Introduction
Chlamydia trachomatis is responsible for the
largest proportion of all sexually transmitted diseases (STDs)
reported to the CDC (1). In the USA,
1,598,354 chlamydial infections were reported in 2016,
corresponding to a rate of 497.3 cases per 100,000 individuals, and
from 2008 to 2015, the rate of reported chlamydial infections
increased from 32.48 to 37.18 cases per 100,000 individuals in
China (2). C.
trachomatis-induced genital tract infections may cause
inflammation, edema and mucosal discharge (3), and ascending uterine infections may
lead to pelvic inflammatory disease, tubal scarring, ectopic
pregnancies and infertility (4).
After entering a cell, C. trachomatis
inclusions absorb nutrients and live in the organelles of the host
cell. A mixture of apoptotic features and atypical cell death
during infection has been demonstrated to occur during the process
of cell death induction (5–7). One of the primary obstacles faced by
chlamydial researchers has been the lack of genetic techniques for
the creation of mutant chlamydial strains, which are necessary for
a thorough exploration of chlamydial pathogenesis. The genetic
tools that are widely used in other bacteria are not applicable to
Chlamydia because of its obligate, intracellular lifestyle
and unique development cycle, hindering the development of vaccines
and therapies (5–7). Until effective vaccines are developed,
screening and treatment procedures appear to be the best approach
for preventing chlamydia-related disease.
All six chlamydiaphages isolated from
Chlamydia (Chp1, Chp2, Chp3, Chp4, φCPG1 and φCPAR39)
(8–12) share similar features, and the high
homology has led to a hypothesis of cross-reactions between
species. Molecular characterization indicates that these six
chlamydiaphages belong to the family Microviridae (8–14). To
date, a C. trachomatis-specific phage has not yet been
detected. φCPG1 is a lytic phage that infects Chlamydia
caviae, a guinea pig inclusion conjunctivitis strain (10,13). The
φCPG1 genome has five open reading frames (ORFs): ORFs 1–3 encode
capsid proteins Vp1, Vp2 and Vp3, and ORFs 4–5 encode proteins VG4
and VG5 (13). The capsid protein
Vp1 plays a crucial role in the adhesion and invasion of
Chlamydia. A genome-wide analysis has revealed a similarity
of 83–95% among the six chlamydiaphage Vp1 capsid proteins
(14).
Vp1 capsid proteins of φCPG1 have been demonstrated
to inhibit C. trachomatis growth and cause a decrease in the
number of C. trachomatis inclusion bodies during infection
(15–17). Vp1 has also been indicated to exert
inhibitory effects on the proliferation of C. trachomatis in
the mouse genital tract (16). In
the present study, whether Vp1 alleviates the cytotoxicity induced
by Chlamydia trachomatis was investigated. C.
trachomatis inclusion bodies were counted under a fluorescence
microscope, and the chlamydial Hsp60 protein levels were evaluated
by western blotting. In addition, the interactions between C.
trachomatis and Vp1 were investigated. C.
trachomatis-induced host cell apoptosis was detected following
Vp1 treatment by flow cytometric analysis. Furthermore, the protein
levels of the host cell proapoptotic p53 protein and the
transcription levels of the antiapoptotic genes Mcl-1 and
cIAP-2 were evaluated by western blotting and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
respectively.
Materials and methods
Vp1 expression, identification and
purification
The Escherichia coli strain BL21 (Tiangen
Biotech Co., Ltd., Beijing, China) with the pET30a(+)/Vp1 plasmid
(EMD Millipore, Billerica, MA, USA) was cultured in Luria-Bertani
medium (Beijing Solarbio Science and Technology Co., Ltd., Beijing,
China) containing kanamycin (50 mg/ml) at 37°C with shaking until
the optical density reached 0.6 at 600 nm.
Isopropyl-β-D-thiogalactoside (Beijing Solarbio Science and
Technology Co., Ltd.) was added to the culture at a concentration
of 0.5 mmol/l, and the mixture was then shaken at 37°C for 3 h.
E. coli cells were then collected by centrifugation at
10,000 × g and 4°C for 5 min. Following resuspension of the cell
pellet in PBS, 3% Triton X-100 and 4 mg/l lysozyme were added. Vp1
protein was released from E. coli following sonication
(ultrasound for 10 sec and pause for 6 sec for a total of 8 min;
all at 0°C). Centrifugation (12,000 × g, 20 min, 4°C) was performed
to remove impurities, then Vp1 protein was purified using a
His.Bind® Purification kit (Merck KGaA, Darmstadt,
Germany). Protein renaturation was performed using gradient
dialysis in PBS. Lipopolysaccharides were neutralized using a
ToxinEraser™ endotoxin removal kit (GenScript,
Piscataway, NJ, USA) and detected using a ToxinSensor Gel Clot
Endotoxin Assay kit (GenScript). The concentration of Vp1 protein
was quantified using a bicinchoninic acid protein assay kit (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). The Vp1 protein was
stored at −80°C until subsequent experiments.
CCK-8 assay of cell viability
The Cell Counting Kit-8 (Dojindo Molecular
Technologies, Inc., Shanghai, China) was used according to the
manufacturer's protocol. The CCK-8 reagent (10 µl) was mixed with
0.1 ml Dulbecco's modified Eagle's medium (DMEM; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) supplemented with 8%
fetal bovine serum (FBS; Tianjin Haoyang Biological Manufacture
Co., Ltd. Tianjin, China). Then, 1×105 HeLa cells (Type
Culture Collection of the Chinese Academy of Sciences, Shanghai,
China) were added and incubated in 96-well plates with 60 µg/ml
Vp1. The absorbance of the medium was read at 450 nm using an ELISA
instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at 24,
48 and 72 h. The experiments were repeated three times.
C. trachomatis infection and Vp1
treatment
Previous studies indicated that both HeLa cells and
normal human cervical cells could be suitable for the experiments
in the current study. For example, González et al (18) and Siegl et al (19) investigated the association between
Chlamydia infection and P53 using HeLa cells and normal
human cells. HeLa cells are often used for the generation and
culture of C. trachomatis (5–7).
Therefore, HeLa cells were selected as a research model. HeLa cells
were grown in DMEM supplemented with 10% FBS and incubated at 37°C
with 5% CO2 prior to chlamydial infection. HeLa cells
were seeded in a 24-well plate and after 24 h, the cells were
pre-treated with Diethylaminoethyl-dextran (Merck KGaA) for 30 min
to increase their susceptibility to C. trachomatis
infection. C. trachomatis strain E (ATCC, Manassas, VA, USA)
was subjected to two freeze-thaw cycles, followed by vortex at 3,
200 rpm for 1 min, room temperature.. Next, C. trachomatis
cells were pre-incubated with 60 µg/ml purified Vp1 in PBS or 60
µg/ml bovine serum albumin (BSA; Beijing Solarbio Science and
Technology Co., Ltd.) in PBS for 1 h. According to our previous
study, 60 µg/ml was selected as the optimal concentration of Vp1
and BSA (15). Following Vp1
pretreatment, C. trachomatis cells were used to infect HeLa
cells at a multiplicity of infection (MOI) of 1. C.
trachomatis adhesion was facilitated by centrifugation at 32°C
and 500 × g for 1 h. Then, DMEM (without cycloheximide) was added,
and the HeLa cells were subsequently incubated at 37°C with 5%
CO2 for 40 h. Mock-infected cells were subjected to the
same procedure without C. trachomatis.
Immunofluorescence microscopy
C. trachomatis-infected or uninfected HeLa
cells grown on glass coverslips were washed with PBS and fixed with
ice-cold methanol for 15 min. Fixed cells were washed three times
and treated with 0.1% Triton-X-100 for 8 min at room temperature.
The cells were then washed three times in PBS and blocked in 10%
BSA in PBS for 1 h at 37°C. The cells were then washed three times
in PBS and reacted with an antibody against C. trachomatis E
serotype (obtained from Professor Guangming Zhong, University of
Texas Health Science Center at San Antonio, San Antonio, USA)
diluted at 1:2,000 in 10% BSA in PBS at 4°C overnight. Following
three washes in PBS, the primary antibody-stained monolayers were
co-reacted with Cyc3-conjugated goat anti-rabbit antibodies (red;
Abcam, Cambridge, MA, USA; cat. no. ab6939; 1:70) in 10% BSA and
Hoechst 32258 (blue) for 50 min at 37°C. Images were acquired using
a fluorescence microscope. The single-color images were merged
using Adobe Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA, USA).
The experiments were repeated three times.
SDS-PAGE and western blotting
Cellular protein was extracted from HeLa cells using
radioimmunoprecipitation assay buffer (Beijing Solarbio Science and
Technology Co., Ltd.). Lysates were centrifuged at a speed of 500 ×
g for 5 min at 4°C, and sample buffer was added to the sediment.
Lysates were heated to 100°C for 5 min, then analyzed by 10%
SDS-PAGE. The protein was quantified using a BCA assay kit (Thermo
Fisher Scientific, Inc.). A total of 15 µl protein was loaded per
lane. Following electrophoresis, the proteins were transferred onto
PVDF membranes (EMD Millipore, Billerica, MA, USA). The membranes
were blocked with 3% BSA in TBS for 2 h at room temperature. The
membranes were subsequently incubated with antibodies against P53
(Wanleibio Co., Ltd., Shanghai, China; cat. no. WL103333) at a
dilution of 1:2,000, overnight at 4°C. The horseradish peroxidase
conjugated anti-mouse immunoglobulin G antibody (Cell Signalin g
Technology, Inc., Danvers, MA, USA; cat. no. 14709) was added at a
dilution of 1:10,000 and incubated for 2 h at room temperature.
Using an enhanced chemiluminescence kit (Merk KGaA), the membranes
were photographed and densitometry was performed using Image J
software (V 1.8.0; National Institutes of Health, Bethesda, MD,
USA). The level of chlamydial Hsp60 as an indicator of the
infection load was detected at different time points following
infection (14, 20, 24, 30 and 36 h). The p53 protein levels were
normalized to β-actin, which was used as an internal control. The
experiments were repeated three times.
RT-qPCR
Total RNA was extracted from HeLa cells at 48 h post
infection using an RNeasy Mini kit (Qiagen, Inc., Valencia, CA,
USA) according to the manufacturer's protocol. Then, 2 µg of total
RNA was used to synthesize first-strand cDNA in a 20-µl reaction
using a M-MLV Reverse Transcriptase kit (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
cDNA product (1 µl) was used for qPCR with an ABI 7500 Fast system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with
SYBR® Premix Ex Taq™ II (Takara Bio, Inc.,
Otsu, Japan) using the specified primer sets. The primers were
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China; Table I). The PCR procedure was as follows:
95°C for 3 min, followed by 40 cycles of 94°C for 5 sec and 60°C
for 30 sec. Using the 2−∆∆Cq method (20), the transcription levels of target
genes were analyzed using the β-actin gene as an internal control.
All experiments were repeated three times.
 | Table I.Primers used for quantitative
polymerase chain reaction. |
Table I.
Primers used for quantitative
polymerase chain reaction.
| Gene | Primer (5′-3′) |
|---|
| β-actin | F,
CCTGGCACCCAGCACAAT |
|
| R,
CTGATCCACATCTGCTGGAA |
| Puma | F,
CGACCTCAACGCACAGTACGA |
|
| R,
AGGCACCTAATTGGGCTCCAT |
| Mcl-1 | F,
GCCAAGGACACAAAGCCAAT |
|
| R,
CCGTCGCTGAAAACATGGAT |
| cIAP-2 | F,
CTGTGATGGTGGACTCAGGT |
|
| R,
TTCATCTCCTGGGCTGTCTG |
Flow cytometric analysis
Following incubation and infection, adherent and
floating cells were collected by trypsinization followed by
centrifugation at a speed of 300 × g for 3 min at room temperature.
HeLa cells were then washed with PBS and resuspended in Annexin
V-fluorescein isothiocyanate (FITC) binding buffer. Annexin V-FITC
was then added, and the cells were incubated for 10 min in the dark
according to the manufacturer's protocol (BD Biosciences, Franklin
Lakes, NJ, USA). Then, propidium iodide was added, and the cell
suspension was analyzed within 1 h on a flow cytometer using
CellQuest software V 5.1 (BD Biosciences).
Statistical analysis
The data were analyzed using SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). Graphs were generated using GraphPad Prism v6.01
(GraphPad Software, Inc., La Jolla, CA, USA). Two-tailed Student's
t-test was used to analyze datasets containing two groups. One-way
analysis of variance with Dunnett's multiple comparison test was
used to analyze datasets containing multiple groups. Data are
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
In vitro cell viability following Vp1
exposure
Recombinant His-tagged Vp1 was expressed in E.
coli and purified using a His-Bind Purification kit. Total
protein was extracted from E. coli, and the purified Vp1
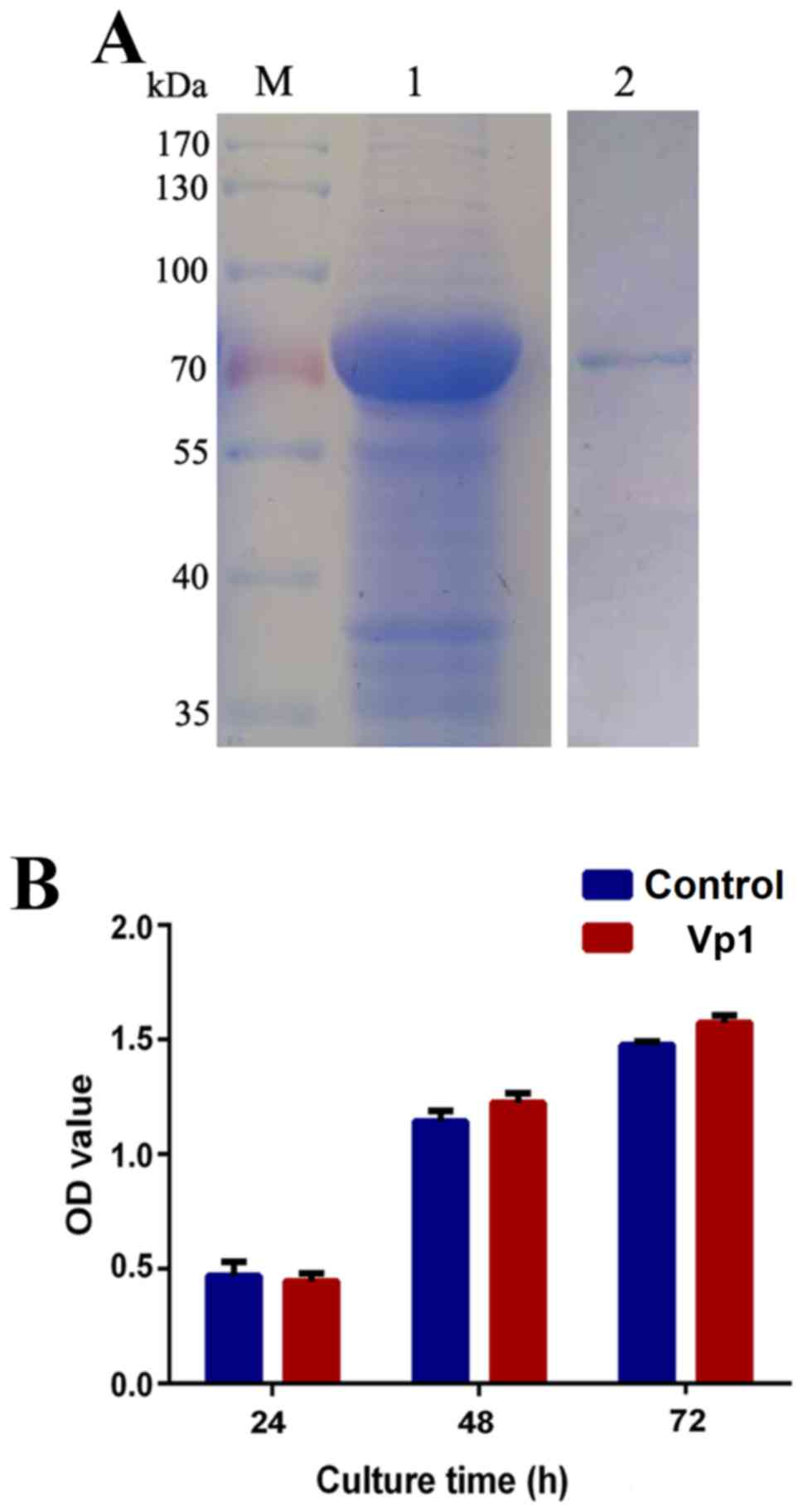
protein was separated by SDS-PAGE (Fig.
1A). Lipopolysaccharides were neutralized using a ToxinEraser
endotoxin removal kit and detected using a ToxinSensor Gel Clot
Endotoxin Assay kit. Purified Vp1 protein was stored at −80°C until
subsequent experimentation. HeLa cells were treated with Vp1 at a
concentration of 60 µg/ml for various durations (24 to 72 h), and a
CCK-8 assay was performed at the end of each exposure. Mock cells
cultured in Vp1-free medium were tested simultaneously. The in
vitro cell viability assay indicated that compared with
untreated HeLa cells, cells exposed to Vp1 for various times (24,
48 or 72 h) exhibited no cytotoxic effects (Fig. 1B). This finding suggests that Vp1 may
not have cytotoxic effects in in vivo studies.
C. trachomatis inclusion numbers
decrease following Vp1 treatment
To detect the inhibitory effect of Vp1 on C.
trachomatis development, C. trachomatis cells were
pre-incubated with BSA or purified Vp1 prior to infection. HeLa
cells infected with C. trachomatis at an MOI of 1 were
incubated with Vp1 or BSA, fixed with methanol at 48 h
post-infection (hpi) and reacted with the corresponding antibodies.
Irregular-shaped C. trachomatis cells with a bright-red
fluorescence were observed in the HeLa cell monolayers at 48 hpi,
and blue fluorescence indicated the cellular DNA. The number of
C. trachomatis inclusions exhibited a notable decrease
following 48 h of incubation in the Vp1-treated group compared with
the BSA-treated group (Fig. 2A and
B). C. trachomatis was quantified by counting the
inclusion bodies per visual field for 20 separate fields. The
average number per visual field of the BSA- and Vp1-treated groups
was 74.90±1.852 and 21.60±1.268, respectively (Fig. 2C). The inhibition rate reached 71% in
the Vp1-treated group, demonstrating that the addition of Vp1
protein during the C. trachomatis culture process could
significantly reduce the number of inclusion bodies. The total
protein from infected cells of the Vp1-treated group was extracted
at different time points, and chlamydial Hsp60 protein was detected
by western blotting as an indicator of the infection load. The
amount of Hsp60 protein gradually decreased after 24 hpi,
indicating that Vp1 may exert an inhibitory action during the later
stages of infection (Fig. 2D). These
results indicated that Vp1 exerts a notable inhibitory effect on
C. trachomatis.
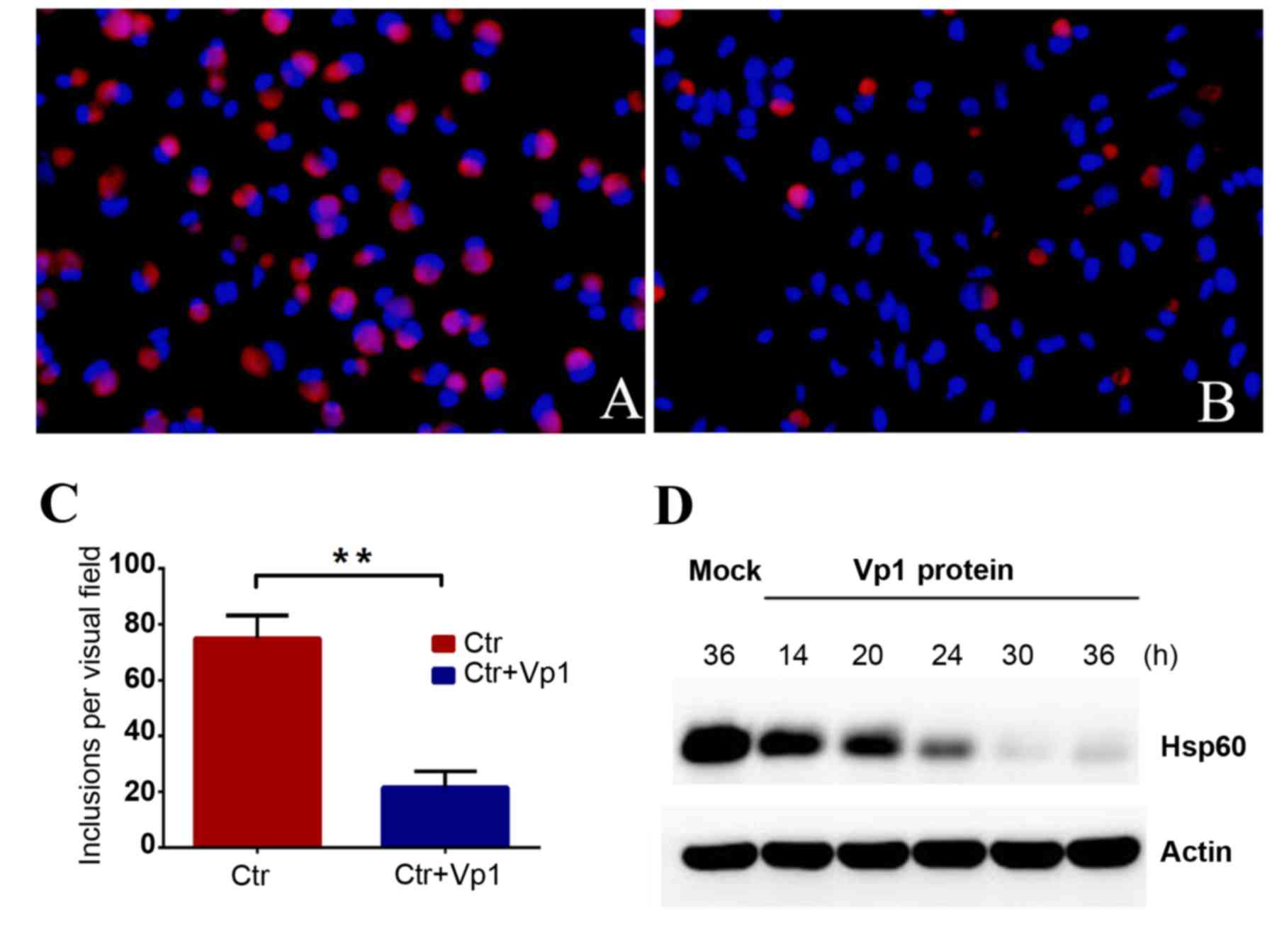 | Figure 2.Chlamydia trachomatis
inclusion numbers decreased in the Vp1-treated group. C.
trachomatis cells were pre-treated with BSA or purified Vp1. (A
and B) Bright-red, irregular-shaped fluorescing cells are
chlamydial organisms and blue fluorescence indicates DNA.
Magnification, × 200. (A) BSA-treated C.
trachomatis-infected group at 48 hpi. (B) Vp1-treated C.
trachomatis-infected group at 48 hpi. (C) Under a fluorescence
microscope, the C. trachomatis inclusion bodies were counted
for each visual field in 20 separate fields. The average number of
inclusions per visual field of the BSA-treated C.
trachomatis-infected group was 74.90±1.852, and the average
number of inclusions in the Vp1-treated C.
trachomatis-infected group was 21.60±1.268. The inhibition rate
reached 71%. **P<0.01. (D) Using western blotting, the protein
levels of chlamydial Hsp60 as an indicator of infection load in the
Vp1-treated C. trachomatis-infected group were detected at
different time points following infection (14, 20, 24, 30 and 36
h). The amount of Hsp60 protein gradually decreased after 24 hpi.
BSA, bovine serum albumin; Ctr, BSA-treated control; hpi, h
post-infection. |
Cytotoxicity induced by C. trachomatis
infection is alleviated by Vp1
Since C. trachomatis is cytotoxic to host
cells, it was explored whether the cytotoxicity induced by C.
trachomatis infection was alleviated after cells were incubated
with Vp1. HeLa cells were incubated with C. trachomatis for
48 h. Following Annexin V/propidium iodide staining, a flow
cytometric analysis was performed. After 48 h of C.
trachomatis infection, early apoptotic cells, late apoptotic
cells and necrotic cells were observed in the Q3, Q2 and Q1
fractions, respectively. In the C. trachomatis-infected
group, HeLa cells exhibited a higher rate of early apoptosis
(Annexin V+ PI−) compared with the
BSA-treated group (Fig. 3). By
contrast, a significantly lower early apoptosis rate was observed
in the Vp1-treated C. trachomatis-infected group compared
with C. trachomatis-infected group (Fig. 3). These results indicated that C.
trachomatis infection-induced apoptosis of HeLa cells was
reduced following Vp1 pretreatment.
Protein levels of p53 and mRNA levels
of Puma, Mcl-1 and cIAP-2 in Vp1-treated C
trachomatis-infected cells recover to normal control
levels. Consistent with previous studies, C. trachomatis
infection resulted in the degradation of p53, which is associated
with the infection dose and duration of infection in host cells
(18,19). In the current study, an obvious
decrease of p53 was observed following 48 h of incubation with
BSA-treated C. trachomatis compared with the mock-infected
cells as controls (Fig. 4A). In
addition, infection-induced p53 degradation was reduced by treating
the C. trachomatis cells with Vp1 prior to infection
(Fig. 4A). The restoration of p53
function was further evaluated by analyzing the proapoptotic gene
Puma, an important transcriptional target of p53 (Fig. 4B). As expected, the mRNA levels of
Puma significantly increased following Vp1 treatment,
consistent with a functional upregulation of p53. Our preliminary
experiments indicated no change in the p53 protein levels when Vp1
interacted with HeLa cells without C. trachomatis (data not
shown). Therefore, these results indicated that the inhibitory
effect of Vp1 is caused by a direct interaction with C.
trachomatis.
The expression levels of Mcl-1 and
cIAP-2 were also evaluated at 48 hpi (Fig. 4C and D). The expression levels of
these two genes in the BSA-treated group indicated a significant
increase compared with the mock-infected cells. Furthermore, the
expression levels of Mcl-1 and cIAP-2 in the
Vp1-treated group revealed a significant decrease compared with the
BSA-treated group.
Discussion
Chlamydial STDs in women are a serious health
problem because infection may lead to infertility, life-threatening
ectopic pregnancy and pelvic inflammatory disease (1). Although C. trachomatis
resistance is a rare occurrence, a single 1 g dose of azithromycin
is not sufficient for the treatment of urogenital and anorectal
C. trachomatis infections (21). The development of a vaccine faces
great challenges, and the immunological mechanisms responsible for
immune protection and immunopathology remain unclear (21). Exploring the etiology and pathogenic
mechanism of C. trachomatis will contribute to the
development of chlamydia infection treatments and preventative
measures. C. trachomatis is able to complete its replication
and development cycle by inhibiting the apoptosis of host cells
through a number of mechanisms, including regulating host cell
mitogen-activated protein kinase signaling pathways, inhibiting
mitochondrial cytochrome c release, degrading pro-apoptotic
proteins and upregulating inhibitor of apoptosis proteins (IAPs)
(22–26). Research by Siegl et al
(19) and González et al
(18) reported that C.
trachomatis promotes p53 proteolysis to inhibit apoptosis,
which leads to persistent infection via an interaction between p53
and murine double minute 2. Mcl-1 and cIAP-2 are well-known key
regulators of apoptosis resistance in C.
trachomatis-infected cells, and activation of the
phosphoinositide 3-kinase pathway in C. trachomatis-infected
cells also stabilizes the anti-apoptotic proteins Mcl-1 and cIAP-2
(27,28).
The current results identified that Vp1 exerts a
clear inhibitory effect on C. trachomatis growth. In
addition, the induction of cytotoxicity in C.
trachomatis-infected host cells was inhibited. The protein
levels of p53 and the expression levels of Mcl-1 and
cIAP-2 recovered to normal levels in the Vp1-treated group
compared with the BSA-treated group. The specific mechanism through
which Vp1 acts on C. trachomatis remains to be elucidated.
To date, six chlamydiaphages have been identified (29), and all of these belong to the family
Microviridae. An amino acid sequence analysis of the φCPG1 Vp1
protein revealed the presence of two major areas of significant
divergence from other chlamydiaphages, namely, amino acids 216–299
(IN5 loop) and 462–467 (INS loop) (10). These two loops are exposed on the
virion surface and likely interact with the host. Using far-western
blotting, our previous study revealed that the φCPG1 Vp1 protein
could bind to the C. trachomatis polymorphic membrane
protein I (PmpI) (30), suggesting
that the binding site of Vp1 is on the surface of C.
trachomatis. In our previous study, 117 differentially
expressed proteins of C. trachomatis treated with Vp1 were
identified by a label-free test and the mRNA levels of several
differentially expressed proteins were assessed using qPCR
(17). According to these results,
it was hypothesized that the combination of Vp1 and PmpI leads to
changes in the function and structure of PmpI. PmpI may transduce
the Vp1 signal in C. trachomatis, and this stimulation may
result in the differential expression of C. trachomatis
proteins. Eventually, this process may cause the cell cycle or
other important C. trachomatis cell processes to be
disrupted.
The increasing rate of C. trachomatis
infection and treatment failure are acknowledged public health
problems. To date, there is no effective C. trachomatis
vaccine, and the effect of a single dose of azithromycin is
unsatisfactory in certain cases. A greater understanding of basic
chlamydial biology and pathogenic mechanisms are important for the
prevention and treatment of C. trachomatis. As the Vp1
protein has been indicated to suppress the growth of C.
trachomatis, the current study provides support for this
potential clinical therapy for C. trachomatis infection.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 31370211 and 31500157).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JR and QL conceived and designed the experiments; JR
performed the experiments; YG analyzed the data; JR wrote the
manuscript; and LS and YL were involved in the cell culture and
protein purification.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Centers for Disease Control and
Prevention: sexually transmitted diseases surveillance 2016. U.S.
Department of Health and Human Services. Atlanta, GA: 2017,
https://www.cdc.gov/std/October
25–2017
|
|
2
|
Yue XL, Gong XD, Teng F, Jiang N, Li J,
Men PX and Wang J: Epidemiologic features of genital Chlamydia
trachomatis infection in national sexually transmitted disease
surveillance sites in China from 2008 to 2015. Chin J Dermatol.
49:308–313. 2016.(In Chinese).
|
|
3
|
Darville T: Recognition and treatment of
chlamydial infections from birth to adolescence. Adv Exp Med Biol.
764:109–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haggerty CL, Gottlieb SL, Taylor BD, Low
N, Xu F and Ness RB: Risk of sequelae after Chlamydia trachomatis
genital infection in women. J Infect Dis. 201 Suppl 2:S134–S155.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brown HM, Knowlton AE and Grieshaber SS:
Chlamydial infection induces host cytokinesis failure at
abscission. Cell Microbiol. 14:1554–1567. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carabeo RA, Grieshaber SS, Fischer E and
Hackstadt T: Chlamydia trachomatis induces remodeling of the actin
cytoskeleton during attachment and entry into HeLa cells. Infect
Immun. 70:3793–3803. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kumar Y and Valdivia RH: Actin and
intermediate filaments stabilize the Chlamydia trachomatis vacuole
by forming dynamic structural scaffolds. Cell Host Microbe.
4:159–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garner SA, Everson JS, Lambden PR, Fane BA
and Clarke IN: Isolation, molecular characterisation and genome
sequence of a bacteriophage (Chp3) from Chlamydophila pecorum.
Virus Genes. 28:207–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoestgaard-Jensen K, Christiansen G,
Honoré B and Birkelund S: Influence of the Chlamydia pneumoniae
AR39 bacteriophage φCPAR39 on chlamydial inclusion morphology. FEMS
Immunol Med Microbiol. 62:148–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsia R, Ohayon H, Gounon P, Dautry-Varsat
A and Bavoil PM: Phage infection of the obligate intracellular
bacterium, Chlamydia psittaci strain guinea pig inclusion
conjunctivitis. Microbes Infect. 2:761–772. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu BL, Everson JS, Fane B, Giannikopoulou
P, Vretou E, Lambden PR and Clarke IN: Molecular characterization
of a bacteriophage (Chp2) from Chlamydia psittaci. J Virol.
74:3464–3469. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Storey CC, Lusher M and Richmond SJ:
Analysis of the complete nucleotide sequence of Chp1, a phage which
infects avian Chlamydia psittaci. J Gen Virol. 70:3381–3390. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hsia RC, Ting LM and Bavoil PM: Microvirus
of chlamydia psittaci strain guinea pig inclusion conjunctivitis:
Isolation and molecular characterization. Microbiology.
146:1651–1660. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sait M, Livingstone M, Graham R, Inglis
NF, Wheelhouse N and Longbottom D: Identification, sequencing and
molecular analysis of Chp4, a novel chlamydiaphage of Chlamydophila
abortus belonging to the family Microviridae. J Gen Virol.
92:1733–1737. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo Y, Guo R, Zhou Q, Sun C, Zhang X, Liu
Y and Liu Q: Chlamydiaphage φCPG1 capsid protein Vp1 inhibits
Chlamydia trachomatis growth via the mitogen-activated protein
kinase pathway. Viruses. 8:992016. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang S, Guo R, Guo YL, Shao LL, Liu Y, Wei
SJ, Liu YJ and Liu QZ: Biological effects of chlamydiaphage phiCPG1
capsid protein Vp1 on Chlamydia trachomatis in vitro and in vivo. J
Huazhong Univ Sci Technolog Med Sci. 37:115–121. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma J, Sun Y, Sun C, Zhou Q, Qi M, Kong J,
Wang J, Liu Y and Liu Q: Identification of proteins differentially
expressed by Chlamydia trachomatis treated with chlamydiaphage
capsid protein VP1 during intracellular growth. Arch Microbiol.
199:1121–1131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
González E, Rother M, Kerr MC, Al-Zeer MA,
Abu-Lubad M, Kessler M, Brinkmann V, Loewer A and Meyer TF:
Chlamydia infection depends on a functional MDM2-p53 axis. Nat
Commun. 5:52012014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Siegl C, Prusty BK, Karunakaran K,
Wischhusen J and Rudel T: Tumor suppressor p53 alters host cell
metabolism to limit Chlamydia trachomatis infection. Cell Rep.
9:918–929. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hafner LM, Wilson DP and Timms P:
Development status and future prospects for a vaccine against
Chlamydia trachomatis infection. Vaccine. 32:1563–1571. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bastidas RJ, Elwell CA, Engel JN and
Valdivia RH: Chlamydial intracellular survival strategies. Cold
Spring Harb Perspect Med. 3:a0102562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fan T, Lu H, Hu H, Shi L, McClarty GA,
Nance DM, Greenberg AH and Zhong G: Inhibition of apoptosis in
chlamydia-infected cells: Blockade of mitochondrial cytochrome c
release and caspase activation. J Exp Med. 187:487–496. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Galluzzi L, Brenner C, Morselli E, Touat Z
and Kroemer G: Viral control of mitochondrial apoptosis. PLoS
Pathog. 4:e10000182008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heuer D, Rejman Lipinski A, Machuy N,
Karlas A, Wehrens A, Siedler F, Brinkmann V and Meyer TF: Chlamydia
causes fragmentation of the Golgi compartment to ensure
reproduction. Nature. 457:731–735. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ying S, Pettengill M, Latham ER, Walch A,
Ojcius DM and Häcker G: Premature apoptosis of Chlamydia-infected
cells disrupts chlamydial development. J Infect Dis. 198:1536–1544.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rajalingam K, Sharma M, Lohmann C, Oswald
M, Thieck O, Froelich CJ and Rudel T: Mcl-1 is a key regulator of
apoptosis resistance in Chlamydia trachomatis-infected cells. PLoS
One. 3:e31022008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rajalingam K, Sharma M, Paland N, Hurwitz
R, Thieck O, Oswald M, Machuy N and Rudel T: IAP-IAP complexes
required for apoptosis resistance of C. trachomatis-infected cells.
PLoS Pathog. 2:e1142006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Śliwa-Dominiak J, Suszyńska E, Pawlikowska
M and Deptuła W: Chlamydia bacteriophages. Arch Microbiol.
195:765–771. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Y, Sun YN, Yao W, Li Y, Li Z, Wei J
and Liu QZ: The detection of the binding protein of chlamydiaphage
phiCPGl capsid protein Vpl on chlamydial outer membrane of serotype
D. Chin J Infect Dis. 32:583–586. 2012.(In Chinese).
|