Introduction
The mammalian Toll-like receptors (TLRs) serve key
roles in innate immunity and can react to different microbes,
including gram-positive and -negative bacteria, mycobacteria and
fungi (1–3). TLR2 was previously revealed to
transduce signals from gram-positive bacteria and fungi (3–5), while
TLR4 responded to bacterial lipopolysaccharide (LPS) (6). In addition, TLR4 was previously
revealed to be a key regulator of the innate immune response
following respiratory syncytial virus (RSV) infection (7,8).
During winter epidemics the prevalence of RSV is
higher than any other virus and RSV-associated infections are a
common reason for hospitalization worldwide (9,10). It
has also been identified that RSV infections may develop into
illnesses as mild as common colds to illnesses as serious as severe
lower respiratory tract infections, and the transition between the
mild and severe disease only requires h (11,12).
Thus, it is important for clinicians to effectively screen patients
with an RSV infection.
MicroRNAs (miRs) are small non-coding RNA sequences
that suppress the translation of mRNA through the incomplete base
pairing mechanism (13,14). Previous studies have demonstrated
that miRs are widely involved in the innate and adaptive immune
systems (15,16). For instance, miR-221 was demonstrated
to regulate RSV replication in the human bronchial epithelium
through suppressing β-nerve growth factor expression (16). The primary focus of the present study
was miR-140-5p, which was reported to be differentially expressed
in numerous tumor types, including breast cancer, gastric cancer,
biliary tract cancer (13,14,17).
However, to the best of our knowledge, the specific role of
miR-140-5p in RSV infections has never been explored.
In the current study, the level of miR-140-5p was
identified to be reduced in the nasopharyngeal airway (NPA) and
peripheral blood samples of patients with an RSV infection compared
with normal controls. Additional study revealed that TLR4 was a
target gene of miR-140-5p, suggesting a potential therapeutic
target in the treatment of patients with RSV infection.
Materials and methods
Study design
The heparinized blood and NPA samples were collected
from patients with a bronchiole RSV infection, causing
bronchiolitis, within 24 h of first contact with the hospital
(6.8±3.9 years, n=104; 45% male), and from healthy controls
(6.5±4.1 years, n=40; 55% male) during their annual physical
examination, at Taizhou People's Hospital (Taizhou, China), between
December 2015 and May 2016. The exclusion criteria were as
previously described, including corticosteroid use 48 h prior to
recruitment, serious congenital heart or lung disease and
immunodeficiency, and the presence of ≥15 viral pathogens (18). Based on the severity of the RSV
infection, the patients were divided into three groups. The mild
group consisted of those who were without hypoxia or severe feeding
problems (n=42). The moderate group consisted of those who required
hospitalization to receive supplemental oxygen (oxygen saturation,
<93%) and/or nasogastric feeding (n=34). The severe group
consisted of those who required mechanical ventilation (n=28).
Written informed consent was obtained from the parents of all
children. Human studies were approved by the Taizhou People's
Hospital Ethics Committee.
NPA samples for miRNA analysis
NPA samples were separated from nostrils by deep
nasal suctioning. An RSV infection was determined using a rapid
antigen test (Alere™ BinaxNOW® RSV Card;
Alere, Inc., Waltham, MA, USA) according to the manufacturer's
protocol and in-house reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) analysis.
Total RNA was isolated with RNAzol LS (Vigorous
Biotechnology, Beijing, China) according to the specific
instructions to isolate small RNAs. Quality, quantity and integrity
of RNA were monitored using a NanoDrop spectrophotometer (ND-1,000,
Nanodrop Technologies; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The commercially available TaqMan® Fast Virus
1-Step Master mix (cat no. 4444432; Thermo Fisher Scientific, Inc.)
was used to detect RSV. In brief, 5 µl extracted nucleic acids was
added in a reaction tube containing 5 µl primer probe mixture and
10 µl TaqMan® Fast Virus 1-Step Master mix in order to
perform cDNA synthesis and qPCR. Subsequently, the PCR
amplification was performed using 1 mg of cDNA and SYBR Green
Master mix (Roche Diagnostics, Basel, Switzerland) on a Roche
Lightcycler 480 (Roche Diagnostics). For qPCR, the mixture was
incubated at 50°C for 10 min. Denaturation followed at 95°C for 30
sec, then 10 cycles of PCR (15 sec at 95°C, 30 sec at 53°C and 30
sec at 60°C) and 30 additional cycles of PCR for the detection of
fluorescence signals (15 sec at 95°C, 30 sec at 53°C and 30 sec at
60°C). Relative expression was normalized against the endogenous
control, GAPDH, using the 2−∆∆Ct method (19). The primers used were as follows:
RSV-A subtype-F: AGATCAACTTCTGTCATCCAGCAA; RSV-A-subtype-R:
TTCTGCACATCATAATTAGGAG; RSV-B-subtype-F:
AAGATGCAAATCATAAATTCACAGGA; RSV-B-subtype-R:
TGATATCCAGCATCTTTAAGTA; GAPDH-F: GAGAAGGCTGGGGCTCATTT; GAPDH-R:
AGTGATGGCATGGACTGTGG.
Then, the respiratory secretions, including phlegm
and mucus, were removed from NPA samples by tracheal suction, and
nasal epithelial cells were collected from each nostril by rotating
a cytology brush (Ningbo Finer Medical Instruments Co., Ltd.,
Ningbo, China) over the anterior nasal mucosa. Following that, the
brushes were immersed in the RNA stabilization reagent,
RNAlater® (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). Epithelial cells were detached from the cytology brushes
by placing the brush in a 15 ml conical tube containing 8 ml of
DMEM (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and
transported on ice. The tube was then centrifuged at 4°C at 400 × g
for 5 min and the supernatant was removed. Then, cells were stored
at −80°C in RNAlater for miRNA analysis.
ELISA
To evaluate the role of miR-140-5p in
pro-inflammatory responses, miR-140-5p mimics or inhibitors were
transfected into BEAS-2B cells for 48 h in the presence of 10 µg/ml
LPS and the supernatant was collected for ELISA assay. In addition,
BEAS-2B cells were also incubated with 0.1, 0.5 or 1.0 µg/ml IFNα
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 48 h and
the supernatant was collected for further assay. Dimethyl sulfoxide
(DMSO) was used as a control for IFNα. BEAS-2B cells were treated
in a lysis buffer (50 mmol/l Tris-HCl, 300 mmol/l NaCl, 5 mmol/l
EDTA, 1% Triton X-100, 0.02% sodium azide) containing a protease
inhibitor cocktail (Roche, Mannheim, Germany). The lysates were
centrifuged at 16,000 × g for 15 min at 4°C and supernatants were
used to quantify the levels of TNF-α (cat no. DTA00C; Human TNF-α
Quantikine ELISA kit), IL-6, (cat no. D6050; Human IL-6 Quantikine
ELISA kit), IL-1β (cat no. DLB50; Human IL-1 beta/IL-1F2 Quantikine
ELISA kit), and IL-8 (cat no. D8000C; Human IL-8/CXCL8 Quantikine
ELISA kit) by way of a sandwich ELISA following the manufacturers'
protocols (R&D Systems, Minneapolis, Minnesota, USA). Samples
were read at a 450 nm wavelength using a microplate reader (Model
3550; Thermo Fisher Scientific, Inc.).
Cell culture
Human bronchial epithelial cells, BEAS-2B were
purchased from the American Type Culture Collection (Manassas, VA,
USA) and cultured in Dulbecco's modified Eagle's medium (DMEM)
(Hyclone; GE Healthcare Life Sciences) supplemented with 10% (v/v)
horse serum (Hyclone; GE Healthcare Life Sciences), 100 U/ml
penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) and 0.1
mg/ml streptomycin (Hyclone; GE Healthcare Life Sciences) at 37°C
in a humidified atmosphere with 5% CO2.
RNA extraction
Total RNA (10 µg) from the whole blood samples (5
ml), in tubes containing EDTA, or BEAS-2B cells was isolated using
RNAVzol LS (Vigorous Biotechnology Beijing Co., Ltd., Beijing,
China) in accordance with the manufacturer's protocol. The
concentration and purity of the RNA samples were determined by the
OD260/OD280 ratio using a microplate reader (Model 3550; Thermo
Fisher Scientific, Inc.).
RT-qPCR analysis
For synthesis of cDNA of the specific miR, 1 µg of
the total RNA was reverse transcribed using TaqMan™ MicroRNA
Reverse Transcription kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with specific primers for miR-140-5p and U6
(Shanghai Sangon Technology Co., Ltd., Shanghai, China). To
quantify the miR-140-5p, a qPCR assay was performed using iQ™
SYBR® Green Supermix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) in an iCycler iQ™ qPCR detection system (both
Bio-Rad Laboratories, Inc). The PCR amplifications were performed
in a 10 µl reaction system containing 5 µl SYBR-Green Supermix, 0.4
µl forward primer, 0.4 µl reverse primer, 2.2 µl double distilled
H2O and 2 µl template cDNA. The thermal cycling
conditions were as follows: A hot start step at 95°C for 10 min,
followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min,
annealing at 55°C for 30 sec and elongation at 72°C for 3 min. The
relative level of miR-140-5p was determined using the
2−ΔΔCq analysis method (19). U6 was selected as the internal
control. The primers used in the present study were as follows:
miR-140-5p-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCA-3′; U6-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATG-3′
miR-140-5p, forward, 5′-GCGCGCAGUGGUUUUACCCUA-3′; U6, forward,
5′-GCGCGTCGTGAAGCGTTC-3′; universal reverse primer,
5′-GTGCAGGGTCCGAGGT-3′. RT stands for stem loop primer here.
Transient transfection
Firstly, 6×105 cells were equally seeded
in the 6-well plates with 2 ml DMEM culture medium containing serum
and antibiotics. miR-140-5p mimics, inhibitors or miR negative
controls were purchased from GenePharma (Shanghai, China).
Simultaneously, miR-140-5p mimics, inhibitors or miR negative
controls were mixed with HiperFect transfection reagent (Qiagen
GmbH, Hilden, Germany) and incubated at room temperature for 10
min. Then, each complex was transfected into two wells containing
the BEAS-2B cells for 48 h at a final concentration of 10 nM. The
specific sequences were as follows: miR-140-5p mimic,
CAGUGGUUUUACCCUAUGGUAG; miR-140-5p inhibitor:
CUACCAUAGGGUAAAACCACUG; miR NC for mimic: UUCUCCGAACGUGUCAACGUTT;
miR NC for inhibitor: CAGUACUUUGUGUAGUACAA.
Bioinformatics analysis
TargetScan 7.1 (www.targetscan.org/vert_71/) was applied to predict
the possible target genes of miR-140-5p.
Dual luciferase reporter assay
The 3untranslated region (UTR) of TLR4, which
contains the predicted target site for miR-140-5p, was cloned into
the pmirGLO luciferase reporter vector (Promega Corporation,
Madison, WI, USA), which was cleaved at SacI and XhoI
sites. The two different 3UTR regions containing the potential
binding sites of TLR4 were cloned into the luciferase reporter
system, which were designated pmirGLO-TLR4-3UTR-1 and
pmirGLO-TLR4-3UTR-2. The former contained the first binding site,
while the latter included the second and the third binding sites.
Details of PCR procedures are described as follows: A hot start
step at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec,
55°C for 45 sec and 72°C for 30 sec.
Prior to conducting the dual reporter assay,
5×104 BEAS-2B cells/well were seeded in 24-well plates
with 500 µl DMEM medium and cultured for 18 h. The cells were
transfected with the modified firefly luciferase reporter vector
(500 ng/µl) mixed with Vigofect transfection reagent (Vigorous,
Beijing, China) according to the manufacturer's protocol. After
continuous exposure of miR-140-5p
(CAGUGGUUUUACCCUAUGGUAG)/pmirGLO-TLR4-3′UTR-1 or
pmirGLO-TLR4-3′UTR-2 or NC (UUCUCCGAACGUGUCAACGUTT)/pmirGLO blank
vector for 48 h, the luciferase activities of firefly and Renilla
were measured with the Dual-Luciferase® Reporter Assay
system (Promega Corporation) according to the manufacturer's
protocol. Firefly luciferase activity was normalized to
Renilla luciferase activity.
Western blotting
Cell protein was extracted using
radioimmunoprecipitation lysis buffer (Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China) and was collected
following centrifugation at 12,000 × g for 30 min at 4°C. A
bicinchoninic protein assay kit (Pierce; Thermo Fisher Scientific,
Inc.) was used to determine the protein concentration. A total of
15 µg protein was loaded per lane, separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes. The membranes
were blocked with 8% non-fat dry milk at 4°C overnight. Following
three washes with PBS with Tween 20 (5 min/wash), the membranes
were incubated with the following primary antibodies at 4°C
overnight: TLR4 (cat no. 14358; 1:1,000; Cell Signaling Technology,
Inc., Danvers, MA, USA) and GAPDH (cat. no. 5174; 1:1,000; Cell
Signaling Technology, Inc.). Following several washes with TBST,
the membranes were incubated with horseradish-peroxidase
(HRP)-conjugated goat anti-rabbit and anti-mouse Immunoglobulin G
(IgG) or HRP-conjugated mouse anti-goat IgG (all 1:5,000; Zhongshan
Gold Bridge Biological Technology Co., Beijing, China) for 2 h at
room temperature and then washed. The blots were then incubated
with horseradish peroxidase (HRP)-conjugated anti-IgG secondary
antibody (1:5,000; OriGene Technologies, Inc., Beijing, China) for
2 h at room temperature and then washed followed by detection with
enhanced chemiluminescent substrate (EMD Millipore, Billerica, MA,
USA). GAPDH was used as an internal control. ImageJ software
(National Institutes of Health, Bethesda, MD, USA) was used for
density analysis.
Interferon-α (IFNα) treatment
In brief, BEAS-2B cells were seeded at a density of
5×105 cells/well in a 6-well plate for 24 h. Then,
BEAS-2B cells were incubated with 0.1, 0.5 or 1.0 µg/ml Recombinant
Human IFNα (CYT-283, ProSpec, Rehovot, Israel) or the corresponding
volume of distilled water (control) for 48 h. Then, the RNA was
isolated and the level of miR-140-5p was quantified using RT-qPCR
according to the aforementioned method. The primers used were as
follows: MiR-140-5p-Stem:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTACCA; U6-Stem:
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAATATG; miR-140-5p-F:
GCCAGTGGTTTTACCCTAT; U6-F: GCGCGTCGTGAAGCGTTC; universal reverse:
GTGCAGGGTCCGAGGT.
Statistical analysis
Data are presented as mean ± standard deviation. To
compare two groups, the two-tailed unpaired Student's t test was
performed. For multiple groups comparisons, one-way analysis of
variance followed by Tukey's post hoc test was used. Statistical
tests were performed using SPSS software (version 13.0; SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-140-5p levels decrease in the NPAs
and peripheral blood of patients with an RSV infection
The clinical characteristics of patients with an RSV
infection are demonstrated in Table
I. To determine the level of miR-140-5p in the NPA samples, the
RNA was extracted from the patients with RSV infections and the
healthy controls. Compared with that of the healthy controls,
miR-140-5p expression was significantly lower in patients with RSV
infections (Fig. 1A). The lowest
miR-140-5p level in the NPA samples was exhibited by patients with
severe RSV infections, followed by patients with moderate and mild
infections, which were significantly lower compared with the
controls. Furthermore, the level of miR-140-5p in the peripheral
blood samples of patients with RSV infections and healthy control
was analyzed. Compared with the healthy controls, the level of
miR-140-5p was significantly decreased in patients with RSV
infections, with the biggest decrease in severe RSV, followed by
the moderate and mild disease subgroups (Fig. 1B).
 | Table I.Clinicopathological characteristics of
controls and patients with RSV included in the reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Clinicopathological characteristics of
controls and patients with RSV included in the reverse
transcription-quantitative polymerase chain reaction analysis.
|
|
| Severity of RSV |
|---|
|
|
|
|
|---|
| Characteristic | Control | Mild | Moderate | Severe |
|---|
| n | 40 | 42 | 34 | 28 |
| Age, years | 6.5 (4.1) | 4.6 (3.8) | 5.8 (3.7) | 5.2 (3.9) |
| Male, n (%) | 22 (55) | 18 (43) | 16 (47) | 13 (46) |
| Weight, g | 5,873±1,647 | 6,998±2,350 | 6,435±1,392 | 5,967±1,845 |
| Duration of symptoms,
days |
| 4.6
(3–6) | 4.8 (4–5) | 4.3
(3–5) |
| Admitted to a ward, n
(%) |
| 6
(14) | 34
(100) | 28
(100) |
| Length of stay,
days |
| 0
(0–0) | 3
(1–4) | 4
(2–6) |
| Length of stay >3
days, n (%) |
| 0 (0) | 7
(21) | 16 (57) |
Decreased miR-140-5p expression levels
induce pro-inflammatory responses
The overexpression of miR-140-5p significantly
decreased the levels of pro-inflammatory factors, tumor necrosis
factor (TNF)α (Fig. 2A), interleukin
(IL)-1β (Fig. 2B), IL-6 (Fig. 2C) and IL-8 (Fig. 2D). By contrast, inhibition of
miR-140-5p significantly enhanced the production of TNFα (Fig. 2E), IL-1β (Fig. 2F), IL-6 (Fig. 2G) and IL-8 (Fig. 2H).
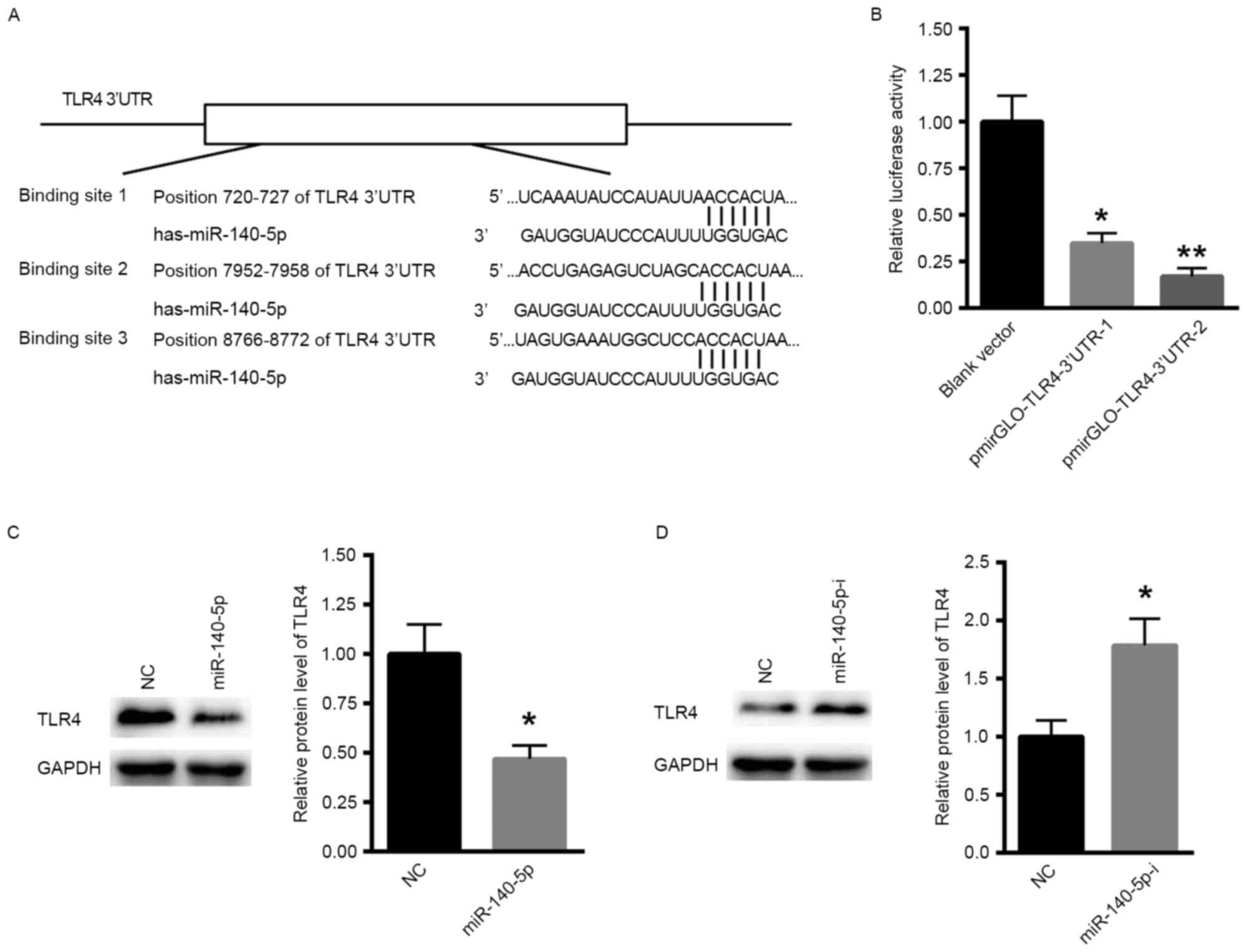
TLR4 may be a direct target of
miR-140-5p
The mechanism underlying the involvement of
miR-140-5p in RSV infections was explored. Bioinformatic analysis
(targetscan.org/vert_71/) demonstrated
that one conserved and two poorly conserved (binding site 1 was
conserved, while binding site 2 and 3 were poorly conserved)
binding sites were identified in the 3UTR of TLR4 (Fig. 3A). The dual luciferase reporter assay
demonstrated that miR-140-5p significantly suppressed the relative
luciferase activity of pmirGLO-TLR4-3UTR-1 and pmirGLO-TLR4-3UTR-2
(Fig. 3B), suggesting that TLR4 is a
direct target gene of miR-140-5p. In addition, the overexpression
of miR-140-5p significantly suppressed the protein level of TLR4 in
BEAS-2B cells (Fig. 3C), while
inhibition of miR-140-5p significantly enhanced the expression of
TLR4 in BEAS-2B cells (Fig. 3D).
IFNα treatment increases the level of
miR-140-5p
BEAS-2B cells were incubated with 0.1, 0.5 or 1.0
µg/ml IFNα for 48 h and the level of miR-140-5p was demonstrated to
increase in a dose-dependent manner following IFNα incubation
(Fig. 4A). miR-140-5p levels
significantly increased with 0.5 and 1.0 µg/ml IFNα compared with
the control. In addition, the supernatant of the BEAS-2B cells
incubated with IFNα was collected for ELISA assay. Following the
incubation of 1.0 µg/ml IFNα, the production of TNFα (Fig. 4B), IL-1β (Fig. 4C), IL-6 (Fig. 4D) and IL-8 (Fig. 4E) in BEAS-2B cells was significantly
suppressed compared with the controls. By contrast, inhibition of
miR-140-5p significantly attenuated the IFNα-mediated
downregulation of TNFα, IL-1β, IL-6 and IL-8 in BEAS-2B cells
compared with the IFNα-treated cells (Fig. 4B-E).
 | Figure 4.INFα treatment increased the level of
miR-140-5p and decreased the levels of TNFα, IL-1β, IL-6 and IL-8.
(A) Quantitative polymerase chain reaction was performed to analyze
miR-140-5p levels following IFNα incubation. Stimulation of IFNα
significantly suppressed the production of (B) TNFα, (C) IL-1β, (D)
IL-6 and (E) IL-8 in BEAS-2B cells. *P<0.05, ***P<0.001 vs.
control; #P<0.05, ##P<0.01 as
indicated. INF, interferon; DMSO, dimethyl sulfoxide; TNF, tumor
necrosis factor; IL, interleukin; miR, microRNA; miR-140-5p-i,
miR-140-5p inhibitor. |
Discussion
The abnormal expression of miRNAs in different
diseases has been widely reported (15,16,20). For
instance, studies have demonstrated that miR-221, let-7i and
miR-30b regulate RSV replication in the human bronchial epithelium
(16,21). In the current study, the expression
of miR-140-5p in the NPA and whole peripheral blood samples of
patients with acute RSV disease was explored. Compared with the
healthy controls, miR-140-5p was lowest in the NPA samples and
peripheral blood of patients with a severe RSV infection, followed
by patients with moderate and mild RSV infections. These in
vivo results indicated that miR-140-5p may function as a
potential biomarker for RSV infections and that there is a negative
association between miR-140-5p levels and the severity of RSV
infections. Notably, the overexpression and inhibition of
miR-140-5p decreased and increased the levels of inflammation
factors, respectively. The in vitro results from the present
study revealed the anti-inflammatory effects of miR-140-5p,
suggesting that inhibition of miR-140-5p contributes to
inflammatory responses and that it may possibly serve a protective
role in preventing RSV infections.
An RSV infection primarily affects the epithelial
cells of the respiratory mucosa (22). By binding to TLR4, RSV triggers the
abnormal activation of the nuclear factor-κB signaling pathway
(23,24). Thus far, the regulation of TLR4
following RSV infection remains unknown. The present study
demonstrates that TLR4 may be a target gene of miR-140-5p. In
vitro investigations revealed that decreased miR-140-5p levels
in the NPA samples contributed to the immune response to RSV, this
may primarily be due to the targeting of TLR4 by miR-140-5p.
Upon RSV infection, type I IFNs are secreted through
the IFNα/β receptor to stimulate the expression of key proteins
that modulate the process of viral replication and immune responses
(25,26). Then, IFNs trigger cell intrinsic
antiviral responses, stimulate natural killer cells, macrophages
and dendritic cells, and modulate the innate and adaptive immune
responses (25,27). IFNα/β expression is induced in
bronchial epithelial cells following the infection of various
viruses, including the influenza virus, RSV and human
metapneumovirus (26,28,29). To
investigate whether miR-140-5p was involved in IFNα-mediated
antiviral therapy in the airways, the expression of miR-140-5p in
BEAS-2B cells treated with IFNα was investigated. It was
demonstrated that treatment with 0.5 or 1.0 µg/ml IFNα
significantly increased miR-140-5p levels in BEAS-2B cells.
Notably, the suppression of miR-140-5p significantly attenuated the
IFNα-mediated downregulation of TNFα, IL-1β, IL-6 and IL-8 in
BEAS-2B cells. These data suggest that miR-140-5p is involved in
INFα-mediated anti-RSV infection therapy.
In conclusion, the current study demonstrated that
miR-140-5p was reduced in the NPA and peripheral blood samples of
patients with RSV infections compared with that of the healthy
controls. Also, inhibiting miR-140-5p enhanced the pro-inflammatory
responses, primarily by targeting TLR4. IFNα treatment was
demonstrated to enhance the level of miR-140-5p in BEAS-2B cells.
Thus, miR-140-5p may be a potential therapeutic target in patients
with RSV infection.
References
|
1
|
Hirschfeld M, Kirschning CJ, Schwandner R,
Wesche H, Weis JH, Wooten RM and Weis JJ: Cutting edge:
Inflammatory signaling by Borrelia burgdorferi lipoproteins is
mediated by toll-like receptor 2. J Immunol. 163:2382–2386.
1999.PubMed/NCBI
|
|
2
|
Means TK, Wang S, Lien E, Yoshimura A,
Golenbock DT and Fenton MJ: Human toll-like receptors mediate
cellular activation by Mycobacterium tuberculosis. J Immunol.
163:3920–3927. 1999.PubMed/NCBI
|
|
3
|
Schwandner R, Dziarski R, Wesche H, Rothe
M and Kirschning CJ: Peptidoglycan- and lipoteichoic acid-induced
cell activation is mediated by toll-like receptor 2. J Biol Chem.
274:17406–17409. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takeuchi O, Hoshino K, Kawai T, Sanjo H,
Takada H, Ogawa T, Takeda K and Akira S: Differential roles of TLR2
and TLR4 in recognition of gram-negative and gram-positive
bacterial cell wall components. Immunity. 11:443–451. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hua F, Ma J, Ha T, Kelley JL, Kao RL,
Schweitzer JB, Kalbfleisch JH, Williams DL and Li C: Differential
roles of TLR2 and TLR4 in acute focal cerebral ischemia/reperfusion
injury in mice. Brain Res. 1262:100–108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ingalls RR, Lien E and Golenbock DT:
Differential roles of TLR2 and TLR4 in the host response to
Gram-negative bacteria: Lessons from a lipopolysaccharide-deficient
mutant of Neisseria meningitidis. J Endotoxin Res. 6:411–415. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Underhill DM and Gantner B: Integration of
Toll-like receptor and phagocytic signaling for tailored immunity.
Microbes Infect. 6:1368–1373. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Underhill DM, Ozinsky A, Smith KD and
Aderem A: Toll-like receptor-2 mediates mycobacteria-induced
proinflammatory signaling in macrophages. Proc Natl Acad Sci USA.
96:14459–14463. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Henrickson KJ, Hoover S, Kehl KS and Hua
W: National disease burden of respiratory viruses detected in
children by polymerase chain reaction. Pediatr Infect Dis J. 23 1
Suppl:S11–S18. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Black CP: Systematic review of the biology
and medical management of respiratory syncytial virus infection.
Respir Care. 48:209–233. 2003.PubMed/NCBI
|
|
11
|
Berger TM, Aebi C, Duppenthaler A and
Stocker M: Swiss Pediatric Surveillance Unit: Prospective
population-based study of RSV-related intermediate care and
intensive care unit admissions in Switzerland over a 4-year period
(2001–2005). Infection. 37:109–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mansbach JM, Clark S, Christopher NC,
LoVecchio F, Kunz S, Acholonu U and Camargo CA Jr: Prospective
multicenter study of bronchiolitis: Predicting safe discharges from
the emergency department. Pediatrics. 121:680–688. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salem O, Erdem N, Jung J, Münstermann E,
Wörner A, Wilhelm H, Wiemann S and Körner C: The highly expressed
5′isomiR of hsa-miR-140-3p contributes to the tumor-suppressive
effects of miR-140 by reducing breast cancer proliferation and
migration. BMC Genomics. 17:5662016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zou J and Xu Y: MicroRNA-140 inhibits cell
proliferation in gastric cancer cell line HGC-27 by suppressing
SOX4. Med Sci Monit. 22:2243–2252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bakre A, Mitchell P, Coleman JK, Jones LP,
Saavedra G, Teng M, Tompkins SM and Tripp RA: Respiratory syncytial
virus modifies microRNAs regulating host genes that affect virus
replication. J Gen Virol. 93:2346–2356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Othumpangat S, Walton C and Piedimonte G:
MicroRNA-221 modulates RSV replication in human bronchial
epithelium by targeting NGF expression. PLoS One. 7:e300302012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu J, Zhang W, Tang H, Qian H, Yang J, Zhu
Z, Ren P and Lu B: Septin 2 accelerates the progression of biliary
tract cancer and is negatively regulated by mir-140-5p. Gene.
589:20–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Templeton KE, Scheltinga SA, Beersma MF,
Kroes AC and Claas EC: Rapid and sensitive method using multiplex
real-time PCR for diagnosis of infections by influenza a and
influenza B viruses, respiratory syncytial virus, and parainfluenza
viruses 1, 2, 3, and 4. J Clin Microbiol. 42:1564–1569. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mi QS, Xu YP, Wang H, Qi RQ, Dong Z and
Zhou L: Deletion of microRNA miR-223 increases Langerhans cell
cross-presentation. Int J Biochem Cell Biol. 45:395–400. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thornburg NJ, Hayward SL and Crowe JE Jr:
Respiratory syncytial virus regulates human microRNAs by using
mechanisms involving beta interferon and NF-κB. MBio. 3:pii:
e00220. 122012. View Article : Google Scholar
|
|
22
|
Schröder NW and Arditi M: The role of
innate immunity in the pathogenesis of asthma: Evidence for the
involvement of toll-like receptor signaling. J Endotoxin Res.
13:305–312. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abe T, Hemmi H, Miyamoto H, Moriishi K,
Tamura S, Takaku H, Akira S and Matsuura Y: Involvement of the
toll-like receptor 9 signaling pathway in the induction of innate
immunity by baculovirus. J Virol. 79:2847–2858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haynes LM, Moore DD, Kurt-Jones EA,
Finberg RW, Anderson LJ and Tripp RA: Involvement of toll-like
receptor 4 in innate immunity to respiratory syncytial virus. J
Virol. 75:10730–10737. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Randall RE and Goodbourn S: Interferons
and viruses: An interplay between induction, signalling, antiviral
responses and virus countermeasures. J Gen Virol. 89:1–47. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Durbin RK, Kotenko SV and Durbin JE:
Interferon induction and function at the mucosal surface. Immunol
Rev. 255:25–39. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
González-Navajas JM, Lee J, David M and
Raz E: Immunomodulatory functions of type I interferons. Nat Rev
Immunol. 12:125–135. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kumagai Y, Takeuchi O, Kato H, Kumar H,
Matsui K, Morii E, Aozasa K, Kawai T and Akira S: Alveolar
macrophages are the primary interferon-alpha producer in pulmonary
infection with RNA viruses. Immunity. 27:240–252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim D, Martinez-Sobrido L, Choi C, Petroff
N, García-Sastre A, Niewiesk S and Carsillo T: Induction of type I
interferon secretion through recombinant Newcastle disease virus
expressing measles virus hemagglutinin stimulates antibody
secretion in the presence of maternal antibodies. J Virol.
85:200–207. 2011. View Article : Google Scholar : PubMed/NCBI
|