Introduction
Hypertrophic scar (HS) is fibrotic abnormality that
is commonly observed after wound healing of human skin (1). Treatments of wounds mainly focus on
promoting wound healing and inhibiting excessive scar formation
(2–4). However, certain contradictions remain
between the two; for instance, growth factors that promote wound
healing may also increase the risk of scar formation (5–7).
Treatments of scars are still empirical (8,9), and are
not always effective (10).
Furthermore, certain scar treatments have severe side effects that
may affect healing (11). Therefore,
there is currently no effective way to promote wound healing and
simultaneously inhibit the formation of scar.
Transforming growth factor (TGF)-β1 is a member of
the TGF superfamily that is generated by multiple types of cell.
TGF-β1 regulates the growth, differentiation and immune functions
of cells. It regulates the production of interleukin (IL)-6 by
human fibroblasts, and facilitates the growth of fibroblasts and
osteoblasts (12). In HS, TGF-β1
exerts its effects via its downstream signal transduction factors,
such as Smad3 and 2. Through this TGF-β1/Smad pathway, scar
formation is promoted (13,14). Another study has demonstrated that in
Smad3 knockout mice, the wound healing rate was significantly
accelerated (15), suggesting that
inhibition of TGF-β1-dependent pathways may achieve the goal of
promoting wound healing and attenuating scar formation.
MicroRNAs (miRNAs or miRs) are a class of small
non-coding RNAs of ~22 nucleotides in length, which have functions
in RNA silencing and post-transcriptional regulation of gene
expression (16). Upregulated
microRNA-20a expression has been reported to inhibit the expression
of TGF-β1 and finally suppresses the proliferation of fibroblasts
(17). Overexpression of miRNAs
upstream of TGF-β1 may be an effective method for the treatment of
scars. For instance, miR-663 has been demonstrated to affect the
proliferation, migration and invasion of tumor cells by regulating
TGF-β1 (18,19). However, to the best of our knowledge,
the effect of miR-663 on fibroblasts in HS has remained to be
demonstrated. The present study therefore investigated the
expression of TGF-β1 mRNA and protein as well as miRNA-663 in HS
and HS-adjacent tissues as well as isolated fibroblasts in order to
elucidate the association between TGF-β1 and miRNA-663 in HS.
Patients and methods
Patients
A total of 51 patients diagnosed with HS at the
Department of Plastic and Cosmetic Surgery of the Second Affiliated
Hospital of Soochow University (Suzhou, China) between December
2013 and February 2016 were included in the present study. HS
tissues (experimental group) and HS-adjacent tissues (control
group) were collected. Among the 51 patients, 18 were male and 33
were female, with an age range of 18–58 years and a median age of
41.6 years. None of the patients had a history of use of hormones,
intake of Chinese medicine or radiochemotherapy. HS of all patients
were formed by scalds or burns, and met the criteria of the Patient
and Observer Scar Assessment Scale classification (20,21). All
procedures were approved by the Ethics Committee of Soochow
University (Suzhou, China). Written informed consent was obtained
from all patients or their families.
Cells
To obtain primary fibroblasts, HS and HS-adjacent
tissues were washed with 0.1 M PBS (pH 7.4) for three times. The
tissues were then digested with 0.25% dispase (Roche Diagnostics,
Basel, Switzerland) at 4°C overnight to remove the skin. After
homogenization, the tissues were digested with 0.1% type I
collagenase (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at
37°C with agitation at 40 × g for 180 min. Digestion was then
terminated by mixing the samples with an equal volume of
low-glucose Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Thermo
Fisher Scientific, Inc.). After centrifugation at 600 × g for 10
min, the supernatant was discarded and the fibroblasts were
resuspended in high-glucose DMEM (Thermo Fisher Scientific, Inc.).
The fibroblasts were seeded onto a culture plate with a diameter of
100 mm at a density of 4×104/cm2 and cultured
at 37°C in an atmosphere containing 5% CO2 with 100%
humidity. The medium was first replenished after 24 h, and replaced
every other day thereafter. The cells were passaged when reaching
confluence.
One day prior to transfection, 3×105
cells in the logarithmic growth phase (second passage) were seeded
onto 24-well plates containing DMEM/F12 medium (Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum, but
without antibiotics. For the silencing of TGF-β1, the
small-interfering RNA (siRNA) of TGF-β1 forward,
5′-ACCCACAACGAAAUCUAUGdTdT-3′ and reverse,
5′-CAUAGAUUUCGUUGUGGGUdTdT-3′ was used to transfect fibroblasts.
Scrambled siRNA (sc-37007; both Sangon Biotech, Co., Ltd.,
Shanghai, China) was used as negative control. For transfection of
fibroblasts with miR-663 mimics (agomiR-663), the cells were
divided into control group (untreated), negative control (NC;
transfected with empty plasmids) group and miR-663 mimics group.
When reaching 70% confluence, 1.25 µl miR-663 mimics plasmid
(Sangon Biotech, Co., Ltd.) and 1 µl Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.) were added into two individual vials each
containing 50 µl OptiMEM medium (Thermo Fisher Scientific, Inc.).
Five minutes later, the liquids in the two vials were mixed and
left to stand for another 20 min. The mixture was then added onto
the cells for an incubation of 6 h. After replacing the medium with
fresh DMEM/F12 supplemented with 10% fetal bovine serum, the
fibroblasts were cultured under normal conditions for 48 h prior to
use.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
Tissues (100 mg) were ground into powder using
liquid nitrogen, followed by addition of 1 ml TRIzol (10606ES60;
Yeasen, Shanghai, China) for lysis. Total RNA was the extracted
using the phenol chloroform method. The purity of RNA was
determined by measuring the 260/280 nm absorbance using ultraviolet
spectrophotometry (Nanodrop ND1000; Thermo Fisher Scientific,
Inc.). The complementary (c)DNA was obtained by RT using the
TIANScript II cDNA first-strand synthesis kit (cat. no. KR107;
Tiangen, Beijing, China) from 1 µg RNA and stored at −20°C. The
primers for TGF-β1 were: Forward, 5′-GGACACCAACTATTGCTTCAG-3′ and
reverse, 5′-TCCAGACTCCAAATGTAG-3′. The primers for internal
reference, β-actin, were as follows: Forward,
5′-TTCCAGCCTTCCTTCCTGG-3′ and reverse, 5′-TTGCGCTCAGGAGGAGGAAT-3′.
The PCR reaction system (25 µl) was composed of 10 µl SuperReal
PreMix (SYBR-Green; cat. no. FP204; Tiangen), 0.5 µl forward
primer, 0.5 µl reverse primer, 2 µl cDNA and 7 µl double-distilled
H2O. The thermocycling conditions were as follows:
Initial denaturation at 95°C for 2 min, followed by 30 cycles of
denaturation at 94°C for 45 sec, annealing at 55°C for 55 sec and
elongation at 72°C for 1 min, and one final elongation step at 72°C
for 10 min (iQ5; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The 2−ΔΔCq method (22)
was used to calculate the relative expression of TGF-β1 mRNA
against β-actin.
To assess the expression of miR-663, the miRcute
miRNA qPCR detection kit (FP401; Tiangen) was selected, using U6 as
an internal reference. The primers for miR-663 were as follows:
Forward, 5′-GCGAGCACAGAATTAATACGAC-3′ and reverse,
5′-AGGCGGGGCGCCGCG-3′. The primers for U6 were: Forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The PCR thermocycling conditions were as follows: Initial
denaturation at 95°C for 5 min; 40 cycles of denaturation at 95°C
for 15 sec and annealing at 60°C for 30 sec; and final elongation
at 72°C for 20 sec (iQ5). The 2−ΔΔCq method was used to
calculate the relative expression of miR-663 against U6.
Western blot analysis
Tissues (100 mg) were ground using liquid nitrogen
and mixed with 200 µl pre-cooled radioimmunoprecipitation assay
lysis buffer (600 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1%
sodium dodecyl sulfate, 1% TritonX-100, 1% sodium deoxycholate;
Beyotime Institute of Biotechnology, Shanghai, China) for lysis on
ice for 30 min. The mixture was then centrifuged at 12,000 × g and
4°C for 15 min. The supernatant was used to determine the protein
concentration by using a bicinchoninic acid protein concentration
determination kit (cat no. RTP7102; Real-Times Biotechnology Co.,
Ltd., Beijing, China). Protein samples (20 µg) were then mixed with
sodium dodecyl sulfate loading buffer prior to denaturation in a
boiling water bath for 5 min. Afterwards, the samples were
subjected to 10% SDS-PAGE. The resolved proteins were transferred
onto polyvinylidene difluoride membranes (Merck KGaA; Darmstadt,
Germany) on ice (100 V, 2 h) and blocked with 5% skimmed milk at
room temperature for 1 h. The membranes were then incubated with
rabbit anti-human TGF-β1 polyclonal primary antibody (1:500
dilution; cat. no. ab92486) and rabbit anti-human β-actin primary
antibody (1:5,000 dilution; cat. no. ab129348; both Abcam,
Cambridge, UK) at 4°C overnight. After extensive washing with PBS
containing Tween-20 (PBST) 3 times for 15 min each, the membranes
were incubated with polyclonal goat anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:3,000 dilution; cat.
no. ab6721; Abcam) for 1 h at room temperature prior to washing
with PBST 3 times for 15 min each. The membrane was developed with
an enhanced chemiluminescence detection kit (Sigma-Aldrich; Merck
KGaA) for imaging with GelDoc XR+ (Bio-Rad Laboratories, Inc.).
Image lab v3.0 software (Bio-Rad Laboratories, Inc.) was used to
acquire and analyze imaging signals. The relative content of TGF-β1
protein was expressed as the TGF-β1/β-actin ratio.
MTT assay
After transfection, cells were seeded into 96-well
plates at a density of 2×103 cells per well. Each
condition was tested in triplicate wells. At 24, 48 and 72 h
following transfection, 20 µl MTT (5 g/l; JRDC000003, JRDUN
Biotechnology, Shanghai, China) was added to each well, followed by
incubation for 4 h at 37°C. Following the aspiration of medium,
DMSO (150 µl per well) was added to dissolve purple crystals. Then,
the absorbance of each well was measured at 490 nm with a
microplate reader (Bio-Rad Laboratories, Inc.) and cell
proliferation curves were plotted.
Dual luciferase reporter assay
Bioinformatics prediction is a powerful tool for the
study of the functions of miRNAs. To understand the regulatory
mechanism of TGF-β1 in HS, miRanda (http://www.microma.org/rnicroma/home.do), TargetScan
(http://www.targetscan.org), PiTa
(http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
and PICTA (http://pictar.mdc-berlin.de/) were used to predict
miRNA molecules that regulate TGF-β1, and miR-663 identified as a
potential regulator of TGF-β1 (Fig.
1). According to these bioinformatics results, wild-type (WT)
and mutant seed regions of miR-663 of the 3′-untranslated region
(3′UTR) of TGF-β1 mRNA were chemically synthesized as described
previously (23). SpeI and HindIII
restriction sites were added, and the sequences were cloned into
pMIR-REPORT luciferase reporter plasmids (Ambion; Thermo Fisher
Scientific, Inc.). Plasmids (0.8 µg) with WT or mutant 3′-UTR DNA
sequences were co-transfected with agomiR-663 (100 nM; Sangon
Biotech, Co., Ltd.) into 293T cells (Type Culture Collection of the
Chinese Academy of Sciences, Shanghai, China). The cells in
negative control (NC) group were transfected with the
aforementioned empty plasmid. After cultivation for 24 h, the cells
were lysed using a dual luciferase reporter assay kit according to
the manufacturer's manual, and the fluorescence intensity was
measured using a GloMax 20/20 luminometer (both Promega Corp.,
Madison, WI, USA). Using Renilla fluorescence activity as an
internal reference, the fluorescence values of each group of cells
were measured.
Statistical analysis
The results were analyzed using SPSS 18.0
statistical software (SPSS, Inc., Chicago, IL, USA). Values are
expressed as the mean ± standard deviation. Data were tested for
normality. In the case of homogeneity of variance, the Least
Significant Differences and Student-Newman-Keuls tests were used,
while in the case of heterogeneity of variance, Tamhane's T2 or
Dunnett's T3 method was used. P<0.05 was considered to indicate
a statistically significant difference.
Results
TGF-β1 has a regulatory role in
HS
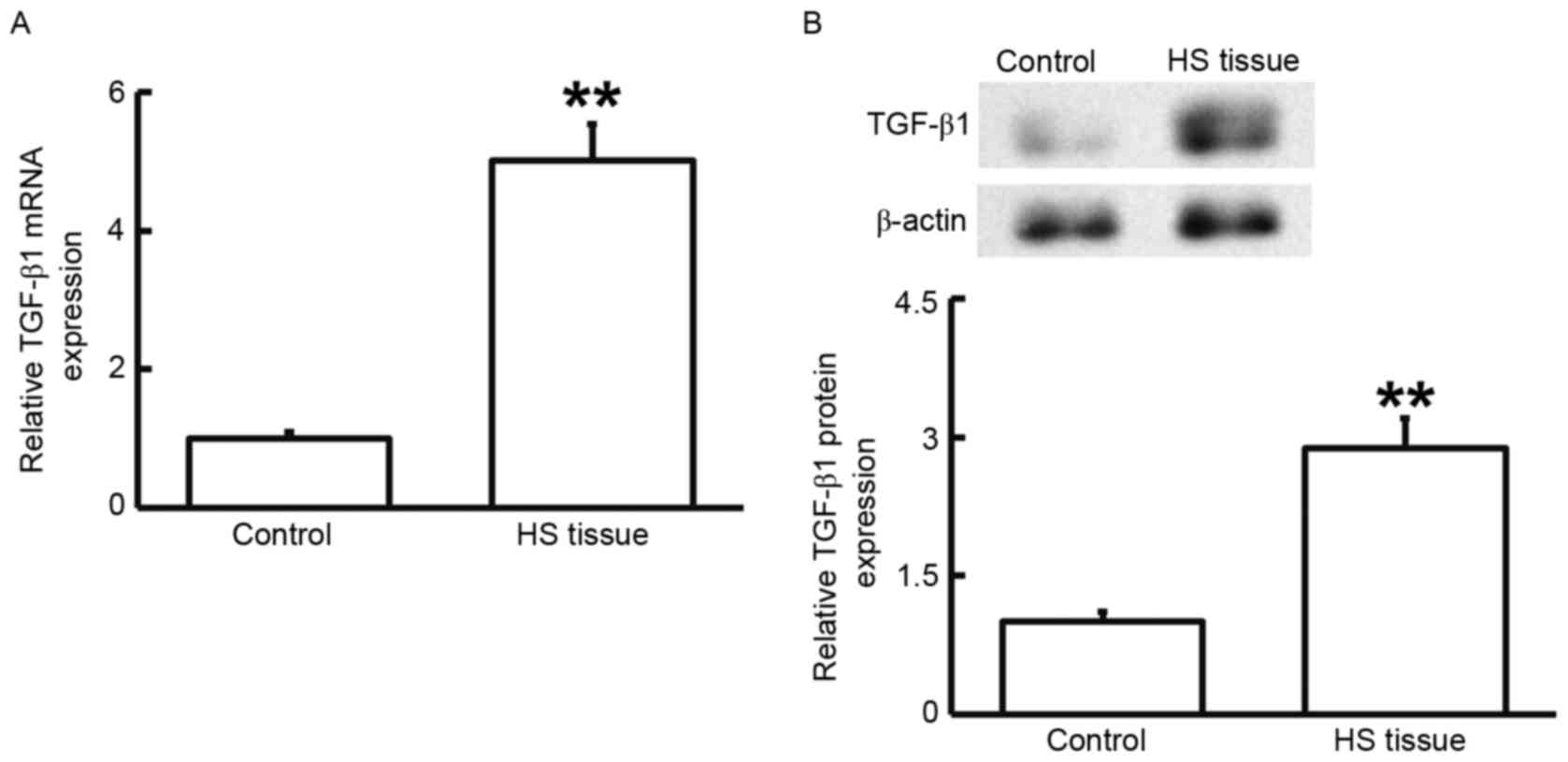
To measure the mRNA and protein expression of
TGF-β1, RT-qPCR and western blot analysis were performed. As
presented in Fig. 2, the levels of
TGF-β1 mRNA and protein in HS tissues were significantly higher
than those in HS-adjacent tissues (P<0.01). These results
suggested that TGF-β1 has a regulatory role in HS.
Reduced TGF-β1 expression leads to
inhibited proliferation of fibroblasts
To assess the effect of TGF-β1 on the proliferation
of fibroblasts, the cells were transfected with siRNA targeting
TGF-β1 and subjected to an MTT assay. RT-qPCR and western blot
analysis demonstrated that the expression of TGF-β1 mRNA and
protein was significantly reduced after transfection with the siRNA
targeting TGF-β1 (P<0.05; Fig.
3). In addition, an MTT assay demonstrated that the
proliferation of fibroblasts transfected with siRNA targeting
TGF-β1 was significantly reduced at 72 h compared with that in the
control group (P<0.05; Fig. 4).
The results indicated that reduced TGF-β1 expression leads to
inhibited proliferation of fibroblasts.
Expression of miR-663 is downregulated
in HS
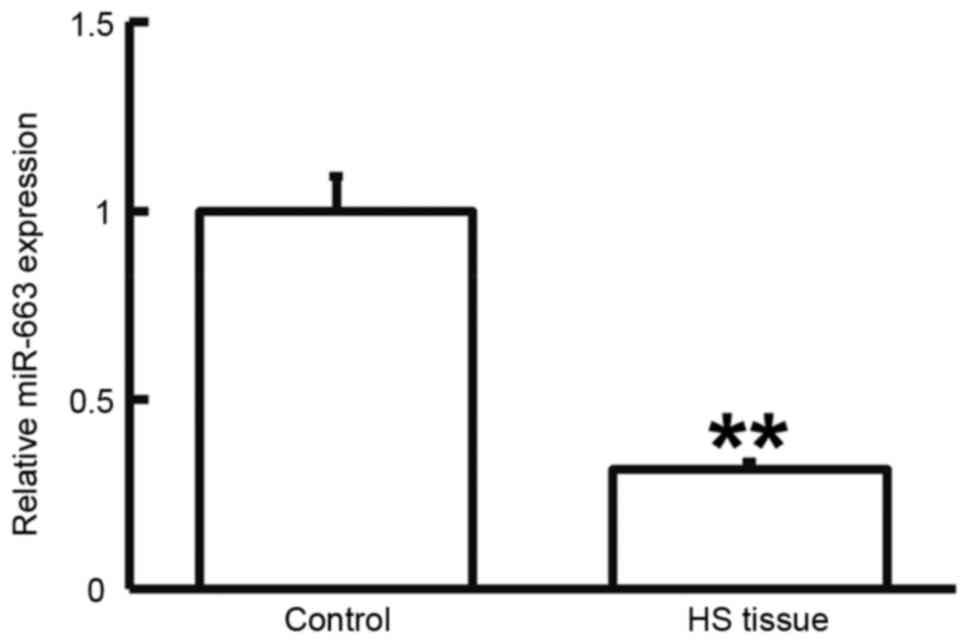
To determine the expression of miR-663, RT-qPCR was
performed. As presented in Fig. 5,
miR-663 levels in HS tissues were significantly lower than those in
HS-adjacent tissues (P<0.05). This result suggested that the
expression of miR-663 is downregulated in HS.
miR-663 regulates the expression of
TGF-β1 by binding with the 3′-UTR of TGF-β1
To test whether miR-663 targets TGF-β1, a dual
luciferase reporter assay was performed. As displayed in Fig. 6, transfection with agomiR-663 and
pMIR-REPORT in the WT group resulted in a significantly reduced
fluorescence intensity compared with that in the negative control
group (P<0.05), while fluorescence intensity in the mutant group
was not significantly different from that in the negative control
group (P>0.05). This result indicated that miR-663 regulates the
expression of TGF-β1 by binding with the 3′-UTR of TGF-β1.
Elevated expression of miR-663
inhibits the proliferation of fibroblasts by regulating TGF-β1
expression
To examine how miR-663 affects the proliferation of
fibroblasts, the cells were transfected with agomiR-663 and
subjected to an MTT assay. RT-qPCR analysis revealed that the
levels of miR-663 in fibroblasts were significantly enhanced after
transfection with agomiR-663 (P<0.01; Fig. 7A). In addition, the expression of
TGF-β1 mRNA was significantly reduced after transfection with
agomiR-663 (P<0.05; Fig. 7B). Of
note, the MTT assay demonstrated that the proliferation of
fibroblasts transfected with agomiR-663 was significantly reduced
at 48 and 72 h compared with that in the control group (P<0.05;
Fig. 8). These results suggested
that elevated expression of miR-663 inhibits the proliferation of
fibroblasts by regulating TGF-β1 expression.
Discussion
The pathological essence of HS is the excessive
proliferation of cells (mainly fibroblasts) and the excessive
deposition of extracellular matrix (mainly collagen). However, the
pathogenesis of HS has remained to be fully elucidated. The
mechanism of HS formation is the metabolic imbalance between the
synthesis and decomposition of extracellular matrix components such
as collagen. Excessive contracture caused by scar hyperplasia often
leads to a variety of abnormalities, leading to pain or functional
disorders of the human body (24).
Several studies have reported the inhibition of cell proliferation
by TGF-β1, with the underlying mechanism mainly comprising the
regulation of Smad3 by TGF-β1 (25,26).
Certain studies have revealed that reduced expression of TGF-β1 is
a significant characteristic of non-scar healing of fetuses,
leading to a faster healing rate, more active cell proliferation,
lower type I collagen content and no scar formation compared with
the skin healing in adults (27–29).
Therefore, non-scar healing of fetuses is an ideal tissue repair
status, and reduced expression of TGF-β1 has an important role in
achieving this ideal healing (30).
In the present study, TGF-β1 expression was found to be elevated in
HS tissues, and the proliferation of primary fibroblasts
transfected with siRNA targeting TGF-β1 was inhibited. These
results suggested that abnormally elevated expression of TGF-β1 may
have an important role in HS.
The regulatory mechanisms of TGF-β1 have remained to
be fully elucidated. Studies have demonstrated that Nemo-like
kinase and Smad proteins regulate the expression of TGF-β1
(31,32). In addition, miRNAs have been
identified to target the mRNA of TGF-β1 to regulate the translation
of TGF-β1, and this process is important in normal development and
physiology, as well as disease conditions. Certain studies have
shown that miR-663 regulates TGF-β1 by direct targeting. Li et
al (18) reported that miR-663
affects the proliferation, migration and invasion of glioma cells
via TGF-β1. Hong et al (33)
discovered that miR-663 is upregulated in epithelial cells in a
high uric acid environment, and affects the phosphatase and tensin
homolog gene by targeting TGF-β1. Another study reported that
miR-663 regulates TGF-β1 expression in a pattern of negative
feedback and reduces the radiation-induced bystander effect of
cells (34). The results of the
present study demonstrated that expression of miR-663 was
downregulated in HS, while TGF-β1 was upregulated, revealing an
inverse association. After stimulation with miR-663 mimics, the
proliferation of fibroblasts from HS tissues was reduced, and
TGF-β1 mRNA and protein levels were decreased, suggesting that
miR-663 regulates TGF-β1. The dual luciferase reporter assay
confirmed this regulatory association. Taken into consideration the
various factors associated with HS other than TGF-β1 (6,7), a vast
variety of miRNA molecules other than miR-663 regulate these
factors.
In conclusion, miRNA-663 was found to negatively
regulate the expression of TGF-β1 in HS, which may provide a novel
approach for the prevention and treatment of HS. However, the
actual effect and mechanism of action of miR-663 still require
further study in cells, animals and clinical experiments.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Development Project of Suzhou City (grant no.
SYSD2015094). The authors would also like to thank Dr Joel Thomson
from Department of Dermatology, Bestway Medical Company, Taiwan for
English-proofreading of this manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QC and TZ designed the present study; QC, XX, DY,
LW, WY and WS performed the experiments; QC, TZ and XX analyzed the
data. The final version of the manuscript was read and approved by
all authors and each collaborated to interpret results and develop
the manuscript.
Ethics approval and consent to
participate
All procedures were approved by the Ethics Committee
of Soochow University (Suzhou, China). Written informed consent was
obtained from all patients or their families.
Patient consent for publication
Written informed consent for publication of any
associated data and accompanying images were obtained from all
patients or their parents, guardians or next of kin.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Niessen FB, Spauwen PH, Schalkwijk J and
Kon M: On the nature of hypertrophic scars and keloids: A review.
Plast Reconstr Surg. 104:1435–1458. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Werner S and Grose R: Regulation of wound
healing by growth factors and cytokines. Physiol Rev. 83:835–870.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Berman B, Viera MH, Amini S, Huo R and
Jones IS: Prevention and management of hypertrophic scars and
keloids after burns in children. J Craniofac Surg. 19:989–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Menke MN, Menke NB, Boardman CH and
Diegelmann RF: Biologic therapeutics and molecular profiling to
optimize wound healing. Gynecol Oncol. 111 2 Suppl:S87–S91. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Younai S, Venters G, Vu S, Nichter L,
Nimni ME and Tuan TL: Role of growth factors in scar contraction:
An in vitro analysis. Ann Plast Surg. 36:495–501. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilgus TA, Ferreira AM, Oberyszyn TM,
Bergdall VK and Dipietro LA: Regulation of scar formation by
vascular endothelial growth factor. Lab Invest. 88:579–590. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grella A, Kole D, Holmes W and Dominko T:
FGF2 overrides TGFβ1-driven integrin ITGA11 expression in human
dermal fibroblasts. J Cell Biochem. 117:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferguson MW and O'Kane S: Scar-free
healing: From embryonic mechanisms to adult therapeutic
intervention. Philos Trans R Soc Lond B Biol Sci. 359:839–850.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reish RG and Eriksson E: Scar treatments:
Preclinical and clinical studies. J Am Coll Surg. 206:719–730.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mofikoya BO, Adeyemo WL and Abdus-salam
AA: Keloid and hypertrophic scars: A review of recent developments
in pathogenesis and management. Nig Q J Hosp Med. 17:134–139.
2007.PubMed/NCBI
|
|
11
|
Leventhal D, Furr M and Reiter D:
Treatment of keloids and hypertrophic scars: A meta-analysis and
review of the literature. Arch Facial Plast Surg. 8:362–368. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Janda K, Krzanowski M, Dumnicka P,
Kuśnierz-Cabala B, Kraśniak A and Sułowicz W: Transforming growth
factor beta 1 as a risk factor for cardiovascular diseases in
end-stage renal disease patients treated with peritoneal dialysis.
Clin Lab. 60:1163–1168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Massagué J: Wounding Smad. Nat Cell Biol.
1:E117–E119. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu W, Wang DR and Cao YL: TGF-beta: A
fibrotic factor in wound scarring and a potential target for
anti-scarring gene therapy. Curr Gene Ther. 4:123–136. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashcroft GS, Yang X, Glick AB, Weinstein
M, Letterio JL, Mizel DE, Anzano M, Greenwell-Wild T, Wahl SM, Deng
C and Roberts AB: Mice lacking Smad3 show accelerated wound healing
and an impaired local inflammatory response. Nat Cell Biol.
1:260–266. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zou M, Wang F, Gao R, Wu J, Ou Y, Chen X,
Wang T, Zhou X, Zhu W, Li P, et al: Autophagy inhibition of
hsa-miR-19a-3p/19b-3p by targeting TGF-β R II during TGF-β1-induced
fibrogenesis in human cardiac fibroblasts. Sci Rep. 6:247472016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Correia AC, Moonen JR, Brinker MG and
Krenning G: FGF2 inhibits endothelial-mesenchymal transition
through microRNA-20a-mediated repression of canonical TGF-β
signaling. J Cell Sci. 129:569–579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Q, Cheng Q, Chen Z, Peng R, Chen R, Ma
Z, Wan X, Liu J, Meng M, Peng Z and Jiang B: MicroRNA-663 inhibits
the proliferation, migration and invasion of glioblastoma cells via
targeting TGF-β1. Oncol Rep. 35:1125–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Y, Liu J, Fan L, Wang F, Yu H, Wei W
and Sun G: miR-663 overexpression induced by endoplasmic reticulum
stress modulates hepatocellular carcinoma cell apoptosis via
transforming growth factor beta 1. Onco Targets Ther. 9:1623–1633.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van de Kar AL, Corion LU, Smeulders MJ,
Draaijers LJ, van der Horst CM and van Zuijlen PP: Reliable and
feasible evaluation of linear scars by the patient and observer
Scar assessment scale. Plast Reconstr Surg. 116:514–522. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoogewerf CJ, van Baar ME, Middelkoop E
and van Loey NE: Patient reported facial scar assessment:
Directions for the professional. Burns. 40:347–353. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen
T, Chen Z, Huang S, Gu J, Li J, et al: MiR-199a-5p is negatively
associated with malignancies and regulates glycolysis and lactate
production by targeting hexokinase 2 in liver cancer. Hepatology.
62:1132–1144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gorney M: Scar: The trigger to the claim.
Plast Reconstr Surg. 117:1036–1037. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hong F, Wu N, Ge Y, Zhou Y, Shen T, Qiang
Q, Zhang Q, Chen M, Wang Y, Wang L and Hong J: Nanosized titanium
dioxide resulted in the activation of TGF-β/Smads/p38MAPK pathway
in renal inflammation and fibration of mice. J Biomed Mater Res A.
104:1452–1461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou B, Zeng S, Li L, Fan Z, Tian W, Li M,
Xu H, Wu X, Fang M and Xu Y: Angiogenic factor with G patch and FHA
domains 1 (Aggf1) regulates liver fibrosis by modulating TGF-β
signaling. Biochim Biophys Acta. 1862:1203–1213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Colwell AS, Longaker MT and Lorenz HP:
Fetal wound healing. Front Biosci. 8:s1240–s1248. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Colwell AS, Longaker MT and Lorenz HP:
Mammalian fetal organ regeneration. Adv Biochem Eng Biotechnol.
93:83–100. 2005.PubMed/NCBI
|
|
29
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Beanes SR, Dang C, Soo C, Wang Y, Urata M,
Ting K, Fonkalsrud EW, Benhaim P, Hedrick MH, Atkinson JB and
Lorenz HP: Down-regulation of decorin, a transforming growth
factor-beta modulator, is associated with scarless fetal wound
healing. J Pediatr Surg. 36:1666–1671. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shi Y, Ye K, Wu H, Sun Y, Shi H and Huo K:
Human SMAD4 is phosphorylated at Thr9 and Ser138 by interacting
with NLK. Mol Cell Biochem. 333:293–298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiao Z, Zhang J, Peng X, Dong Y, Jia L, Li
H and Du J: The Notch γ-secretase inhibitor ameliorates kidney
fibrosis via inhibition of TGF-β/Smad2/3 signaling pathway
activation. Int J Biochem Cell Biol. 55:65–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hong Q, Yu S, Geng X, Duan L, Zheng W, Fan
M, Chen X and Wu D: High concentrations of uric acid inhibit
endothelial cell migration via miR-663 which regulates phosphatase
and tensin homolog by targeting transforming growth factor-β1.
Microcirculation. 22:306–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu W, Xu S, Yao B, Hong M, Wu X, Pei H,
Chang L, Ding N, Gao X, Ye C, et al: MiR-663 inhibits
radiation-induced bystander effects by targeting TGFB1 in a
feedback mode. RNA Biol. 11:1189–1198. 2014. View Article : Google Scholar : PubMed/NCBI
|