Introduction
Lumbar disc degeneration (LDD) is a worldwide
orthopedic disease. In particular, 80–90% of the population aged
over 60 years are suffering from LDD. LDD is a common disease
affecting the health of the elderly (1,2). The
pathogenesis of LDD is relatively complicated, and the risk factors
are not only related to the external environment and labor
involved, but also closely associated with the genetic factors of
the body. Recent studies have reported that multiple human genetic
mutations can make lumbar disc more susceptible to disease
(3). Growth differentiation factor 5
(GDF5) is a transforming growth factor, which is a key protein
factor in the growth and development of bone and cartilage and
plays an important role in the formation of bone, especially joint
(4,5). Some findings have revealed that GDF5
can affect the expression of isotypic collagenase gene by virtue of
the multiple differentiation and proliferation abilities of stem
cells, thereby improving the structure of the lumbar disc in rats
(6). As a single nucleotide
polymorphism (SNP) site in GDF5 gene, rs143383 is located in
5′ non-coding region of GDF5 gene, and a mutation at this
site is sure to result in downregulated gene expression, decreasing
GDF5 gene expression in the body, thereby increasing the
onset risk of LDD (7,8). This study investigated the correlation
between SNP rs143383 in GDF5 and LDD.
Materials and methods
General data
A total of 210 patients with LDD diagnosed and
treated in Shanghai General Hospital of Nanjing Medical University
(Shanghai, China) from August 2013 to March 2017 were randomly
selected as observation group, and 320 patients without lumbar
diseases diagnosed and treated in the hospital during the same
period were randomly selected as control group. In the observation
group, there were 120 males and 90 females aged 39–81 years, with
mean age of 64.2±19.3 years. Among them, in terms of Thompson's
pathological grading, there were 21 cases of Grade I, 45 cases of
Grade II, 38 cases of Grade III, 56 cases of Grade IV and 50 cases
of Grade V. Control group had 190 males and 130 females aged 37–83
years, whose mean age was 65.5±19.7 years. All patients were aware
of this study and signed the informed consent, and this study was
approved by the Ethics Committee of Shanghai General Hospital of
Nanjing Medical University. There were no statistically significant
differences in sex, age, living habits, between he two groups
(P>0.05) (Table I), and the
results were comparable.
 | Table I.General clinical data. |
Table I.
General clinical data.
| Parameters | Observation group
(n=210) | Control group
(n=320) | P-value |
|---|
| Age (years) | 64.2±19.3 | 65.5±19.7 | 0.462 |
| Sex |
|
| 0.733 |
| Male | 120 (57.1%) | 190 (59.4%) |
|
|
Female | 90
(42.9%) | 130 (40.6%) |
|
| Body mass index (BMI)
(kg/m2) | 23.4±3.6 | 23.6±3.8 | 0.754 |
| Smoking |
|
| 0.415 |
| Yes | 86
(41%) | 137 (42.8%) |
|
| No | 124 (59%) | 183 (57.2%) |
|
| Drinking |
|
| 0.536 |
| Yes | 121 (57.6%) | 191 (59.7%) |
|
| No | 89 (42.4%) | 129 (40.3%) |
|
Extraction of genomic deoxyribonucleic
acid (DNA)
Whole blood (5 ml) was collected from each patient
with an anticoagulant tube containing ethylenediamine tetraacetic
acid dipotassium (EDTAK2). Then, genomic DNA was extracted from the
blood using an Omega Mag-Binds Forensic DNA kit (Omega Bio-Tek,
Inc., Norcross, GA, USA). After that, the concentration and purity
of DNA were determined by NanoDrop, and DNA was stored at
−20°C.
SNP typing via polymerase chain
reaction (PCR)
Primer sequences and their Taqman probe sequences at
SNP site (Table II) designed by
Oligo6.0 were used. Primer synthesis was accomplished by Sangon
Biotech Co., Ltd. (Shanghai, China). DNA solution (1 µl) and 1.2 µl
prepared primer solution (including 0.4 µl upstream primer, 0.4 µl
downstream primer and 0.4 µl probe primer) were added to 17.8 µl
pre-prepared TransStart Probe qPCR SuperMix (Beijing TransGen
Biotech Co., Ltd., Beijing, China), slightly shaken to mix, and
placed into a CFX96 fluorescent quantitative PCR instrument
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Reaction
conditions: i) 94°C for 3 min, for 1 cycle; ii) 94°C for 15 sec and
60°C for 60 sec, for 42 cycles. After each cycle, the fluorescence
value was read once. The experimental results were generated by
built-in software of the instrument. Three replicate wells were
made for each sample, diethyl pyrocarbonate (DEPC) water was used
as negative control, and positive plasmid containing the sequence
(synthesized by Sangon Biotech) was used as positive control.
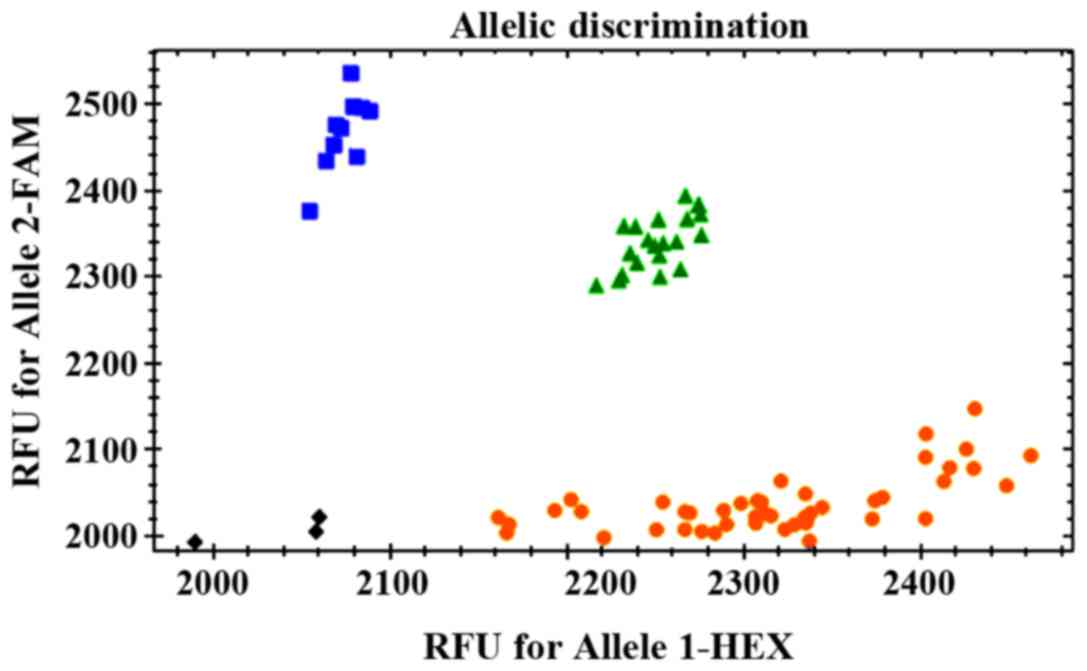
Determination of genotypes: The wild homozygous genotype was near
the FAM abscissa, the mutant homozygous genotype was near the VIC
ordinate, and the heterozygous genotype was near the 45° line.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| SNP | Primer sequence | Probe sequence |
|---|
| rs143383 | Upstream:
5′-CAGGCAGCATTACGCCATTCTTC-3′ | FAM:
5′-CGGTCGGCTTTCTCCTTTCAAG-3′ |
|
| Downstream:
5′-CACCGTCTCCAGTCAGCAGCTG-3′ | VIC:
5′-CGGTTGGCTTTCTCCTTTCAAG-3′ |
Statistical analysis
Statistical Product and Service Solutions (SPSS; IBM
Corp., Armonk, NY, USA) 19.0 software was used for statistical
analysis. Chi-square test was employed for statistical analyses of
genotype distribution differences between case group and control
group. Logistic regression analysis was adopted for the
associations between various genotypes and the risk of LDD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Distributions of genotypes at rs143383
in two groups
A total of 210 patients with LDD and 320 patients in
control group obtained clear genotyping results (Fig. 1). There were no statistically
significant differences in distribution frequency of two genotypes,
namely, TT and TC, at site rs143383 between LDD patient group and
control group (P>0.05), but the distribution frequency of CC
genotype at rs143383 in LDD patient group showed a statistically
significant difference compared with that in control group
(P<0.05) (Table III). The
distributions in two groups were in line with Hardy-Weinberg
equilibrium [P(control)=0.31 and P(observation)=0.35].
 | Table III.Distribution frequency at rs143383 (n,
%). |
Table III.
Distribution frequency at rs143383 (n,
%).
|
| Observation group
(n=210) | Control group
(n=320) |
|
|
|---|
|
|
|
|
|
|
|---|
| Genotype | n | % | n | % | χ2
value | P-value |
|---|
| TT | 125 | 59.5 | 204 | 63.8 | 0.391 | 0.532 |
| TC | 64 | 30.5 | 108 | 33.7 | 0.235 | 0.628 |
| CC | 21 | 10 | 8 | 2.5 | 4.8 | 0.028 |
| T | 314 | 74.8 | 516 | 80.6 | 0.971 | 0.324 |
| C | 106 | 25.2 | 124 | 19.4 |
|
|
Onset risks of LDD analyzed in
different models
In dominant models, odds ratio (OR) of (TC+CC/TT)
was 1.195 (P=0.532). In recessive models, OR of (CC/TT+TC) was
4.333 (P=0.028). In co-dominant models, ORs of (TC/TT) and (CC/TT)
were 0.967 and 4.43, respectively (P=0.09) (Table IV).
 | Table IV.Onset risks of LDD analyzed in
different models. |
Table IV.
Onset risks of LDD analyzed in
different models.
| Model type | Genotype | Observation group
(n=210) | Control group
(n=320) | OR value [95%
confidence interval (CI)] | P-value |
|---|
| Dominant model | TT | 125 | 204 | 1 | 0.532 |
|
| TC+CC | 85 | 116 | 1.195
(0.732–1.532) |
|
| Recessive model | TT+TC | 189 | 312 | 1 | 0.028 |
|
| CC | 21 | 8 | 4.333
(2.321–7.786) |
|
| Co-dominant
model | TT | 125 | 204 | 1 | 0.09 |
|
| TC | 64 | 108 | 0.967
(0.657–1.214) |
|
|
| CC | 21 | 8 | 4.43
(2.451–7.698) |
|
Association between genotypes at
rs143383 and clinicopathologic grade of LDD
There were no statistically significant differences
in three genotypes (TT, TC and CC) among different pathological
grades (Grade I–V) (χ2=1.034, P=0.998), and the
differences in T and C also showed no statistical significance
(χ2=0.012, P=0.999) (Table
V).
 | Table V.Genotypes at rs143383 and
distributions of clinicopathologic grades in LDD patients (n,
%). |
Table V.
Genotypes at rs143383 and
distributions of clinicopathologic grades in LDD patients (n,
%).
|
|
| rs143383 |
|---|
|
|
|
|
|---|
| Grade | n (210) | TT (n=125) | TC (n=64) | CC (n=21) | T (n=314) | C (n=106) |
|---|
| I | 36 | 21 (16.8) | 12 (18.8) | 3 (14.3) | 54 (17.2) | 18 (17) |
| II | 48 | 29 (23.2) | 14 (21.9) | 5 (23.8) | 72 (22.9) | 24 (22.6) |
| III | 41 | 24 (19.2) | 13 (20.3) | 4 (19) | 61 (19.4) | 21 (19.8) |
| IV | 45 | 27 (21.6) | 13 (20.3) | 5 (23.8) | 67 (21.3) | 23 (21.7) |
| V | 40 | 24 (19.2) | 12 (18.8) | 4 (19) | 60 (19.1) | 20 (18.9) |
| χ2 |
|
| 1.034 |
| 1.012 |
|
| P-value |
|
| 0.998 |
| 0.999 |
|
Associations between different models
and clinicopathologic grades of LDD using logistic regression
analysis
Pathological grades in dominant models, recessive
models and co-dominant models were analyzed, and the results showed
that there were no statistically significant differences among
pathological grades in dominant models, as well as in recessive
models and co-dominant models. (P>0.05) (Table VI).
 | Table VI.Associations between different models
and clinicopathologic grades of LDD (n, %). |
Table VI.
Associations between different models
and clinicopathologic grades of LDD (n, %).
| Model type | Genotype | Grade I (36) | Grade II (48) | Grade III (41) | Grade IV (45) | Grade V (40) | P-value |
|---|
| Dominant model | TT | 21 (16.8) | 29 (23.2) | 24 (19.2) | 27 (21.6) | 24 (19.2) | 0.999 |
|
| TC+CC | 15 (17.6) | 19 (22.4) | 17 (20) | 18 (21.2) | 16 (18.8) |
|
| Recessive
model | TT+TC | 33 (17.5) | 43 (22.8) | 37 (19.6) | 40 (21.2) | 36 (19) | 0.975 |
|
| CC | 3
(14.3) | 5
(23.8) | 4 (19) | 5
(23.8) | 4 (19) |
|
| Co-dominant
model | TC | 12 (18.8) | 14 (21.9) | 13 (20.3) | 13 (20.3) | 12 (18.8) | 0.990 |
|
| TT+CC | 24 (16.4) | 34 (23.3) | 28 (19.2) | 32 (21.9) | 28 (19.2) |
|
Discussion
The expression function of genes may be influenced
by many factors including the effects of non-coding regions and
various regulators and the changes of gene structure caused by SNP
in gene, affecting the action of translation function, indirectly
influencing health, and leading to a variety of diseases (9,10).
Therefore, predicting the risks of associated diseases via
statistical analyses of SNPs in the body becomes very meaningful.
rs143383 is located in 5′ non-coding region of GDF5 gene,
where the promoter of gene manipulation system is located, so the
regulation of GDF5 gene expression will inevitably be
affected (11). Moreover, the main
function of GDF5 gene is to code transforming factors that
are closely related to the regeneration and formation of bone.
Therefore, rs143383 polymorphism is certainly associated with
bone-related diseases (12,13). The incidence of LDD is mainly
correlated with following factors: age is increased, the rate of
bone formation is lower than that of bone loss, osteoporosis
occurs, the elasticity of bone deteriorates, the ability to
withstand load declines, and fracture and dislocation occur easily
due to external pressure (14).
Recent studies have found that mutations in GDF5 gene are
highly correlated with osteoarthritis, developmental dysplasia of
hip and lumbar disc-related diseases (15).
Yin et al (16) used a variety of SNP typing methods
and reported that Taqman probe method has an irreplaceable
advantage in terms of the accuracy of typing. In this study, this
method was used, and good genotyping results were obtained in
patients with LDD and patients in control group. Tsezou (17) demonstrated that C/T at rs143383 is
highly associated with LDD in White Europeans. This study found
that CC genotype at rs143383 had very high association and risk
with LDD in Chinese Han population with LDD. Among them, OR of
(CC/TT+TC) was 4.333 (P=0.028) in recessive models, and OR of
(CC/TT) was 4.43 (P=0.021) in co-dominant models. A study by
Huétink et al (18) suggested
that there is a certain association between GDF5 gene and
the severity of osteoarthritis in White Europeans. However, no
significant differences were found in CC, CT and TT genotypes among
different pathological grades (Grade I–V) in Chinese Han population
(χ2=1.034, P=0.998), and there were also no
statistically significant differences in T and C
(χ2=0.012, P=0.999). In addition, no significant
differences were found in dominant models, recessive models and
over dominant models among different pathological grades
(P>0.05). It is possible that SNP mutations in GDF5 gene
have various effects on different bone diseases in different ethnic
groups. Therefore, it is necessary to analyze the correlations
between base mutations and LDD in different populations.
Furthermore, there may be some differences in the results of the
associations between SNP and diseases due to diverse sample sizes,
so a larger sample size is more meaningful (19,20).
In conclusion, CC mutant type at rs143383 in
GDF5 gene is strongly associated with the incidence of LDD
and has a higher prevalence risk, but it is not significantly
correlated with pathological grade.
Acknowledgements
The authors thank all the study participants and the
participating general practitioners contributing to all the study
cohorts.
Funding
This study is granted by National Natural Science
Fund of China (no. 81572169).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
ZW and YL collected and analyzed the general data of
patients. YW and XW extracted genomic deoxyribonucleic acid. JZ and
JT performed PCR. All authors read and approved the final version
of the manuscript
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Shanghai General Hospital of Nanjing Medical University (Shanghai,
China). All patients were aware of this study and signed the
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kanna RM, Shetty AP and Rajasekaran S:
Patterns of lumbar disc degeneration are different in degenerative
disc disease and disc prolapse magnetic resonance imaging analysis
of 224 patients. Spine J. 14:300–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colombier P, Clouet J, Hamel O, Lescaudron
L and Guicheux J: The lumbar intervertebral disc: From embryonic
development to degeneration. Joint Bone Spine. 81:125–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma T, Guo CJ, Zhao X, Wu L, Sun SX and Jin
QH: The effect of curcumin on NF-κB expression in rat with lumbar
intervertebral disc degeneration. Eur Rev Med Pharmacol Sci.
19:1305–1314. 2015.PubMed/NCBI
|
|
4
|
Zheng CJ and Chen J: Disc degeneration
implies low back pain. Theor Biol Med Model. 12:242015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li YF, Tang XZ, Liang CG, Hui YM, Ji YH,
Xu W, Qiu W and Cheng LM: Role of growth differentiation factor-5
and bone morphogenetic protein type II receptor in the development
of lumbar intervertebral disc degeneration. Int J Clin Exp Pathol.
8:719–726. 2015.PubMed/NCBI
|
|
6
|
Eskola PJ, Lemmelä S, Kjaer P, Solovieva
S, Männikkö M, Tommerup N, Lind-Thomsen A, Husgafvel-Pursiainen K,
Cheung KM, Chan D, et al: Genetic association studies in lumbar
disc degeneration: A systematic review. PLoS One. 7:e499952012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Williams FM, Popham M, Hart DJ, de
Schepper E, Bierma-Zeinstra S, Hofman A, Uitterlinden AG, Arden NK,
Cooper C, Spector TD, et al: GDF5 single-nucleotide polymorphism
rs143383 is associated with lumbar disc degeneration in Northern
European women. Arthritis Rheum. 63:708–712. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reynard LN, Bui C, Syddall CM and Loughlin
J: CpG methylation regulates allelic expression of GDF5 by
modulating binding of SP1 and SP3 repressor proteins to the
osteoarthritis susceptibility SNP rs143383. Hum Genet.
133:1059–1073. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mu J, Ge W, Zuo X, Chen Y and Huang C:
Analysis of association between IL-1β, CASP-9, and GDF5 variants
and low-back pain in Chinese male soldier: Clinical article. J
Neurosurg Spine. 19:243–247. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deng B, Ren JZ, Meng XQ, Pang CG, Duan GQ,
Zhang JX, Zou H, Yang HZ and Ji JJ: Expression profiles of MMP-1
and TIMP-1 in lumbar intervertebral disc degeneration. Genet Mol
Res. 14:19080–19086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei F, Zhong R, Wang L, Cui S, Liu S, Zou
X, Zhou Z and Liang Z: Relationship between bone mineral density
and lumbar intervertebral disc degeneration in rhesus macaques.
Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 28:718–722. 2014.(In
Chinese). PubMed/NCBI
|
|
12
|
Gologorsky Y and Chi J: Genetic
predisposition to lumbar disc degeneration. Neurosurgery.
74:N10–N11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou H, Zhu F, Qiu Y, Zhu Z, Liu Z, Bao H,
He S and Qiao J: Effect of intervertebral disc degeneration on
spinal flexibility in patients with degenerative lumbar scoliosis.
Zhonghua Wai Ke Za Zhi. 52:739–744. 2014.(In Chinese). PubMed/NCBI
|
|
14
|
Syddall CM, Reynard LN, Young DA and
Loughlin J: The identification of trans-acting factors that
regulate the expression of GDF5 via the osteoarthritis
susceptibility SNP rs143383. PLoS Genet. 9:e10035572013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan F, Tian J, Winzenberg T, Ding C and
Jones G: Association between GDF5 rs143383 polymorphism and knee
osteoarthritis: An updated meta-analysis based on 23,995 subjects.
BMC Musculoskelet Disord. 15:4042014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin Y and Wang Y: Association of BMP-14
rs143383 ploymorphism with its susceptibility to osteoarthritis: A
meta-analysis and systematic review according to PRISMA guideline.
Medicine (Baltimore). 96:e74472017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsezou A: Osteoarthritis year in review
2014: Genetics and genomics. Osteoarthritis Cartilage.
22:2017–2024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huétink K, van der Voort P, Bloem JL,
Nelissen RG and Meulenbelt I: Genetic contribution to the
development of radiographic knee osteoarthritis in a population
presenting with nonacute knee symptoms a decade earlier. Clin Med
Insights Arthritis Musculoskelet Disord. 9:57–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ratnayake M, Plöger F, Santibanez-Koref M
and Loughlin J: Human chondrocytes respond discordantly to the
protein encoded by the osteoarthritis susceptibility gene GDF5.
PLoS One. 9:e865902014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mu J, Ge W, Zuo X, Chen Y and Huang C: A
SNP in the 5′UTR of GDF5 is associated with susceptibility to
symptomatic lumbar disc herniation in the Chinese Han population.
Eur Spine J. 23:498–503. 2014. View Article : Google Scholar : PubMed/NCBI
|