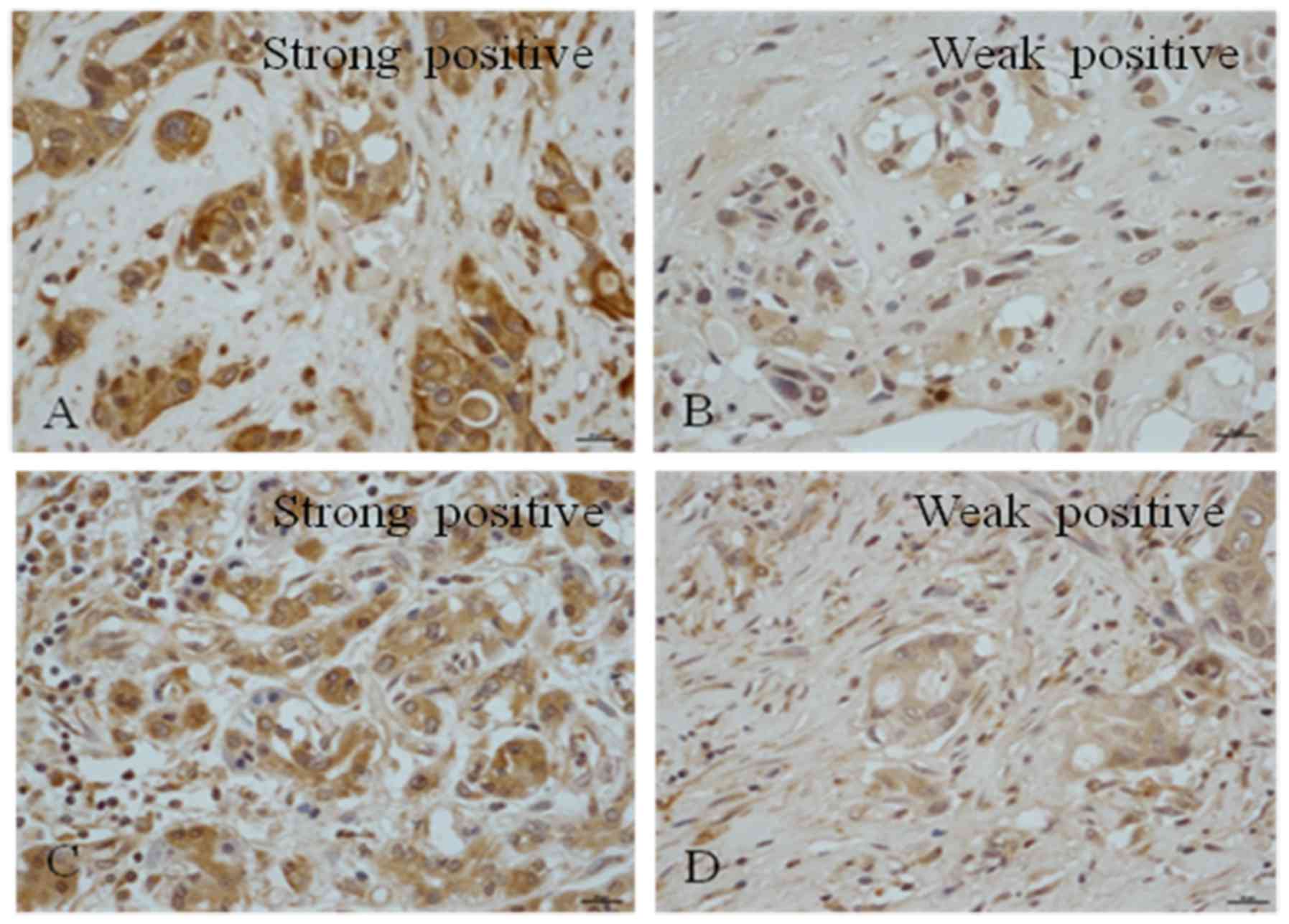
(Fig. 1). It was observed that
Notch1 was expressed in all PDAC samples; 91.7% of the tissues
exhibited strong positive staining and 8.3% demonstrated weak
positive staining; none of the samples were negative for Notch1
(Table III). A total of 41.7% of
the samples were negative for Notch2, 37.5% exhibited weak
positivity and 20.8% of the samples exhibited positive nuclear
staining (Table III). It was
observed that 41.7% of the samples had positive staining for
Notch3, among which 12.5% were strongly positive (Table III). Among the samples, 45.8% were
weakly positive for Notch3 and 12.5% were negative (Table III). A total of 41.7% of the
samples were negative for Notch4, 20.8% were weakly positive, 37.5%
exhibited positive staining and 4.2% exhibited strong positive
expression (Table III).
 | Table III.Positive rate and intensity of Notch
receptors and ligands expression. |
Table III.
Positive rate and intensity of Notch
receptors and ligands expression.
| Antigen | 0 (%) | 1 (%) | 2 (%) | 3 (%) | Total positive rate
(%) |
|---|
| Notch1 |
0.0 |
8.3 | 25.0 | 66.7 | 91.7 |
| Notch2 | 41.7 | 37.5 | 20.8 |
0.0 | 20.8 |
| Notch3 | 12.5 | 45.8 | 29.2 | 12.5 | 41.7a |
| Notch4 | 41.7 | 20.8 | 33.3 |
4.2 | 37.5b |
| JAGGED1 | 62.5 | 20.9 |
8.3 |
8.3 | 16.6b |
| JAGGED2 | 45.8 | 16.7 | 37.5 |
0.0 | 37.5b |
| DLL1 |
0.0 | 20.8 | 25.0 | 54.2 | 79.2a |
| DLL3 | 25.0 | 25.0 | 29.2 | 20.8 | 50a |
| DLL4 |
8.3 | 29.2 | 12.5 | 50.0 | 62.5a |
Positive rate and intensity of Notch
ligand expression
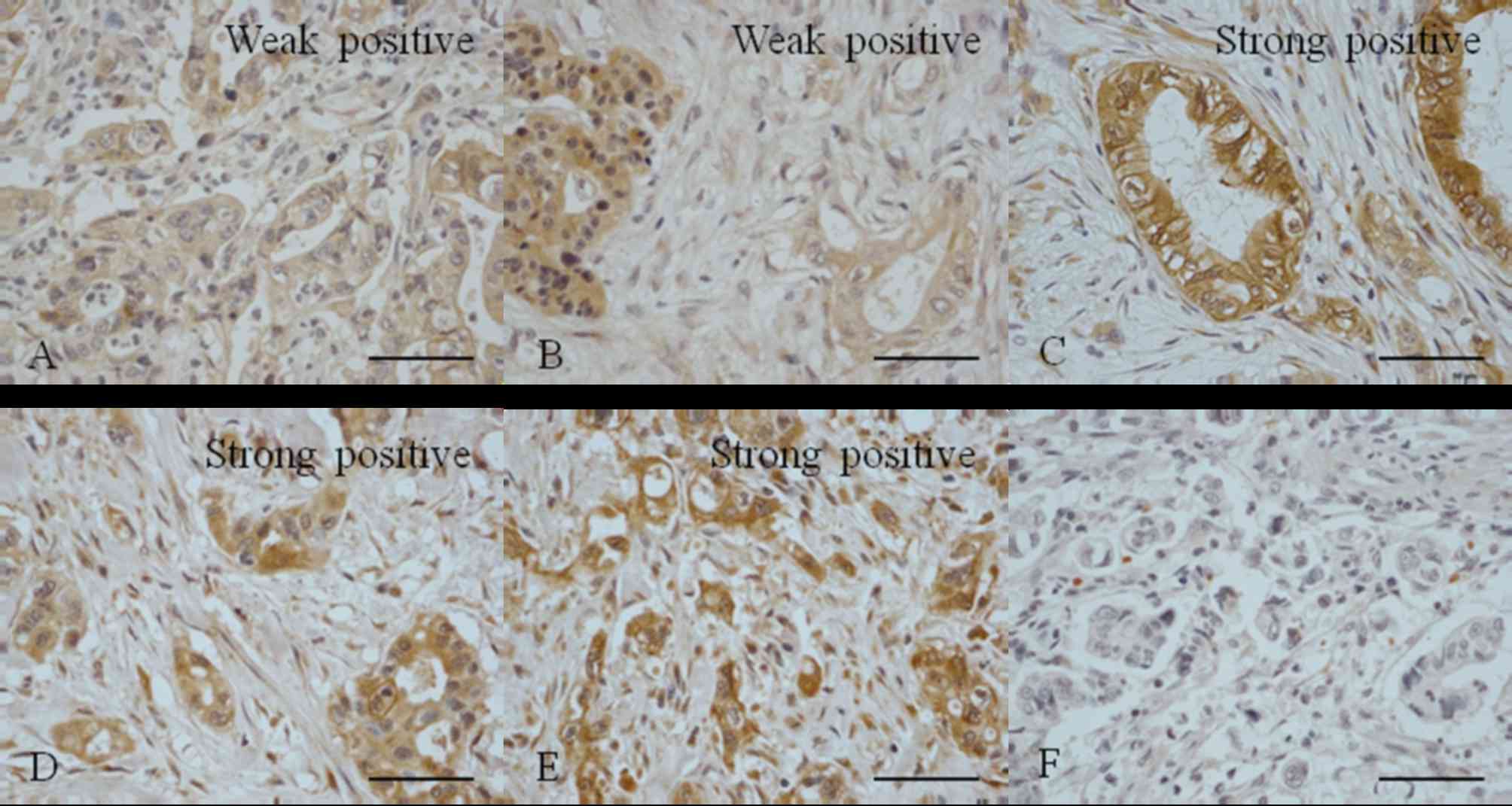
Polyclonal Jagged1 and 2 antibodies were used to
detect the expression of Jagged1 and 2 in PDAC tissues (Fig. 2). It was observed that 62.5% of the
samples were negative for Jagged1 expression and 20.9% were weakly
positive; 16.6% of samples were positive for Jagged1 but only 8.3%
exhibited strong positive expression (Table II). A total of 45.8% of the samples
were negative for Jagged2 expression, 16.7% were weakly positive
and 37.5% exhibited positive staining.
It was observed that only 20.8% of the PDAC tissue
samples exhibited weak positive staining for DLL1; 79.2% were
positive for DLL1 and 54.2% exhibited strong expression (Table III). None of the samples were
negative for DLL1. A total of 25% of the PDAC tissue samples were
negative for DLL3 expression, 25% were weakly positive and 50% were
positive, with 20.8% exhibiting strong positive expression
(Table II). The results revealed
that 8.3% of the PDAC tissue samples were negative for DLL4, 29.2%
were weakly positive and 62.5% were positive, with 50% of the
samples exhibiting strong positive expression (Table III). These results were similar to
the results obtained regarding DLL1 expression.
Correlation between the expression of
Notch receptors and their ligands
As demonstrated in Figs.
1 and 2 and Table III, the expression of Notch
receptors and their ligands were examined. It was observed that the
majority of PDAC tissue samples (91.7%) exhibited high Notch1
expression. A total of 41.7% of samples exhibited positive Notch3
expression. Among the ligands, the majority of PDAC tissues (79.2%)
stained positive for DLL1; 62.5% stained positive for DLL4 and 50%
stained positive for DLL3 (Table
II). A significant positive correlation between Notch1 and DLL1
was observed. Notch1 also exhibited a positive correlation with
DLL4 and DLL3 (r=0.7; P=0.0020). However, Notch1 was negatively
correlated with Notch4, Jagged1 and Jagged2. Notch2 expression was
positively correlated with Notch4, Jagged1 and Jagged2 (r=0.5;
P=0.0087).
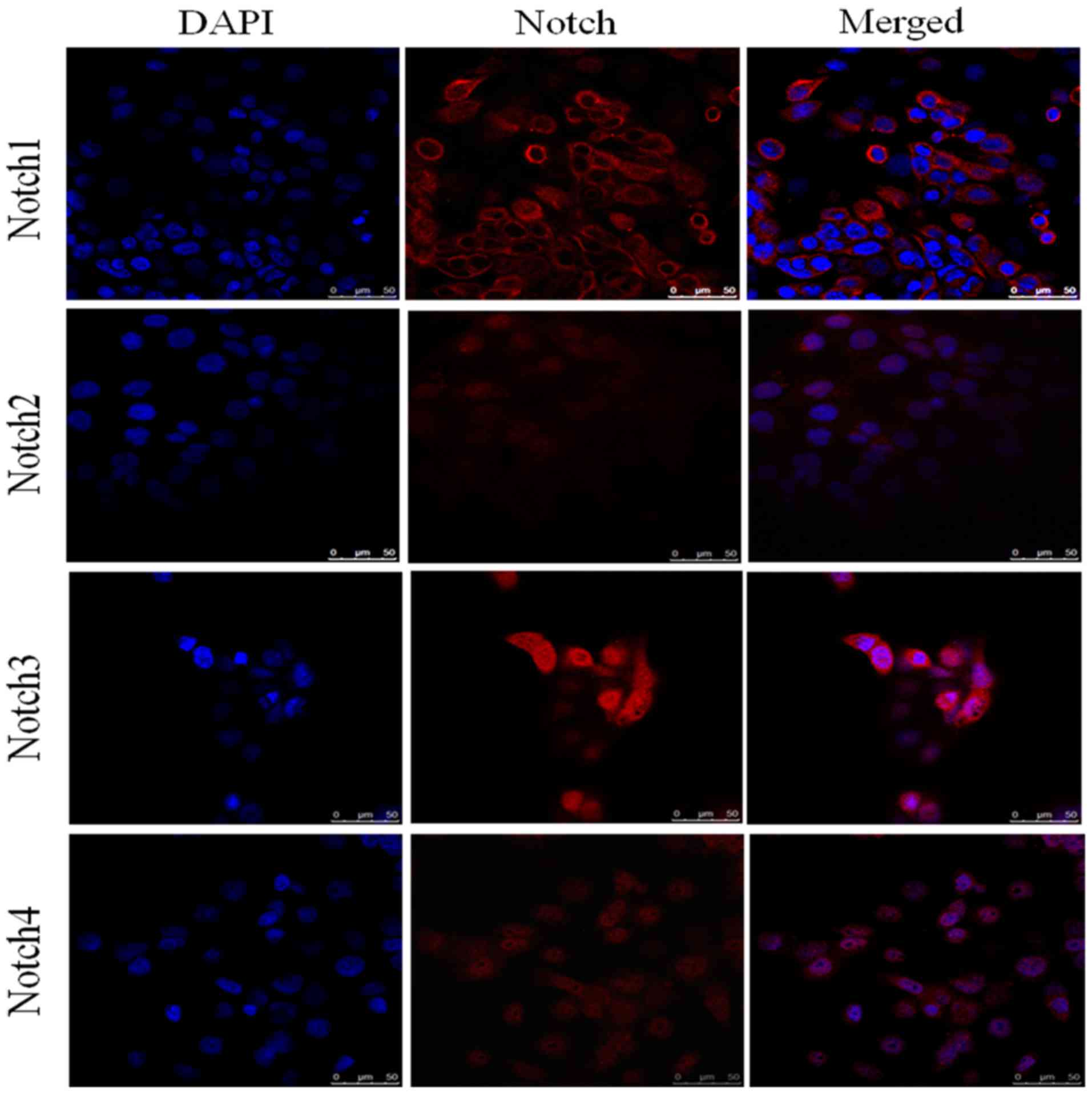
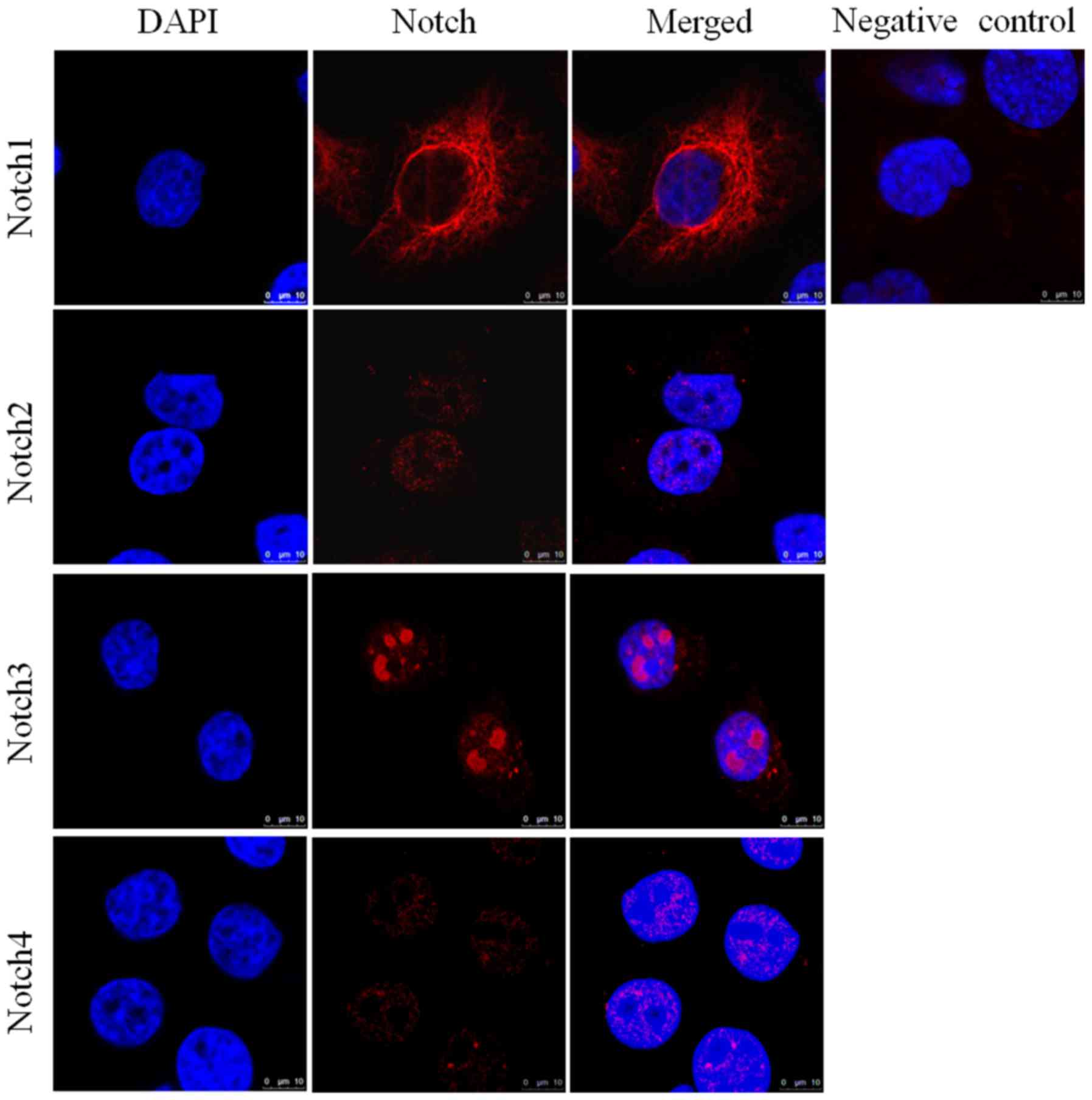
Expression of Notch receptors in
pancreatic cancer cell lines assessed by immunofluorescence
analysis
Immunofluorescence analysis of HPAC (Fig. 3) and PANC-1 (Fig. 4) cell lines assessed the expression
of Notch1, Notch2, Notch3 and Notch4 in each of these cell lines.
DAPI (blue) was used to stain the nucleus. Notch1 exhibited
positive expression in the cytoplasm and around the nucleus,
whereas Notch3 had clear nuclear localization (Fig. 4). The expression of Notch2 and Notch4
was notably lower in each of the cell lines compared with Notch1
and Notch3. The elevated expression of Notch1 and Notch3 in HPAC
and PANC-1 cell lines was in accordance with their pattern of
expression in tissue samples from patients with PDAC. Similarly,
the lower expression of Notch2 and Notch4 in the HPAC and PANC-1
cell lines was consistent with their lower expression in PDAC
tissue samples.
Expression of Notch receptors in
pancreatic cancer cell lines determined by western blot
analysis
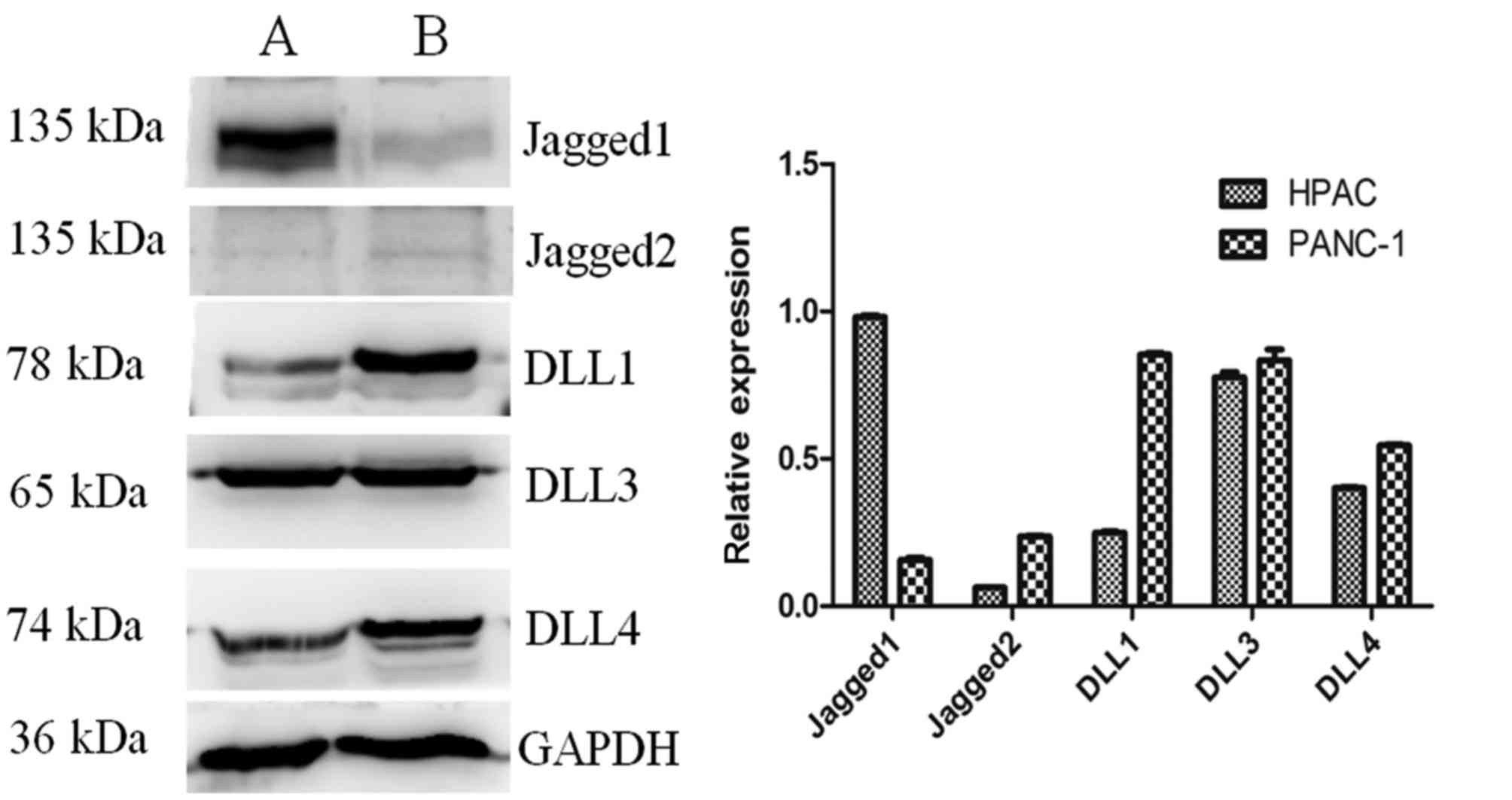
The expression of the Notch receptors in the
pancreatic HPAC and PANC-1 cell lines was assessed (Fig. 5). The results revealed that the
expression of Notch1 was notably increased in HPAC cells compared
with PANC-1 cells, while Notch3 was highly expressed in the two
cell lines. The expression of Notch2 and Notch4 was markedly lower
than Notch1 and Notch3 in the two cell lines. The protein
expression of the Notch ligands in HPAC and PANC-1 cells was also
measured (Fig. 6). Levels of DLL1,
DLL3 and DLL4 expression were higher than those of Jagged2 in each
of the cell lines. The expression of Jagged1 was notably higher in
HPAC cells compared with PANC-1 cells. The specificity of the
antibodies was confirmed in HeLa and 293T cell lines (data not
shown).
Discussion
In the present study, immunohistochemistry was
performed to evaluate the expression of proteins in the Notch
signaling pathway, including various receptors and ligands
associated with PDAC. To the best of our knowledge, the current
study is the first to evaluate the expression of Notch signaling
pathway components and investigate the correlations among them.
The Notch gene was first identified in
Drosophila in 1917 (19) and
Notch1 was revealed to be associated with T-cell acute
lymphoblastic leukemia in 1991 (20). The roles of the Notch signaling
pathway in embryonic development (21,22),
adult differentiation (23,24), and the development tumors (10–13) have
been previously studied and positively confirmed. In eukaryotes,
the Notch signaling pathway is highly conserved and regulates cell
proliferation, differentiation and apoptosis through interactions
between adjacent cells (25–27).
Notch signaling serves a central role in tumors and
during the embryonic development of the pancreas, in which it
controls cellular differentiation (26). However, the expression and functions
of each of the Notch signaling pathway components differ in tumor
development, including in PDAC. Miyamoto et al (14) revealed that the expression of the
Notch1, Notch2, Notch3 and Notch4 receptors and their ligand
Jagged1 was upregulated in resected pancreatic cancer samples. The
expression of the Notch signaling target transcription factor Hes1
(Hes1) was also upregulated in pancreatic cancer cells (14). However, Vo et al (16) reported that among the Notch family
members, Notch3 was primarily overexpressed in pancreatic cancer,
followed by Notch4 and Jagged1, whereas Notch1 was not expressed in
malignant cells. Another previous study demonstrated that Notch3
was significantly overexpressed in the cytoplasm and nucleus in
43.5% of pancreatic adenocarcinoma tumors (28). Mann et al (29) demonstrated that the Notch signaling
pathway components Notch1, Notch3, Notch4, Hes1 and
hairy/enhancer-of-split related with YRPW motif protein 1 were
significantly elevated in pancreatic adenocarcinoma. Therefore, it
remains unclear how the expression of various Notch proteins
changes during the progression of cancer, and to the best of our
knowledge a complete investigation examining the expression of all
Notch receptors and their ligands in PDAC has not yet been
conducted.
The elevated expression of Notch1 in pancreatic
cancer leads to the accumulation of undifferentiated precursor
cells (30), whereas the
downregulation of Notch1 decreases cyclin D1 and B-cell lymphoma 2
expression, which increases the apoptosis of pancreatic cancer
cells (31). It has been
demonstrated that inhibiting the Notch signaling pathway using
Notch1 small interfering RNA triggers apoptosis in the pancreatic
cancer cell lines BxPC-3, MIAPaCa-2 and PANC-1 (32). Blocking Notch2/3 inhibits tumor
growth and tumor-initiating cells (33) and the inhibition of Notch1 and Notch4
expression inhibits tumor growth (34,35).
Mazur et al (11)
demonstrated that Notch2 is a central regulator of pancreatic
intraepithelial neoplasia progression and malignant transformation.
Notch1 has also been reported to function as a tumor suppressor
gene in PDAC (18). A number of
studies have reported conflicting results and the role of Notch
signaling in PDAC remains highly controversial. In addition, there
is little information available concerning the expression pattern
of Notch receptors and their ligands in PDAC.
Therefore, the present study was conducted to
estimate the expression and potential pathological significance of
all Notch receptors and their ligands in human PDAC. In the present
study, Notch1 exhibited increased expression in PDAC tissues, in
which it may serve a role in the development of pancreatic cancer
development by acting as an oncogene. Notch3 was also highly
expressed, suggesting that it serves a similar role to Notch1 in
PDAC. The DLL1, DLL3 and DLL4 ligands were upregulated. By
contrast, levels of Notch2 and Notch4 were decreased in PDAC
tissues. In the cohort of patients assessed in the current study,
the expression of the ligands Jagged1 and Jagged2 were also
decreased compared with DLL1, DLL3 and DLL4.
To determine the expression and potential functions
of these molecules, HPAC and PANC-1 pancreatic cancer cell lines
were selected and immunofluorescence staining and western blot
analysis was performed. The results of immunofluorescence staining
revealed that Notch1 was expressed in the cytoplasm and around the
nucleus. The expression of Notch3 was also positive, however, it
was localized in the nucleus. Levels of Notch2 and Notch4 were
decreased compared with Notch1 and Notch3. Therefore, the
expression of Notch1 and Notch3 in the pancreatic cancer cell lines
corresponded with their expression in PDAC cancer tissues,
confirming that they are highly expressed in PDAC. The expression
of Notch2 and Notch4 in the cancer cell lines was also consistent
with their expression in cancer tissues; Notch2 and Notch4
expression were decreased compared with Notch1 and Notch3. Western
blot analysis also revealed notably elevated expression of Notch1
and Notch3 compared with Notch2 and Notch4 in the pancreatic cancer
cell lines HPAC and PANC-1. The Notch ligands DLL1, DLL3 and DLL4
exhibited markedly higher expression than that of Jagged2.
In the present study, a positive correlation was
observed between the expression of Notch1 and Notch3, and between
Notch1 and the ligands DLL1, DLL3 and DLL4. The results of the
western blot analysis were consistent with those of
immunohistochemistry, suggesting that the Notch1 and Notch3
pathways may be initiated by DLL1, DLL3 or DLL4. However, they do
not seem to be initiated by Jagged2.
Due to the limited number of patients recruited and
the lack of normal controls in the present study, these results may
not be representative of the entire population. Future studies
should be conducted to investigate a greater number of samples to
confirm the results of the present study. Understanding the
molecular characteristics of tumors may provide an important basis
for the clinical diagnosis and treatment of patients with
pancreatic cancer. The present study suggested that Notch1 and
Notch3 may be potential targets for treatments against PC, and may
provide the basis for a novel method of treatment and diagnosis of
PC in the future.
Acknowledgements
Not applicable.
Funding
The study was supported by the National Natural
Science Foundation of China (grant no. 81372156) and the Doctor
Scientific Research Foundation of Xinxiang Medical University
(grant no. XXBSKYZZ201821).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YZ and HS conceived and designed the experiments.
HS, YW and HL conducted the experiments.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated Center Hospital of Xinxiang Medical
University and written informed consent was obtained from all
patients prior to their inclusion within the study.
Consent for publication
All patients provided written consent for the
publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:1039–1049. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bliss LA, Witkowski ER, Yang CJ and Tseng
JF: Outcomes in operative management of pancreatic cancer. J Surg
Oncol. 110:592–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Apelqvist A, Li H, Sommer L, Beatus P,
Anderson DJ, Honjo T, de Angelis MH, Lendahl U and Edlund H: Notch
signalling controls pancreatic cell differentiation. Nature.
400:877–881. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Espinoza I and Miele L: Deadly crosstalk:
Notch signaling at the intersection of EMT and cancer stem cells.
Cancer Lett. 341:41–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang J, Han F, Wu J, Lee SW, Chan CH, Wu
CY, Yang WL, Gao Y, Zhang X, Jeong YS, et al: The role of Skp2 in
hematopoietic stem cell quiescence, pool size, and self-renewal.
Blood. 118:5429–5438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oishi H, Sunamura M, Egawa S, Motoi F,
Unno M, Furukawa T, Habib NA and Yagita H: Blockade of delta-like
ligand 4 signaling inhibits both growth and angiogenesis of
pancreatic cancer. Pancreas. 39:897–903. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mazur PK, Einwächter H, Lee M, Sipos B,
Nakhai H, Rad R, Zimber-Strobl U, Strobl LJ, Radtke F, Klöppel G,
et al: Notch2 is required for progression of pancreatic
intraepithelial neoplasia and development of pancreatic ductal
adenocarcinoma. Proc Natl Acad Sci USA. 107:13438–13443. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizuma M, Rasheed ZA, Yabuuchi S, Omura N,
Campbell NR, de Wilde RF, De Oliveira E, Zhang Q, Puig O, Matsui W,
et al: The gamma secretase inhibitor MRK-003 attenuates pancreatic
cancer growth in preclinical models. Mol Cancer Ther. 11:1999–2009.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu Y, Su H, Li X, Guo G, Cheng L, Qin R,
Qing G and Liu H: The NOTCH ligand JAGGED2 promotes pancreatic
cancer metastasis independent of NOTCH signaling activation. Mol
Cancer Ther. 14:289–297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miyamoto Y, Maitra A, Ghosh B, Zechner U,
Argani P, Iacobuzio-Donahue CA, Sriuranpong V, Iso T, Meszoely IM,
Wolfe MS, et al: Notch mediates TGF alpha-induced changes in
epithelial differentiation during pancreatic tumorigenesis. Cancer
Cell. 3:565–576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomas MM, Zhang Y, Mathew E, Kane KT,
Maillard I and di Magliano Pasca M: Epithelial Notch signaling is a
limiting step for pancreatic carcinogenesis. BMC Cancer.
14:8622014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vo K, Amarasinghe B, Washington K,
Gonzalez A, Berlin J and Dang TP: Targeting notch pathway enhances
rapamycin antitumor activity in pancreas cancers through PTEN
phosphorylation. Mol Cancer. 10:1382011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Long B and Wang Z: Role of Notch
signaling pathway in pancreatic cancer. Am J Cancer Res. 7:173–186.
2017.PubMed/NCBI
|
|
18
|
Hanlon L, Avila JL, Demarest RM, Troutman
S, Allen M, Ratti F, Rustgi AK, Stanger BZ, Radtke F, Adsay V, et
al: Notch1 functions as a tumor suppressor in a model of
K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res.
70:4280–4286. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morgan MM and Mahowald AP: Multiple
signaling pathways establish both the individuation and the
polarity of the oocyte follicle in Drosophila. Arch Insect
Biochem Physiol. 33:211–230. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ellisen LW, Bird J, West DC, Soreng AL,
Reynolds TC, Smith SD and Sklar J: TAN-1, the human homolog of the
Drosophila notch gene, is broken by chromosomal
translocations in T lymphoblastic neoplasms. Cell. 66:649–661.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Piccirilli D, Baldini E, Massimiani M,
Camaioni A, Salustri A, Bernardini R, Centanni M, Ulisse S, Moretti
C and Campagnolo L: Thyroid hormone regulates protease expression
and activation of Notch signaling in implantation and embryo
development. J Endocrinol. 236:1–12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hussain M, Xu C, Ahmad M, Yang Y, Lu M and
Wu X, Tang L and Wu X: Notch signaling: Linking embryonic lung
development and asthmatic airway remodeling. Mol Pharmacol.
92:676–693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Chen SY, Zhao XY, Zhang MC and Xie
HT: Rat limbal niche cells prevent epithelial stem/progenitor cells
from differentiation and proliferation by inhibiting Notch
signaling pathway in vitro. Invest Ophthalmol Vis Sci.
58:2968–2976. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Z, Zou Y, Fan J, Li C and Ma L: Notch1
is associated with the differentiation of human bone marrow-derived
mesenchymal stem cells to cardiomyocytes. Mol Med Rep.
14:5065–5071. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Williams E, Villar-Prados A, Bowser J,
Broaddus R and Gladden AB: Loss of polarity alters proliferation
and differentiation in low-grade endometrial cancers by disrupting
Notch signaling. PLoS One. 12:e01890812017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Irles P, Elshaer N and Piulachs MD: The
Notch pathway regulates both the proliferation and differentiation
of follicular cells in the panoistic ovary of Blattella germanica.
Open Biol. 6:150192016. View Article : Google Scholar
|
|
27
|
Wang L, Song G, Liu M, Chen B, Chen Y,
Shen Y, Zhu J and Zhou X: MicroRNA-375 overexpression influences
P19 cell proliferation, apoptosis and differentiation through the
Notch signaling pathway. Int J Mol Med. 37:47–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Doucas H, Mann CD, Sutton CD, Garcea G,
Neal CP, Berry DP and Manson MM: Expression of nuclear Notch3 in
pancreatic adenocarcinomas is associated with adverse clinical
features, and correlates with the expression of STAT3 and
phosphorylated Akt. J Surg Oncol. 97:63–68. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mann CD, Bastianpillai C, Neal CP, Masood
MM, Jones DJ, Teichert F, Singh R, Karpova E, Berry DP and Manson
MM: Notch3 and HEY-1 as prognostic biomarkers in pancreatic
adenocarcinoma. PLoS One. 7:e511192012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sjolund J, Manetopoulos C, Stockhausen MT
and Axelson H: The Notch pathway in cancer: Differentiation gone
awry. Eur J Cancer. 41:2620–2629. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Z, Zhang Y, Li Y, Banerjee S, Liao J
and Sarkar FH: Down-regulation of Notch-1 contributes to cell
growth inhibition and apoptosis in pancreatic cancer cells. Mol
Cancer Ther. 5:483–493. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Du X, Wang YH, Wang ZQ, Cheng Z, Li Y, Hu
JK, Chen ZX and Zhou ZG: Down-regulation of Notch1 by small
interfering RNA enhances chemosensitivity to gemcitabine in
pancreatic cancer cells through activating apoptosis activity.
Zhejiang Da Xue Xue Bao Yi Xue Ban. 43:313–318. 2014.(In Chinese).
PubMed/NCBI
|
|
33
|
Yen WC, Fischer MM, Axelrod F, Bond C,
Cain J, Cancilla B, Henner WR, Meisner R, Sato A, Shah J, et al:
Targeting Notch signaling with a Notch2/Notch3 antagonist
(tarextumab) inhibits tumor growth and decreases tumor-initiating
cell frequency. Clin Cancer Res. 21:2084–2095. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kunnimalaiyaan S, Trevino J, Tsai S,
Gamblin TC and Kunnimalaiyaan M: Xanthohumol-mediated suppression
of Notch1 signaling is associated with antitumor activity in human
pancreatic cancer cells. Mol Cancer Ther. 14:1395–1403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu Y, Zhu F, Xu S and Liu L: Anti-tumor
effect of the extract from qingyihuaji formula on pancreatic cancer
by down-regulating Notch-4 and Jagged-1. J Tradit Chin Med.
35:77–83. 2015. View Article : Google Scholar : PubMed/NCBI
|