Introduction
Abdominal aortic aneurysm (AAA) is defined as a
maximum infrarenal abdominal aortic aneurysm with a diameter of
≥3.0 cm and is characterized by permanent, localized dilations of
the abdominal aorta (1,2). AAA is one of the most significant cause
of morbidity and mortality in populations aged >65 years
worldwide (3,4). Acute rupture is the most dangerous
clinical consequence of AAA progression and causes ~80% of
associated deaths in the US (5).
Pathologic features of AAA include vascular smooth muscle cell
apoptosis, infiltration of inflammatory cells, loss of integrity of
the arterial wall, increase of oxidative stress and significant
matrix degradation (6). However, the
precise molecular mechanisms underlying the progression of AAA
progression still remain elusive. Therefore, it is important to
elucidate the etiological mechanisms of AAA progression to develop
novel targets for the diagnosis, treatment, and prognostication of
AAA patients.
Long non-coding (lnc)RNAs are a class of non-coding
RNAs of >200 nucleotides in length, which have little or no
protein-coding function (7). It has
been discovered that lncRNAs have important roles in the
progression of numerous human diseases, including AAA (8). For instance, Yang et al
(9) identified 3,688 differentially
expressed lncRNAs between AAA and normal tissues. lncRNAs regulate
the protein expression at epigenetic, transcriptional and
post-translational levels (10–13). One
of the most well-known mechanisms of lncRNAs is their action as
competing endogenous (ce)RNAs (14).
The ceRNA hypothesis, which was proposed by Tay et al
(15), holds that pseudogenes,
lncRNAs, circular RNAs and mRNAs may impair micro (mi)RNA activity
through sequestration, thereby upregulating miRNA target gene
expression. Franco-Zorrilla et al (16) reported for the first time that
non-coding RNA interferon-β promoter stimulator 1 promoted
phosphate metabolism (PHO)2 protein in plants by sequestering
miR-399 and preventing it from inhibiting the stability and
translation of PHO2 mRNA. Poliseno et al (17) also reported that certain
protein-coding genes and their pseudogenes contain the same
evolutionarily conserved miRNA binding sites in their
3′-untranslated regions, and that they regulate their respective
expression levels by competing for miRNA binding. Emerging studies
have indicated that ceRNAs act as important regulators in different
types of disease, including cancer, cardiac fibrosis, rheumatoid
arthritis and type 2 diabetes mellitus (18–20).
Thus, constructing ceRNA networks provides a novel perspective to
explore the function of yet uncharacterized lncRNAs involved in AAA
progression.
In the present study, differentially expressed RNAs,
miRNA and mRNAs in AAA were identified from data provided by the
National Center for Biotechnology Information Gene Expression
Omnibus (NCBI GEO), and lncRNA-miRNA-mRNA networks were
constructed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) analyses were also performed to explore the
potential roles of differentially expressed lncRNAs in AAA. The
present study aimed to provide useful information to identify novel
lncRNAs as biomarkers for AAA.
Materials and methods
Microarray data
In the present study, two public datasets, GSE7084
(21) and that provided by Yang
et al (9) were analyzed to
identify differentially expressed mRNAs in AAA. The GSE7084 dataset
was downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). The dataset from
Yang et al (9) was downloaded
from the supplementary information of their publication. To further
identify differentially expressed lncRNAs in AAA, the dataset by
Yang et al (9) was analyzed,
providing 896 upregulated and 1,197 downregulated lncRNAs.
Differently expressed miRNAs were determined from the GSE24194
dataset from rats (22), which was
downloaded from the NCBI GEO website. A t-test (23) in the Limma package (24) in R (25) was used to identify differentially
expressed genes between normal and AAA samples. The threshold for
the differentially expressed genes (DEGs) was set as a corrected
P-value of <0.05 and |log2 fold-change (FC)|≥1.
GO and KEGG pathway analysis
To identify functions of DEGs in AAA, GO term
enrichment analysis in the categories biological process, cellular
component and molecular function was performed using the Database
for Annotation, Visualization and Integrated Discovery (DAVID;
http://david.ncifcrf.gov/). KEGG pathway
enrichment analysis was also performed to identify pathways
enriched by DEGs in AAA using DAVID. The P-value was calculated by
hypergeometric distribution and a pathway with P<0.05 was
considered as significant.
Construction of the lncRNA-miRNA-mRNA
network
The StarBase dataset was used to identify
potentially dysregulated lncRNA-miRNA pairs. Next, miRcode
(26), StarBase (27) and the Targetscan database (http://www.targetscan.org) were used to identify
miRNA-mRNA pairs. Finally, lncRNA-miRNA-mRNA networks were
constructed. In the present study, only downregulated miRNAs and
upregulated mRNAs were integrated into the ceRNA network for
upregulated lncRNAs, while only upregulated miRNAs and
downregulated mRNAs were integrated into the ceRNA network fir
downregulated lncRNAs.
Results
Identification of differentially
expressed lncRNAs, mRNAs and miRNAs in AAA
In the present study, two public datasets, GSE7084
(21) and that of Yang et al
(9), were analyzed to identify
differentially expressed mRNAs (Fig. 1A
and B). mRNAs with a FC of ≥2 or ≤0.5 and P<0.05 were
selected as differentially expressed mRNAs. A total of 2,292
upregulated and 2,501 downregulated mRNAs were identified from the
GSE7084 dataset. In the dataset from Yang et al (9), 1,501 upregulated and 969 downregulated
mRNAs were identified. In total, the two datasets had 722
upregulated and 497 downregulated mRNAs in common (Fig. 1C and D). To identify differentially
expressed lncRNAs, the dataset from Yang et al (9) was re-analyzed, and 896 upregulated and
1,197 downregulated lncRNAs were obtained (Fig. 1E). Furthermore, miRNAs with a FC of
≥2 or ≤0.5 and P<0.05 were selected as differentially expressed
miRNAs. A total of 21 upregulated miRNAs and 36 downregulated
miRNAs were selected for further ceRNA network construction by
analyzing the GSE24194 dataset obtained from rats (Fig. 1F). Hierarchical clustering provided
systematic variations in the expression of mRNAs, miRNAs and
lncRNAs in AAA.
Functional prediction of
differentially expressed mRNAs in AAA
GO and KEGG pathway analyses were performed to
explore the function of the 1,219 differentially expressed mRNAs
using DAVID. The results indicated that the upregulated mRNAs were
enriched in GO terms including immune response, signal
transduction, inflammatory response, chemotaxis, cell adhesion,
cell-cell signaling, protein amino acid phosphorylation,
proteolysis, cellular defense response, regulation of transcription
and apoptosis (Fig. 2A), while the
downregulated genes were enriched in GO terms including regulation
of transcription, oxidation/reduction, cell adhesion, signal
transduction, ion transport, protein amino acid phosphorylation,
development, nervous system development, muscle development and
cell-matrix adhesion (Fig. 2B).
KEGG pathway analysis revealed that the upregulated
mRNAs were mainly involved in cytokine-cytokine receptor
interaction, hematopoietic cell lineages, the B-cell receptor
signaling pathway, cell adhesion molecules, leukocyte
transendothelial migration, the T-cell receptor signaling pathway,
graft-versus-host disease, Toll-like receptor signaling pathway and
the mitogen-activated protein kinase signaling pathway (Fig. 2C), while the downregulated genes were
involved in regulation of the actin cytoskeleton, calcium signaling
pathway, focal adhesion, neuroactive ligand-receptor interaction,
tight junctions, tryptophan metabolism, and valine, leucine and
isoleucine degradation (Fig.
2D).
Construction of lncRNA-miRNA-mRNA
networks in AAA
To explore the functions of lncRNAs acting as miRNA
sponges in AAA, two lncRNA-mediated ceRNA networks were first
constructed by using bioinformatics analysis. First, the StarBase
dataset was used to identify potentially dysregulated lncRNA-miRNA
pairs. Next, miRcode (26), StarBase
(27) and the Targetscan database
were used to identify miRNA-mRNA pairs. Finally, lncRNA-miRNA-mRNA
networks were constructed. In the present study, only downregulated
miRNAs and upregulated mRNAs were integrated into the ceRNA network
mediated by upregulated lncRNAs, while, only upregulated miRNAs and
downregulated mRNAs were integrated into the ceRNA network mediated
by downregulated lncRNAs.
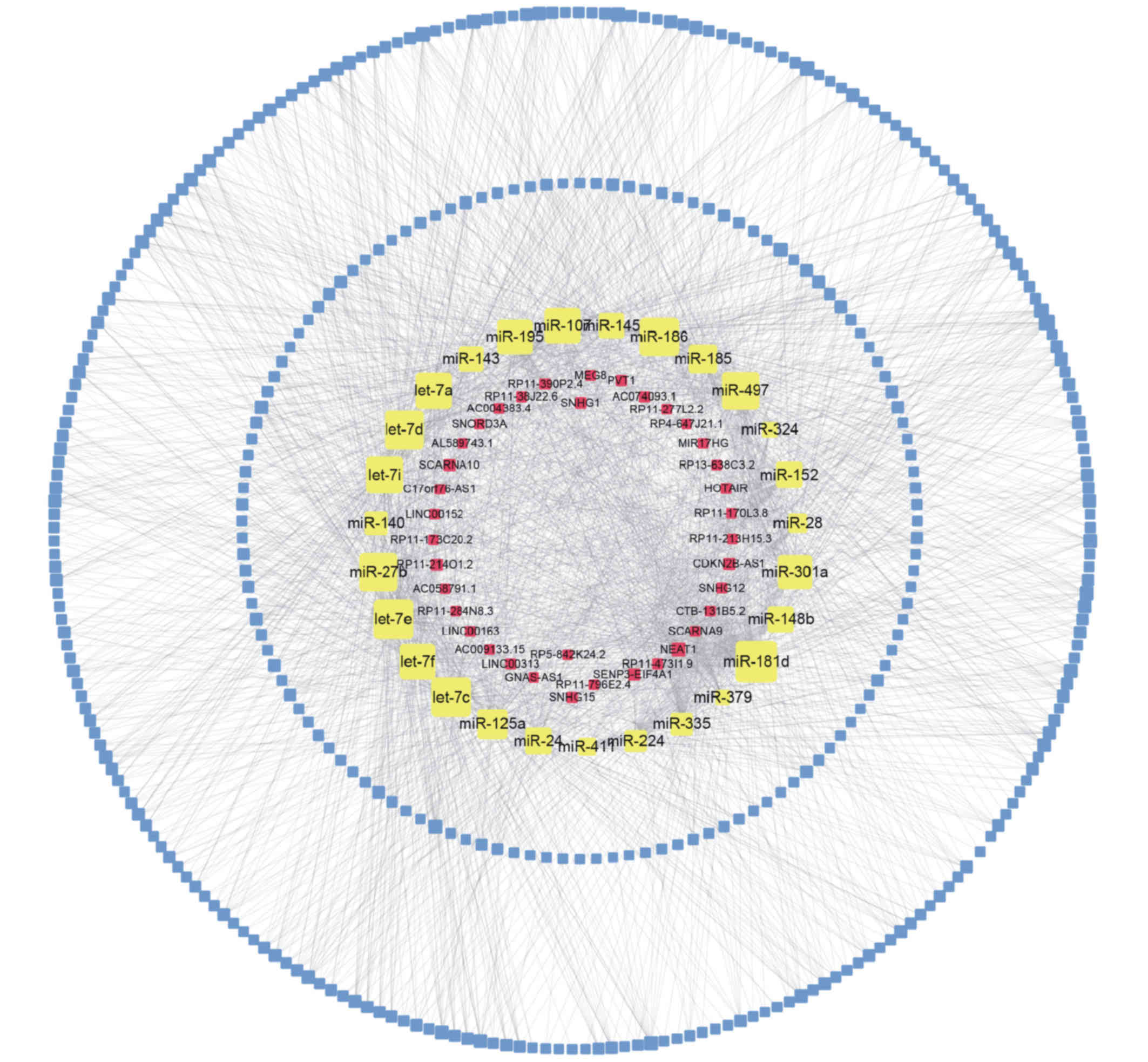
As presented in Fig.
3, the upregulated lncRNA-miRNA-mRNA network contained 38
lncRNA nodes, 374 mRNA nodes, 27 miRNA nodes and 2,021 edges. In
addition, the downregulated lncRNA-miRNA-mRNA network comprised 28
lncRNA nodes, 238 mRNA nodes, 12 miRNA nodes and 729 edges.
Identification of key
lncRNA-mRNA-biological processes sub-network
According to the ceRNA network analysis, all node
degrees were calculated to identify hub lncRNAs, which have
critical roles in biological networks. Five upregulated lncRNAs
[nuclear paraspeckle assembly transcript 1 (NEAT1),
cyclin-dependent kinase inhibitor 2B (CDKN2B)-antisense RNA 1
(AS1), small Cajal body-specific RNA 10 (SCARNA10), AC005224.4 and
SUMO1/sentrin/SMT3-specific peptidase 3 (SENP3)-eukaryotic
translation initiation factor 4A1 (EIF4A1)] and the
downregulated zinc ribbon domain containing 1 (ZNRD1-AS1) were
identified as key lncRNAs in the ceRNA networks and interacted with
>5 different miRNAs (Fig. 4).
Furthermore, key lncRNA-mRNA-biological processes
analysis indicated that NEAT1, CDKN2B-AS1, SCARNA10, AC005224.4 and
SENP3-EIF4A1 were involved in regulating signal transduction,
protein amino acid phosphorylation and immune response (Fig. 5A). ZNRD1-AS1 was associated with the
regulation of transcription, development, and cell differentiation
(Fig. 5B).
 | Figure 5.Identification of key
lncRNA-mRNA-biological processes subnetwork. (A) Key upregulated
lncRNA-mRNA-biological processes analysis indicated that NEAT1,
CDKN2B-AS1, SCARNA10, AC005224.4 and SENP3-EIF4A1 are involved in
regulating signal transduction, protein amino acid phosphorylation
and immune response. (B) The downregulated gene ZNRD1-AS1 was
associated with regulation of transcription, development, and cell
differentiation. lncRNA, long non-coding RNA; NEAT1, nuclear
paraspeckle assembly transcript 1; CDKN2B, cyclin-dependent kinase
inhibitor 2B; AS1, antisense RNA 1; SCARNA10, small Cajal
body-specific RNA 10; SENP3, SUMO1/sentrin/SMT3-specific peptidase
3; EIF4A1, eukaryotic translation initiation factor 4A1; ZNRD1-AS1,
zinc ribbon domain containing 1. |
Discussion
AAA is one of the most significant causes of
morbidity and mortality in populations aged >65 years worldwide
(3,4). Previous studies reported that various
proteins, including mitochondrial uncoupling protein-2, c-Jun
N-terminal kinase and matrix metalloproteinase-9 and miRNAs,
including miRNA-103a and miRNA-516a-5p, were associated with AAA
progression (28). Of note, the
underlying mechanisms of AAA have remained to be fully elucidated.
lncRNAs are a class of non-coding RNAs with no protein-coding
function (7). Several studies have
focused on exploring the roles of lncRNAs in aortic aneurysm
disease. Hypoxia-inducible factor 1α-AS1 was the first lncRNA
reported to be involved in the pathogenesis of thoracic aortic
aneurysm (8). Yang et al
(9) identified 3,688 differentially
expressed lncRNAs between AAA and normal tissues. However, the
molecular functions and detailed mechanisms of lncRNAs in AAA
remain largely elusive. In the present study, DEGs in AAA were
explored by analyzing a series of public datasets, including
GSE7084, the dataset from Yang et al (9) and GSE24194 obtained from rats. A total
of 1,219 mRNAs, 2,093 lncRNAs and 57 miRNAs were identified to
differently express in AAA.
The ceRNA hypothesis was first proposed by Salmena
et al (18) in 2011. This
hypothesis holds that lncRNAs act as miRNA ‘sponges’ to promote the
expression of target genes of miRNAs, which provided a novel basis
for exploring the molecular functions of lncRNAs. Emerging studies
have reported that ceRNAs act as important regulators in different
types of disease, including cancer, cardiac fibrosis and rheumatoid
arthritis (20). For instance,
Karreth (29) reported that the
B-Raf pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. In the present study, deregulated
lncRNA-miRNA-mRNA networks in AAA were constructed. The results
indicated that several key lncRNAs served important roles in
regulating AAA progression by regulating a series of miRNAs and
mRNAs. To the best of our knowledge, the present study was the
first to screen ceRNA networks in AAA.
To date, only few studies have focused on exploring
the roles of lncRNAs in AAA. In the present study, hub lncRNAs with
critical roles in biological networks were identified. The
upregulated lncRNAs NEAT1, CDKN2B-AS1, SCARNA10, AC005224.4 and
SENP3-EIF4A1, and the downregulated ZNRD1-AS1 were identified as
key lncRNAs in the ceRNA networks and interacted with >5
different miRNAs. Furthermore, key lncRNA-mRNA-biological processes
analysis revealed that NEAT1, CDKN2B-AS1, SCARNA10, AC005224.4 and
SENP3-EIF4A1 were involved in regulating signal transduction,
protein amino acid phosphorylation and immune response, while
ZNRD1-AS1 was associated with regulation of transcription,
development and cell differentiation. Of these key lncRNAs, NEAT1
was reported to be overexpressed in different types of solid tumor,
including lung cancer, oesophageal cancer, colorectal cancer and
hepatocellular carcinoma (30–33).
CDKN2B-AS1 was reported to interact with protein regulator of
cytokinesis 1 and −2, leading to epigenetic silencing of CDKN2B
(34). CDKN2B-AS1 has key roles in
the progression of several cancer types (35–37),
intracranial aneurysm (38), type-2
diabetes (39,40), periodontitis (41,42),
Alzheimer's disease (43,44), endometriosis (45), frailty in the elderly (46) and glaucoma (36,47).
Several limitations of the present study should be
noted. First, further validation of key lncRNA expression levels in
AAA samples is required. Although the datasets used in the present
study have been reported and validated, further study to confirm
the differences in lncRNA expression between AAA and normal samples
is required. Another limitation is that the public datasets for
analyzing AAA-associated lncRNAs were limited. An urgent
requirement remains to identify differently expressed lncRNAs in
AAA by using high-throughput methods, including lncRNA expression
array and RNA sequencing. Finally, additional functional
investigations of these lncRNAs on AAA progression are still
required.
Based on the ceRNA hypothesis, the present study was
the first to constructed lncRNA-miRNA-mRNA networks in AAA, to the
best of our knowledge. In the present study, a total of 1,219
mRNAs, 2,093 lncRNAs and 57 miRNAs were identified to be
differently expressed in AAA based on analyzing public datasets
including GSE7084, the dataset by Yang et al (9) and GSE24194. The present study further
identified 5 upregulated lncRNAs (NEAT1, CDKN2B-AS1, SCARNA10,
AC005224.4 and SENP3-EIF4A1) and the downregulated ZNRD1-AS1 as key
lncRNAs in the ceRNA networks. Key lncRNA-mRNA-biological processes
analysis indicated that these key lncRNAs are involved in
regulating signal transduction, protein amino acid phosphorylation,
immune response, transcription, development and cell
differentiation. The present study provides novel clues for
exploring the mechanistic involvement of lncRNA in AAA
progression.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
High-resolution versions of Figs. 3 and 4
are available on request. The datasets used and/or analyzed during
the current study are available from the corresponding author on
reasonable request
Authors' contributions
HZ, LT and DL contributed to the conception and
design of the study. LT and XH developed the methodology. YH
collected the samples. ZW analyzed and interpreted the data. LT, DL
and HZ wrote, reviewed and revised the manuscript. The final
version of the manuscript has been read and approved by all
authors.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Alcorn HG, Wolfson SK Jr, Sutton-Tyrrell
K, Kuller LH and O'Leary D: Risk factors for abdominal aortic
aneurysms in older adults enrolled in the cardiovascular health
study. Arterioscler Thromb Vasc Biol. 16:963–970. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Humphrey JD and Taylor CA: Intracranial
and abdominal aortic aneurysms: Similarities, differences, and need
for a new class of computational models. Annu Rev Biomed Eng.
10:221–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hallett JW Jr, Marshall DM, Petterson TM,
Gray DT, Bower TC, Cherry KJ Jr, Gloviczki P and Pairolero PC:
Graft-related complications after abdominal aortic aneurysm repair:
Reassurance from a 36-year population-based experience. J Vasc
Surg. 25:277–284; discussion 285–276. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johansson G and Swedenborg J: Ruptured
abdominal aortic aneurysms: A study of incidence and mortality. Br
J Surg. 73:101–103. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maegdefessel L, Azuma J, Toh R, Deng A,
Merk DR, Raiesdana A, Leeper NJ, Raaz U, Schoelmerich AM, McConnell
MV, et al: MicroRNA-21 blocks abdominal aortic aneurysm development
and nicotine-augmented expansion. Sci Transl Med. 4:122ra1222012.
View Article : Google Scholar
|
|
6
|
Miyake T and Morishita R: Pharmacological
treatment of abdominal aortic aneurysm. Cardiovasc Res. 83:436–443.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Duggirala A, Delogu F, Angelini TG, Smith
T, Caputo M, Rajakaruna C and Emanueli C: Non coding RNAs in aortic
aneurysmal disease. Front Genet. 6:1252015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang YG, Li MX, Kou L, Zhou Y, Qin YW, Liu
XJ and Chen Z: Long noncoding RNA expression signatures of
abdominal aortic aneurysm revealed by microarray. Biomed Environ
Sci. 29:713–723. 2016.PubMed/NCBI
|
|
10
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibney ER and Nolan CM: Epigenetics and
gene expression. Heredity (Edinb). 105:4–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wan X, Huang W, Yang S, Zhang Y, Pu H, Fu
F, Huang Y, Wu H, Li T and Li Y: Identifcation of
androgen-responsive lncRNAs as diagnostic and prognostic markers
for prostate cancer. Oncotarget. 7:60503–60518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Franco-Zorrilla JM, Valli A, Todesco M,
Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA and
Paz-Ares J: Target mimicry provides a new mechanism for regulation
of microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Rooij E and Olson EN: MicroRNA
therapeutics for cardiovascular disease: opportunities and
obstacles. Nat Rev Drug Discov. 11:860–872. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Li C, Tan C and Liu X: Circular
RNAs: A new frontier in the study of human diseases. J Med Genet.
53:359–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lenk GM, Tromp G, Weinsheimer S, Gatalica
Z, Berguer R and Kuivaniemi H: Whole genome expression profiling
reveals a significant role for immune function in human abdominal
aortic aneurysms. BMC Genomics. 8:2372007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu G, Huang Y, Lu X, Lu M, Huang X, Li W
and Jiang M: Identification and characteristics of microRNAs with
altered expression patterns in a rat model of abdominal aortic
aneurysms. Tohoku J Exp Med. 222:187–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cui X and Churchill GA: Statistical tests
for differential expression in cDNA microarray experiments. Genome
Biol. 4:2102003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gondro C, Porto-Neto LR and Lee SH: R for
genome-wide association studies. Methods Mol Biol. 1019:1–17. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moran CS, McCann M, Karan M, Norman P,
Ketheesan N and Golledge J: Association of osteoprotegerin with
human abdominal aortic aneurysm progression. Circulation.
111:3119–3125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Karreth FA, Reschke M, Ruocco A, Ng C,
Chapuy B, Léopold V, Sjoberg M, Keane TM, Verma A, Ala U, et al:
The BRAF pseudogene functions as a competitive endogenous RNA and
induces lymphoma in vivo. Cell. 161:319–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo S, Chen W, Luo Y, Ren F, Zhong T, Rong
M, Dang Y, Feng Z and Chen G: Clinical implication of long
non-coding RNA NEAT1 expression in hepatocellular carcinoma
patients. Int J Clin Exp Pathol. 8:5395–5402. 2015.PubMed/NCBI
|
|
31
|
Clemson CM, Hutchinson JN, Sara SA,
Ensminger AW, Fox AH, Chess A and Lawrence JB: An architectural
role for a nuclear noncoding RNA: NEAT1 RNA is essential for the
structure of paraspeckles. Mol Cell. 33:717–726. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun C, Li S, Zhang F, Xi Y, Wang L, Wang L
and Li D: Long non-coding RNA NEAT1 promotes non-small cell lung
cancer progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016.PubMed/NCBI
|
|
33
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang XR, Liang XY, Pfeiffer RM, Wheeler W,
Maeder D, Burdette L, Yeager M, Chanock S, Tucker MA and Goldstein
AM: Associations of 9p21 variants with cutaneous malignant
melanoma, nevi, and pigmentation phenotypes in melanoma-prone
families with and without CDKN2A mutations. Fam Cancer. 9:625–633.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Falchi M, Bataille V, Hayward NK, Duffy
DL, Bishop JA, Pastinen T, Cervino A, Zhao ZZ, Deloukas P, Soranzo
N, et al: Genome-wide association study identifies variants at 9p21
and 22q13 associated with development of cutaneous nevi. Nat Genet.
41:915–919. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sherborne AL, Hosking FJ, Prasad RB, Kumar
R, Koehler R, Vijayakrishnan J, Papaemmanuil E, Bartram CR,
Stanulla M, Schrappe M, et al: Variation in CDKN2A at 9p21.3
influences childhood acute lymphoblastic leukemia risk. Nat Genet.
42:492–494. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Helgadottir A, Thorleifsson G, Magnusson
KP, Grétarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT,
Rinkel GJ, Blankensteijn JD, Ronkainen A, et al: The same sequence
variant on 9p21 associates with myocardial infarction, abdominal
aortic aneurysm and intracranial aneurysm. Nat Genet. 40:217–224.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Broadbent HM, Peden JF, Lorkowski S, Goel
A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G,
et al: Susceptibility to coronary artery disease and diabetes is
encoded by distinct, tightly linked SNPs in the ANRIL locus on
chromosome 9p. Hum Mol Genet. 17:806–814. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cugino D, Gianfagna F, Santimone I, de
Gaetano G, Donati MB, Iacoviello L and Di Castelnuovo A: Type 2
diabetes and polymorphisms on chromosome 9p21: A meta-analysis.
Nutr Metab Cardiovasc Dis. 22:619–625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schaefer AS, Richter GM,
Groessner-Schreiber B, Noack B, Nothnagel M, El Mokhtari NE, Loos
BG, Jepsen S and Schreiber S: Identification of a shared genetic
susceptibility locus for coronary heart disease and periodontitis.
Plos Genet. 5:e10003782009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ernst FD, Uhr K, Teumer A, Fanghänel J,
Schulz S, Noack B, Gonzales J, Reichert S, Eickholz P, Holtfreter
B, et al: Replication of the association of chromosomal region
9p21.3 with generalized aggressive periodontitis (gAgP) using an
independent case-control cohort. BMC Med Genet. 11:1192010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yu JT, Yu Y, Zhang W, Wu ZC, Li Y, Zhang N
and Tan L: Single nucleotide polymorphism rs1333049 on chromosome
9p21.3 is associated with alzheimer's disease in han chinese. Clin
Chim Acta. 411:1204–1207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Emanuele E, Lista S, Ghidoni R, Binetti G,
Cereda C, Benussi L, Maletta R, Bruni AC and Politi P: Chromosome
9p21.3 genotype is associated with vascular dementia and
Alzheimer's disease. Neurobiol Aging. 32:1231–1235. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Uno S, Zembutsu H, Hirasawa A, Takahashi
A, Kubo M, Akahane T, Aoki D, Kamatani N, Hirata K and Nakamura Y:
A genome-wide association study identifies genetic variants in the
CDKN2BAS locus associated with endometriosis in Japanese. Nat
Genet. 42:707–710. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Melzer D, Frayling TM, Murray A, Hurst AJ,
Harries LW, Song H, Khaw K, Luben R, Surtees PG, Bandinelli SS, et
al: A common variant of the p16 INK4a genetic region is associated
with physical function in older people. Mech Ageing Dev.
128:370–377. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ramdas WD, van Koolwijk LME, Lemij HG,
Pasutto F, Cree AJ, Thorleifsson G, Janssen SF, Jacoline TB, Amin
N, Rivadeneira F, et al: Common genetic variants associated with
open-angle glaucoma. Hum Mol Genet. 20:2464–2471. 2011. View Article : Google Scholar : PubMed/NCBI
|