Introduction
Breast cancer is one of the most common
gynecological tumor types worldwide (1). The majority of breast cancer-associated
mortalities is caused by local migration and distant metastases of
tumor cells (2). While the current
therapeutic schedules, including surgical techniques, as well as
radiation, chemotherapy and gene targeted therapy, have improved
the overall survival (OS) of breast cancer patients, the outcome
remains generally poor (3,4). Of note, previous studies have indicated
that numerous genes are involved in epigenetic modifications among
patients with breast cancer (5–7). Genetic
analyses and testing for inherited gene mutations in patients with
breast cancer have been reported in clinical studies (8,9).
Therefore, it is essential to explore potential genes for
diagnosing patients with breast cancer in the early stage.
At present, ultrasound, fluorodeoxyglucose-positron
emission tomography (PET)/computed tomography (CT) and magnetic
resonance imaging (MRI) are widely used for diagnosing and staging
of human breast cancer (10–12). Eminently, MRI provides a higher
sensitivity and accuracy in the detection of breast cancer than CT
and ultrasound (13). Application of
dynamic contrast enhancement MRI and post-processing techniques are
helpful for making the correct diagnosis for patients with
suspected breast cancer (14). Youk
et al (15) have suggested
that MRI may be used to identify malignant breast lesions by
analyzing their morphological and kinetic features. In addition,
MRI is more efficient than CT in assessing the response to
neo-adjuvant chemotherapy and is beneficial for identifying the
primary tumor in breast cancer patients (16). Furthermore, comparison between PET/CT
and MRI in the diagnosis, staging and follow-up of breast cancer
patients revealed that MRI is useful for distinguishing between
benign and malignant pulmonary nodules, has a high sensitivity and
specificity for nodal staging, and is helpful for evaluating the
early response to systemic chemotherapy (17). However, the diagnostic accuracy of
single MRI for breast cancer is insufficient (18). In recent years, genetic diagnosis of
breast cancer has been applied, which may be an accurate and novel
diagnostic method for the evaluation of sentinel lymph node
metastasis in breast cancer patients (19–21).
In the present study, the diagnostic efficacy of MRI
in combination with detection of gene expression in breast cancer
patients was evaluated. It was reported that MRI combined with
detection of gene expression not only improves the accuracy, but
also contributes to the selection of efficient treatments for
patients with breast cancer.
Materials and methods
Patients and selection
A total of 84 female patients (median age, 46.2
years; range, 28.4–65.5 years) with suspected breast cancer who
were assessed at the Radiology Department of Beijing Tiantan
Hospital (Beijing, China) between May 2015 and October 2016 were
recruited for the present study. All patients underwent
pre-operative MRI and/or detection of gene expression (Ki-67,
BCL11A, FOXC1, HOXD13, PCDHGB7 and her-2). All patients were
finally diagnosed by histopathology in diagnostic biopsy samples.
Patients with a family history of cancer, chronic renal failure and
heart disease, as well as those with a history of ipsilateral
breast surgery, chest radiotherapy and oncoplastic breast cancer
were excluded from the study. Patients who received neoadjuvant
chemotherapy or radiotherapy were also excluded from the
recruitment. The present study was approved by the Ethics Committee
of Capital Medical University (Beijing, China). All patients
provided written informed consent.
MRI analysis
The MRI protocol was in accordance with that
described in a previous study (22).
In brief, a Siemens Verio 3.0 T magnet MRI machine (Siemens AG,
Munich, Germany) was used to analyze breast tumor lesions and
determine their volume. All patients with suspected breast cancer
were subjected to MRI screening. MR images were analyzed using a
SUN T2000 workstation (Sun Microsystems Inc., Mountain View, CA,
USA) with the Eigentool image analysis software 3.0 (Image Analysis
Lab, Henry Ford Hospital, Detroit, MI, USA). Threshold ranges were
determined using histogram analysis from regions of interest placed
around the identified lesion using 95% confidence intervals
determined from signal intensity.
Detection of gene expression
Total RNA was extracted from tumor cells (1.0 µg) in
biopsy samples using an RNeasy Mini kit (Qiagen, Hilden, Germany)
according to the manufacturer's protocol. Total RNA (1 µg) was
reverse transcribed into complementary (c)DNA using a QuantiTect
Reverse Transcription kit (cat. no. 205310; Qiagen) according to
the manufacturer's protocol. The cDNA (10 ng) was subjected to
quantitative polymerase chain reaction analysis using SYBR Green
Master mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All
primers were synthesized by Invitrogen (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) as described previously (23). The reaction conditions were
accordance with those of a previous study (24). Relative mRNA expression changes were
calculated using the 2−ΔΔCq method (25). The results are expressed as a fold of
the control.
Histopathology
Breast tumor staging was performed based on the
American Joint Committee on Cancer Staging manual, sixth edition
(26). Histological analysis was
performed using the modified Bloom-Richardson classification
(27). Primary breast cancer tissues
were evaluated from formalin-fixed, paraffin-embedded tumor
sections using immunohistochemistry. Tumor sections were incubated
with primary antibodies rabbit anti-human against estrogen receptor
(ER; 1:1,000; cat. no. clone SP1; Neomarkers for Lab Vision,
Fremont, CA, USA) and progesterone receptor (1:1,000, PR; cat. no.
clone PgR 636; Dako, Glostrup, Denmark) for 12 h at 4°C. Tumor
tissues were then incubated with using horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin G monoclonal
antibody (1:1,000, cat. no. PV-6001; Zhongshan Goldenbridge-BIO,
Beijing, China) for 2 h at 37°C. A Ventana Benchmark automated
staining system (Olympus BX51, Olympus; Tokyo, Japan) was used to
assess protein expression in tumor tissues. The staining results
were semi-quantitatively evaluated by multiplying the staining
intensity and the percentage of positively stained cells
(magnification, ×400). The cutoff value for ER and PR positivity
was >10% staining on immunohistochemistry (27).
Treatment
Standard treatments for patients with breast cancer
in the present study were in accordance with those described in a
previous study (28). In brief,
breast cancer patients received breast surgery and breast
radiotherapy.
Statistical analysis
Values are expressed as the mean ± standard
deviation of triplicate experiments. Pearson's correlation analysis
was performed to assess correlations. All data were analyzed with
SPSS version 20 (IBM Corp., Armonk, NY, USA) using one-way analysis
of variance followed by Tukey's multiple comparison post-hoc test.
The OS rates were calculated using the Kaplan-Meier method and the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
MRI diagnosis of patients with breast
cancer
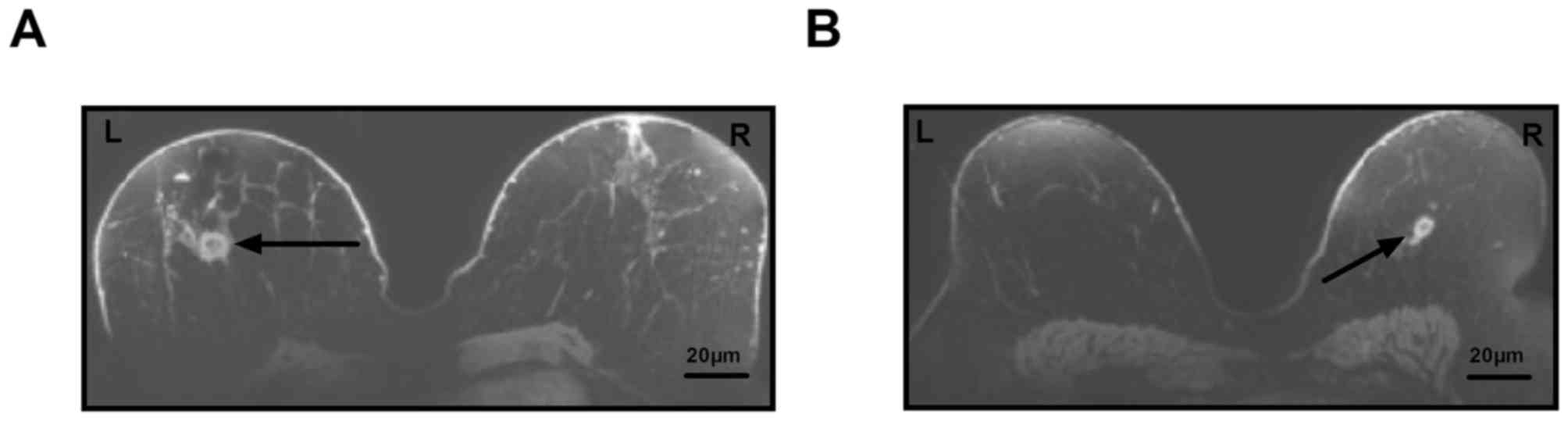
MRI was used to diagnose patients with suspected
breast cancer in a total of 84 cases. In a total of 78 patients, a
breast lump was detected, which required further confirmation by
pathological analysis. Representative MRI images of breast lumps in
patients with suspected breast cancer are displayed in Fig. 1.
Analysis of breast cancer-associated
gene expression in biopsy samples of patients with breast
cancer
To identify differences in gene expression in
patients with breast cancer, the gene expression levels of Ki-67,
B-cell CLL/lymphoma 11a (BCL11A), forkhead box (FOX)C1, homeobox
(HOX)D13; protocadherin γ subfamily B, 7 (PCDHGB7) and her-2 were
detected in patients with breast cancer. The analysis identified 72
patients with elevated gene expression of Ki-67, BCL11A, FOXC1,
HOXD13, PCDHGB7 and her-2, while 32 patients had lower expression
(Table I). Of the 78 patients in
which a breast lump was detected on MRI, 72 had higher gene
expression. The present study demonstrated that there were a total
of 66 patients with and 12 patients without breast tumors (Table I).
 | Table I.Gene expression in patients with
suspected breast cancer. |
Table I.
Gene expression in patients with
suspected breast cancer.
| Gene | Patients with breast
tumors (n=66; %) | Individuals without
tumors (n=12; %) |
|---|
| Ki-67 | 9.74±2.47 | 2.2±1.2 |
| BCL11A | 16.2±4.2 | 1.8±0.7 |
| FOXC1 | 8.4±3.2 | 2.3±1.0 |
| HOXD13 | 7.5±3.0 | 2.6±1.5 |
| PCDHGB7 | 6.8±2.6 | 1.7±0.7 |
| Her-2 | 9.3±3.5 | 2.5±1.5 |
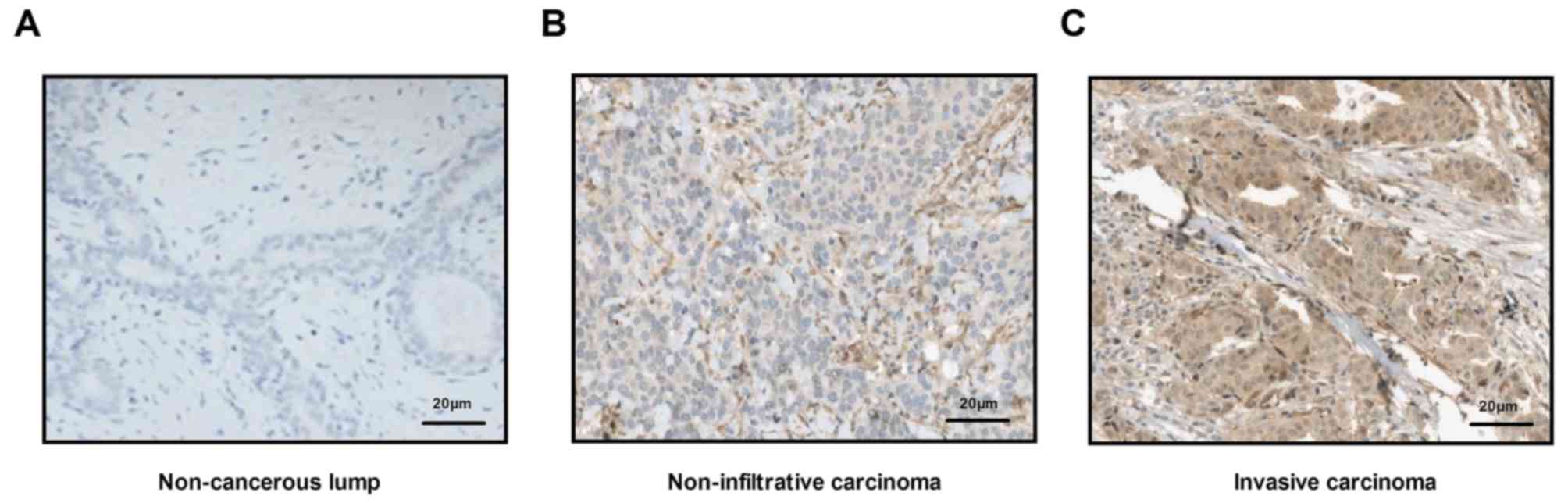
Pathological analysis of breast cancer
tissue
Histopathological analysis was used to confirm
whether patients had breast cancer. Analyses demonstrated that all
cancer tissues were either non-infiltrative carcinoma or early
invasive carcinoma. Representative images of non-cancerous lump, as
well as non-infiltrative carcinoma and invasive carcinoma tissue
sections are displayed in Fig. 2. A
total of 66 patients were finally diagnosed with breast cancer,
including 45 cases of non-infiltrative carcinomas and 21 cases of
invasive carcinoma (Table II).
 | Table II.Characteristics of the groups,
divided by pathological analysis. |
Table II.
Characteristics of the groups,
divided by pathological analysis.
| Type | n | Tumor size
(cm) |
|---|
| Non-infiltrative
carcinoma | 45 | <2 |
| Invasive
carcinoma | 21 | >2 |
| Healthy
individuals | 16 | 0 |
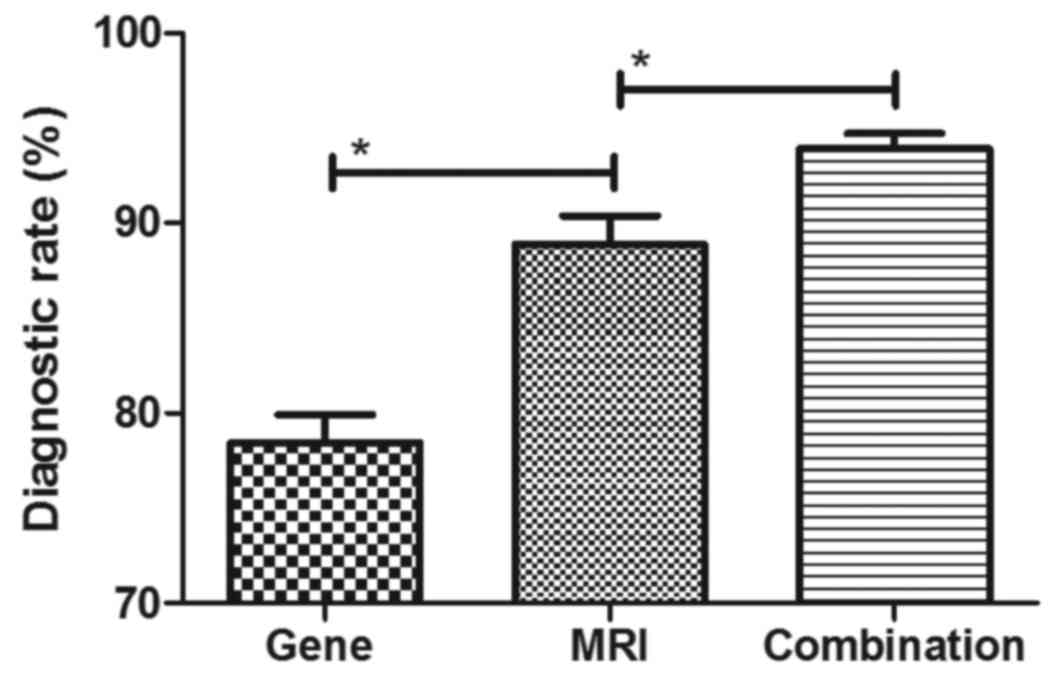
Efficacy of combined diagnosis of MRI
and detection of gene expression for patients with breast
cancer
The efficacy of combined diagnosis of MRI and
detection of gene expression was evaluated in patients with breast
cancer. Pearson's R test was used to assess the correlation between
gene expression and the risk of breast cancer. A significant
positive correlation was observed between expression levels of the
genes Ki-67, BCL11A, FOXC1, HOXD13, PCDHGB7 and her-2, and the risk
of breast cancer (Pearson's R=0.516; P<0.01; Fig. 3). In addition, a significant positive
correlation was observed between gene expression levels and tumor
size in patients with breast cancer (Pearson's R=0.410; P<0.01;
Fig. 4). It was demonstrated that
the efficacy of combined diagnosis by MRI and detection of gene
expression improved the diagnostic rate compared with that achieved
by either MRI or detection of gene expression alone (Fig. 5).
Association between gene expression
levels and breast cancer subtypes
The present study then analyzed the association
between gene expression levels and the breast cancer subtype,
including non-infiltrative carcinoma and early invasive carcinoma.
As presented in Fig. 6, breast
tumor-associated genes were lower expressed in non-infiltrative
carcinoma compared with early invasive carcinoma.
Survival of breast cancer patients
diagnosed by MRI and detection of gene expression
The survival of breast cancer patients diagnosed by
MRI and detection of gene expression enrolled in the present study
was then investigated. A significant improvement of patients'
survival time was observed after diagnosis by MRI and detection of
gene expression compared with the mean survival of breast cancer
patients diagnosed by MRI and detection of gene expression
(Fig. 7). These outcomes suggest
that diagnosis with a combination of MRI and detection of gene
expression contributes to the early implementation of treatment and
therefore patient survival.
Discussion
The efficacy of contrast-enhanced MRI in the
diagnosis of breast cancer has been previously proven (29). In addition, diagnosis of breast
cancer using malignancy-associated biomarkers has been widely
accepted (30). The present study
analyzed the efficacy of the combination of MRI and detection of
gene expression in diagnosing patients with breast cancer in the
early stage. The results indicated that combination of MRI and
detection of gene expression not only markedly improved the
diagnostic accuracy, but also contributed to early tumor
treatments, which further resulted in a long survival time of
breast cancer patients.
Although various methods for the early-stage
diagnosis of breast cancer have been introduced, including
contrast-enhanced MRI, it is difficult to differentiate between
breast tumors and normal lumps in women with suspected breast
cancer (31–33). Molecular diagnosis comprising the
detection of high-frequency mutations in breast cancer patients has
provided a novel strategy for the early diagnosis of human breast
cancer (34). In the present study,
the expression levels of six genes, namely Ki-67, BCL11A, FOXC1,
HOXD13, PCDHGB7 and her-2, was determined to analyze the risk of
breast cancer in a total of 84 patients with suspected breast
cancer. The results indicated a significant positive correlation
between gene expression levels and the risk of breast cancer, as
well as between gene expression levels and tumor size in patients
with breast cancer. In the present study, gene expression analysis
identified 72 patients with an elevated risk of breast cancer among
84 suspicious patients.
Ki-67 has been regarded as a prognostic marker based
on a molecular subtype of breast cancer (35). The present study reported that Ki-67
expression was higher in invasive carcinoma compared with that in
non-infiltrative carcinoma. BCL11A is overexpressed in
triple-negative breast cancer compared with that in normal mammary
epithelial cells (36). In the
present study, BCL11A was 12–20-fold increased in tumor biopsy
samples in patients with breast cancer compared with that
non-malignant breast lumps in healthy individuals. A previous study
has indicated that FOXC1 overexpression is a marker of poor
response to anthracycline-based adjuvant chemotherapy in
triple-negative breast cancer (37).
In addition, the prognostic significance of HOXD13 expression in
human breast cancer has been elucidated and a previous study
indicated that HOXD13 is a potential prognostic marker for patients
with breast cancer (38).
Furthermore, quantification of her-2 expression in
immunohistochemically-identified biopsies can be used to diagnose
breast cancer (39). The present
study indicated that the expression levels of BCL11A, FOXC1,
HOXD13, PCDHGB7 and her-2 were upregulated in patients with breast
cancer. The present study analyzed the diagnostic efficacy of the
combination of MRI and detection of gene expression for patients
with breast cancer in the early stage.
Studies have indicated that MRI is an accurate
diagnostic method for patients with breast cancer (40,41). The
present study reported that out of 78 patients in whom a breast
lump was detected on MRI, 72 were indicated to have breast cancer
according to detection of gene expression. In fact, only 66
patients were confirmed to have breast cancer by pathological
analysis. The results of the present study also identified that MRI
and detection of gene expression alone are not sufficient for
breast cancer diagnosis. The results of the present study also
indicated that breast tumor-associated genes were lower expressed
in non-infiltrative carcinoma compared with those in in early
invasive carcinoma. Of note, the survival time of patients
diagnosed by MRI and detection of gene expression was longer than
the mean survival of breast cancer patients reported previously
(42).
In conclusion, the present study analyzed the
efficacy of MRI and detection of gene expression in diagnosing
breast cancer patients. It was demonstrated that the detection of
the gene expression levels of BCL11A, FOXC1, HOXD13, PCDHGB7 and
her-2 may be regarded as an auxiliary method for MRI in diagnosing
human breast cancer. However, further studies in large populations
of patients with suspected breast cancer are required to determine
the combined diagnostic efficacy of MRI and detection of gene
expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DK, RY, LJ analyzed and interpreted the patient data
regarding the hematological disease and the transplant. DK
performed the histological examination of the kidney, and was a
major contributor in writing the manuscript. All authors read and
approved the final manuscript.
Ethical approval and consent to
participate
This study was approved by the Ethics Committee of
Capital Medical University (Beijing, China). All patients provided
written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Novik AV, Protsenko SA, Baldueva IA,
Ivantsov AO, Nekhaeva TL, Akhaeva ZY, Yanus GA, Iyevleva AG and
Imyanitov EN: Vemurafenib-induced progression of breast cancer: A
case report and review of the literature. Target Oncol. 11:235–238.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Araki K and Ito Y: A review multigene
assays for Clinical Utility in breast cancer. Gan to kagaku ryoho.
Gan To Kagaku Ryoho. 43:1332–1340. 2016.(In Japanese).
|
|
3
|
Plourde N, Brown HK, Vigod S and Cobigo V:
Contextual factors associated with uptake of breast and cervical
cancer screening: A systematic review of the literature. Women
Health. 56:906–925. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Post KE and Flanagan J: Web based
survivorship interventions for women with breast cancer: An
integrative review. Eur J Oncol Nurs. 25:90–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thivya KS, Sakthivel P and Venkata Sai PM:
Analysis of framelets for breast cancer diagnosis. Technol Health
Care. 24:21–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Korkmaz SA, Korkmaz MF and Poyraz M:
Diagnosis of breast cancer in light microscopic and mammographic
images textures using relative entropy via kernel estimation. Med
Biol Eng Comput. 54:561–573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zuo X, Chen L, Liu L, Zhang Z, Zhang X, Yu
Q, Feng L, Zhao X and Qin T: Identification of a panel of complex
autoantigens (LGALS3, PHB2, MUC1, and GK2) in combination with
CA15-3 for the diagnosis of early-stage breast cancer. Tumour Biol.
37:1309–1317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang DY, Done SJ, McCready DR, Boerner S,
Kulkarni S and Leong WL: A new gene expression signature, the
ClinicoMolecular Triad Classification, may improve prediction and
prognostication of breast cancer at the time of diagnosis. Breast
Cancer Res. 13:R922011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miki Y: Gene expression-based diagnosis of
efficacy of chemotherapy for breast cancer. Breast Cancer.
17:97–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Richman I, Asch SM, Bendavid E,
Bhattacharya J and Owens DK: Breast density notification
legislation and breast cancer stage at diagnosis: Early evidence
from the SEER registry. J Gen Intern Med. 32:603–609. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abdoli G, Bottai M, Sandelin K and Moradi
T: Breast cancer diagnosis and mortality by tumor stage and
migration background in a nationwide cohort study in Sweden.
Breast. 31:57–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang S, Zhou F, Sun Y, Wei L, Zhu S, Yang
R, Huang Y and Yang J: CEA in breast ductal secretions as a
promising biomarker for the diagnosis of breast cancer: A
systematic review and meta-analysis. Breast Cancer. 23:813–819.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kruger S, Mottaghy FM, Buck AK, Maschke S,
Kley H, Frechen D, Wibmer T, Reske SN and Pauls S: Brain metastasis
in lung cancer. Comparison of cerebral MRI and 18F-FDG-PET/CT for
diagnosis in the initial staging. Nuklearmedizin. 50:101–106.
2011.
|
|
14
|
Peng KQ, Huang ZL, Xie CM, Chen L, Ouyang
Y, Zheng QS, Zhang Y, He HQ and Wu PH: Application of dynamic
contrast enhancement MRI and post-processing technique for
diagnosis of breast cancer. Ai Zheng. 28:549–554. 2009.(In
Chinese). PubMed/NCBI
|
|
15
|
Youk JH, Son EJ, Kim EK, Kim JA, Kim MJ,
Kwak JY and Lee SM: Diagnosis of breast cancer at dynamic MRI in
patients with breast augmentation by paraffin or silicone
injection. Clin Radiol. 64:1175–1180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morrow M, Waters J and Morris E: MRI for
breast cancer screening, diagnosis, and treatment. Lancet.
378:1804–1811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu T, Cheng T, Xu W, Yan WL, Liu J and
Yang HL: A meta-analysis of 18FDG-PET, MRI and bone scintigraphy
for diagnosis of bone metastases in patients with breast cancer.
Skeletal Radiol. 40:523–531. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kawashima H, Inokuchi M, Furukawa H and
Kitamura S: Accuracy for a diagnosis of breast cancer spread using
3.0T MRI. Nihon rinsho. Nihon Rinsho. 70(Suppl 7): S5306–S5308.
2012.(In Japanese).
|
|
19
|
Kwong A, Chen J, Shin VY, Ho JC, Law FB,
Au CH, Chan TL, Ma ES and Ford JM: The importance of analysis of
long-range rearrangement of BRCA1 and BRCA2 in genetic diagnosis of
familial breast cancer. Cancer Genet. 208:448–454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Woodson AH, Muse KI, Lin H, Jackson M,
Mattair DN, Schover L, Woodard T, McKenzie L, Theriault RL,
Hortobágyi GN, et al: Breast cancer, BRCA mutations, and attitudes
regarding pregnancy and preimplantation genetic diagnosis.
Oncologist. 19:797–804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Derks-Smeets IA, de Die-Smulders CE,
Mackens S, van Golde R, Paulussen AD, Dreesen J, Tournaye H,
Verdyck P, Tjan-Heijnen VC, Meijer-Hoogeveen M, et al: Hereditary
breast and ovarian cancer and reproduction: an observational study
on the suitability of preimplantation genetic diagnosis for both
asymptomatic carriers and breast cancer survivors. Breast Cancer
Res Treat. 145:673–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Heacock L, Melsaether AN, Heller SL, Gao
Y, Pysarenko KM, Babb JS, Kim SG and Moy L: Evaluation of a known
breast cancer using an abbreviated breast MRI protocol: Correlation
of imaging characteristics and pathology with lesion detection and
conspicuity. Eur J Radiol. 85:815–823. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ethier JL and Amir E: The Role of the
21-gene recurrence score in breast cancer treatment. Mol Diagn
Ther. 20:307–313. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hoogland AM, Bottcher R, Verhoef E,
Jenster G and van Leenders GJ: Gene-expression analysis of gleason
grade 3 tumor glands embedded in low- and high-risk prostate
cancer. Oncotarget. 7:37846–37856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Printz C: New AJCC cancer staging manual
reflects changes in cancer knowledge. Cancer. 116:2–3. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park HS, Park S, Kim JH, Lee JH, Choi SY,
Park BW and Lee KS: Clinicopathologic features and outcomes of
metaplastic breast carcinoma: Comparison with invasive ductal
carcinoma of the breast. Yonsei Med J. 51:864–869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cho JH, Park JM, Park HS, Park S, Kim SI
and Park BW: Oncologic safety of breast-conserving surgery compared
to mastectomy in patients receiving neoadjuvant chemotherapy for
locally advanced breast cancer. J Surg Oncol. 108:531–536. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Telegrafo M, Rella L, Stabile Ianora AA,
Angelelli G and Moschetta M: Breast MRI background parenchymal
enhancement (BPE) correlates with the risk of breast cancer. Magn
Reson Imaging. 34:173–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Masood S, El-Gabry E, Zhang C and Wang Z:
The potential of identification of a malignancy-associated
biomarker in breast cancer diagnosis and research: hTERT gene DNA
methylation. Diagn Cytopathol. 44:670–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yoon H, Yoon D, Yun M, Choi JS, Park VY,
Kim EK, Jeong J, Koo JS, Yoon JH, Moon HJ, et al: Metabolomics of
Breast Cancer Using High-Resolution Magic Angle Spinning Magnetic
Resonance Spectroscopy: Correlations with 18F-FDG Positron Emission
Tomography-Computed Tomography, Dynamic Contrast-Enhanced and
Diffusion-Weighted Imaging MRI. PLoS One. 11:e01599492016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu S, Berg WA, Zuley ML, Kurland BF,
Jankowitz RC, Nishikawa R, Gur D and Sumkin JH: Breast MRI contrast
enhancement kinetics of normal parenchyma correlate with presence
of breast cancer. Breast Cancer Res. 18:762016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chung MT, Lourenco AP and Mainiero MB:
Screening Breast MRI in Women with a Personal History of Breast
Cancer. Breast J. 22:252–253. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mannan AU, Singh J, Lakshmikeshava R,
Thota N, Singh S, Sowmya TS, Mishra A, Sinha A, Deshwal S, Soni MR,
et al: Detection of high frequency of mutations in a breast and/or
ovarian cancer cohort: Implications of embracing a multi-gene panel
in molecular diagnosis in India. J Hum Genet. 61:515–522. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Soliman NA and Yussif SM: Ki-67 as a
prognostic marker according to breast cancer molecular subtype.
Cancer Biol Med. 13:496–504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Khaled WT, Choon Lee S, Stingl J, Chen X,
RazaAli H, Rueda OM, Hadi F, Wang J, Yu Y, Chin SF, et al: BCL11A
is a triple-negative breast cancer gene with critical functions in
stem and progenitor cells. Nat Commun. 6:59872015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu YL, Yao R, Li J, Zhou YD, Mao F, Pan B
and Sun Q: FOXC1 overexpression is a marker of poor response to
anthracycline-based adjuvant chemotherapy in sporadic
triple-negative breast cancer. Cancer Chemother Pharmacol.
79:1205–1213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong ZB, Shan M, Qian C, Liu T, Shi QY,
Wang J, Liu Y, Liu Y, Huang YX and Pang D: Prognostic significance
of HOXD13 expression in human breast cancer. Int J Clin Exp Pathol.
8:11407–11413. 2015.PubMed/NCBI
|
|
39
|
Mendoza G, Portillo A and Olmos-Soto J:
Accurate breast cancer diagnosis through real-time PCR her-2 gene
quantification using immunohistochemically-identified biopsies.
Oncol Lett. 5:295–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Swayampakula AK, Dillis C and Abraham J:
Role of MRI in screening, diagnosis and management of breast
cancer. Expert Rev Anticancer Ther. 8:811–817. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tozaki M: Diagnosis of breast cancer: MDCT
versus MRI. Breast Cancer. 15:205–211. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schwentner L, Wolters R, Wischnewsky M,
Kreienberg R and Wockel A: Survival of patients with bilateral
versus unilateral breast cancer and impact of guideline adherent
adjuvant treatment: A multi-centre cohort study of 5292 patients.
Breast. 21:171–177. 2012. View Article : Google Scholar : PubMed/NCBI
|