Introduction
Gastric cancer (GC) has one of the highest mortality
rates of all types of cancer, as most patients are diagnosed at an
advanced stage (1,2). The highest mortality rate of males and
females was 28.3% in Korea and 10.8% in Ecuador, respectively
(3). Cisplatin (DDP)-based
chemotherapy remains the primary strategy for the treatment of GC;
however, DDP resistance is a major problem (4–6). Several
factors contribute to chemotherapeutic resistance, including
genetic or epigenetic changes in cancer cells (7). Previous studies have indicated that the
primary cause of resistance is the overexpression of energy
dependent transporters, including P-glycoprotein and multidrug
resistance-associated protein, which eject antitumor drugs out of
cancer cells (8,9). Further studies have also demonstrated
that a defect in drug-induced cancer cell apoptosis was another
cause (8,9). The clinical use of DDP-based
chemotherapy is thus limited by resistance, meaning that continuous
and multiple DDP treatments may lead to various side effects,
including renal impairment, neurotoxicity and ototoxicity (10). Therefore, improving the sensitivity
of GC cells to DDP may increase the effectiveness of GC
treatment.
It is well-established that microRNAs (miRNAs miRs)
are a class of small non-coding RNAs, comprised of 18–24
nucleotides, which regulate gene expression by binding to target
mRNAs (11). It has been previously
demonstrated that miRNAs are abnormally expressed in various types
of cancer (12–14): Aberrant miRNAs regulate cancer cell
differentiation, proliferation, survival and apoptosis (15). Previous studies have also
demonstrated that miRNAs regulate anti-cancer drug resistance
(16). Therefore, the present study
hypothesized that the combination of miRNA regulation and
chemotherapy may serve important roles in the treatment of certain
types of cancer. A number of previous studies have indicated that
miR-218 is downregulated in many types of cancer, including lung
cancer, prostate cancer, cervical carcinoma and GC (17–20).
However the association between miR-218 and DDP resistance in GC is
yet to be fully elucidated.
The present study demonstrated that the expression
of miR-218 decreased significantly in DDP-resistant human GC
SGC7901/DDP cells and the induction of miR-218 regulated DDP
sensitivity in SGC7901/DDP cells by targeting survivin. The results
of the current study may help to resolve the issue of DDP
resistance in the clinical treatment of GC in the future.
Materials and methods
Cell culture
The human gastric adeno-carcinoma cell line SGC7901
was purchased from the Shanghai Yansheng Industrial Co., Ltd.
(Shanghai, China) and DDP-resistant SGC7901/DDP cells were
purchased from Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China).
GC cells were cultured in RPMI-1640 medium supplemented with 10%
fetal bovine serum (FBS), 100 IU/ml penicillin and 100 IU/ml
streptomycin (Gibco; Thermo Fisher Scientific, Inc.). In order to
maintain the DDP resistant phenotype, SGC7901/DDP cells were
cultured for 1 week at 37°C in medium supplemented with 1 µg/ml DDP
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and then cultured
in medium without DDP for 1 week prior to the study.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of miR-218
Following SGC7901 and SGC7901/DDP cell culture and
harvesting, total RNA was extracted from the cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Extracted RNA was then reverse
transcribed into cDNA using a TaqMan™ MicroRNA Reverse
Transcription kit (cat. no. 4366596; Applied Biosystems; Thermo
Fisher Scientific, Inc.). The temperature protocol of reverse
transcription was 16°C for 30 min, followed by 42°C for 30 min and
85°C for 5 min. miR-218 qPCR was performed using a TaqMan miRNA
assay kit (cat. no. 4426961; Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol and as
previously described (21). U6 RNA
was used as an internal control (cat. no. 4351372; Applied
Biosystems; Thermo Fisher Scientific, Inc.). The 2−ΔΔCq
method was used to evaluate the relative expression of miR-218 in
GC cells (22).
Transfection
SGC7901 and SGC7901/DDP cells were seeded into
6-well plates (2,000,000 cells/well) in RPMI-1640 medium
supplemented with 10% FBS for 24 h at 37°C. The miR-218 inhibitor
(100 nM; cat. no. 4464084; Thermo Fisher Scientific, Inc.) or the
miRNA inhibitor control (100 nM; cat. no. 4464076; Thermo Fisher
Scientific, Inc.) were transfected into SGC7901 cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Additionally, 100 nM
miR-218 mimic (cat. no. 4464066; Thermo Fisher Scientific, Inc.) or
the miRNA mimic control (cat. no. 4464058; Thermo Fisher
Scientific, Inc.) were transfected into SGC7901/DDP cells using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Survivin small
interfering (si)RNA was purchased from Santa Cruz Biotechnology,
Inc. (Dallas, TX, USA; cat. no. sc-29499) and the survivin gene
knockout assay was performed according to the manufacturer's
protocol, untreated cells were used as the control. Following 48 h,
samples were used for further experimentation.
Cell viability assay
SGC7901 and SGC7901/DDP cells transfected with miRNA
mimics or inhibitors were seeded into 96-well plates (10,000
cells/well). Following cellular adhesion, DDP was added into the
cultured cells at concentrations of 0.00, 0.03, 0.10, 0.30, 1.00,
3.00, 10.00, 30.00 and 100.00 µg/ml and incubated for 48 h at 37°C.
At 48 h following cell culture, cell viability was evaluated via an
MTT assay. MTT solution was added to each well at a concentration
of 0.5 mg/ml. DMSO was utilized to dissolve the purple formazan. At
4 h following MTT incubation at 37°C, the absorbance at 570 nm was
measured using a microplate reader. A relative survival curve was
generated and the half maximal inhibitor concentration
(IC50) was defined as the concentration at which DDP
inhibited the 50% cell viability.
MiRNA target predictions
To determine the potential target of miR-218,
potential genes were identified using computer-aided algorithms
from targetscan (http://www.targetscan.org) and mirbase targets
(http://microrna.sanger.ac.uk/cgi-bin/targets/v5/search.pl).
Luciferase assay
The wild type (WT) 3′-untranslated region (UTR) of
survivin mRNA (NM_001168, https://www.ncbi.nlm.nih.gov/gene/?term=NM_001168;
5′-CUACAAUUAAAACUAAGCACAA-3′) containing predicted binding sites of
miR-218 was amplified from Human Genome DNA by Genewiz (Genewiz,
South Plainfield, NJ, USA) and cloned into the downstream region of
the firefly luciferase gene in the pMIR-Report reporter vector
(Ambion; Thermo Fisher Scientific, Inc.). Mutant (Mut;
5′-CUACAAUUAAAACUUGAGCAGG3′) survivin was used as a control.
pGL3-Survivin-3′-UTR and miR-218 mimic or miRNA mimic controls were
co-transfected into SGC7901/DDP cells using Lipofectamine 3000
according to the manufacture's protocol. Following transfected cell
culture for 48 h at 37°C, cells were harvested and submitted to the
luciferase assay. The Dual-Luciferase Reporter Assay System was
performed according to the manufacture's protocol (cat. no. E1910,
Promega Corporation, Madison, WI, USA) and the relative activity
was normalized to the expression of Renilla luciferase.
Flow-cytometric analysis of
apoptosis
Transfected SGC7901/DDP cells were seeded into 6
well plates (600,000 cells/well). Following cellular adhesion, DDP
was added to the cultured cells at a concentration of 10 µg/ml.
Cells were then cultured at 37°C for a further 48 h and submitted
to flow cytometry to determine the relative quantity of apoptotic
cells using the Annexin V-FITC Apoptosis Detection kit (cat. no.
CA1020; Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China). CellQuest Pro software was used for the analysis
of results (Version 5.1; BD Biosciences, Franklin Lakes, NJ,
USA)
Western blot analysis
Transfected SGC7901/DDP and SGC7901 cells were
seeded in 6 well plates (600,000 cells/well) and cultured at 37°C
for 48 h, after which they were harvested. Protein was extracted
using lysis buffer (cat. no. P0013K; Beyotime Institute of
Biotechnology, Haimen, China) and protein concentration was
measured using a bicinchoninic acid assay. Total protein (50 µg)
was the separated using 10% SDS-PAGE gel electrophoresis. Western
blotting was performed as previously described (23). The primary antibodies for survivin
(1:1,000; cat. no. 2808) and GAPDH (1:2,000; cat. no. 2118) were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
GAPDH was used as the internal control. The second antibodies used
were horseradish peroxidase labeled-Goat Anti-Rabbit Immunoglobulin
G (1:1,000; cat. no. A0208; Beyotime Institute of
Biotechnology).
RT-qPCR to detect survivin mRNA
Total RNA was isolated from transfected SGC7901/DDP
or SGC7901 cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
Total RNA (2 µg) was reverse transcribed into cDNA using an iScript
cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The temperature protocol for RT was as follows: 5 min at 25°C,
followed by 30 min at 42°C and 5 min at 85°C. qPCR was performed
using an LC480 machine (Roche Diagnostics, Basel, Switzerland)
using a FastStart Universal SYBR Green Master (cat. no.
04913914001; Roche) and gene-specific primers. The thermocycling
conditions were as follows: 95°C for 10 min; 95°C for 10 sec; 57kC
for 20 sec and 72°C for 10 sec (40 cycles). The primer sequences
utilized were as follows: GAPDH forward,
5′-TCGACAGTCAGCCGCATCTTCTTT-3′ and reverse,
5′-ACCAAATCCGTTGACTCCGACCTT-3′; and Survivin forward,
5′-GGACCACCGCATCTCTACAT-3′ and reverse, 5′-GACAGAAAGGAAAGCGCAAC-3′.
Relative transcript expression of survivin relative to GAPDH was
determined using the 2−ΔΔCq method (22).
Statistical analysis
Each experiment was performed in triplicate. The
data was expressed as the mean + standard deviation. Statistical
analysis was performed using a two-tailed Student's t-test with
SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant result.
Results
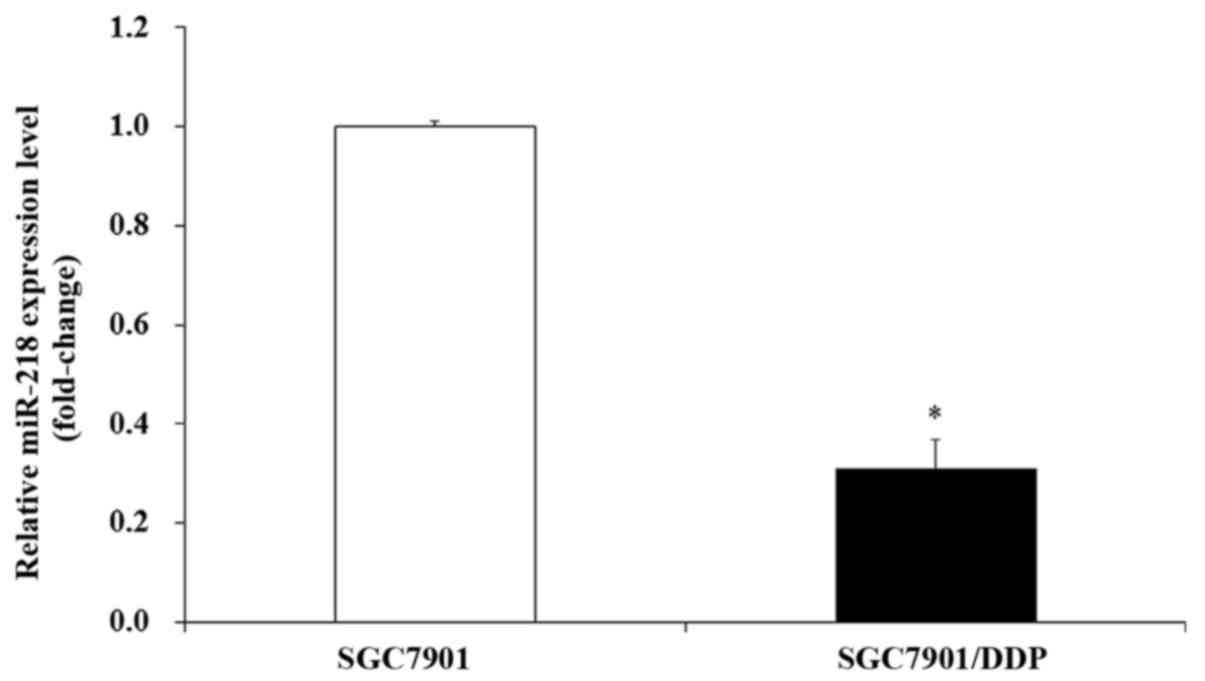
miR-218 is downregulated in
DDP-resistant SGC7901/DDP cells
A previous study has demonstrated that miR-218 is
downregulated in gastric cancer cells (24). The present study therefore assessed
the expression of miR-218 in the DDP-resistant gastric cancer cell
line (SGC7901/DDP) and its mock cell line (SGC7901). RT-qPCR
results indicated that the expression of miR-218 was downregulated
in DDP-resistant gastric cancer SGC7901/DDP cells compared with
SGC7901 cells, with a decreased fold change of ~3.1 (Fig. 1).
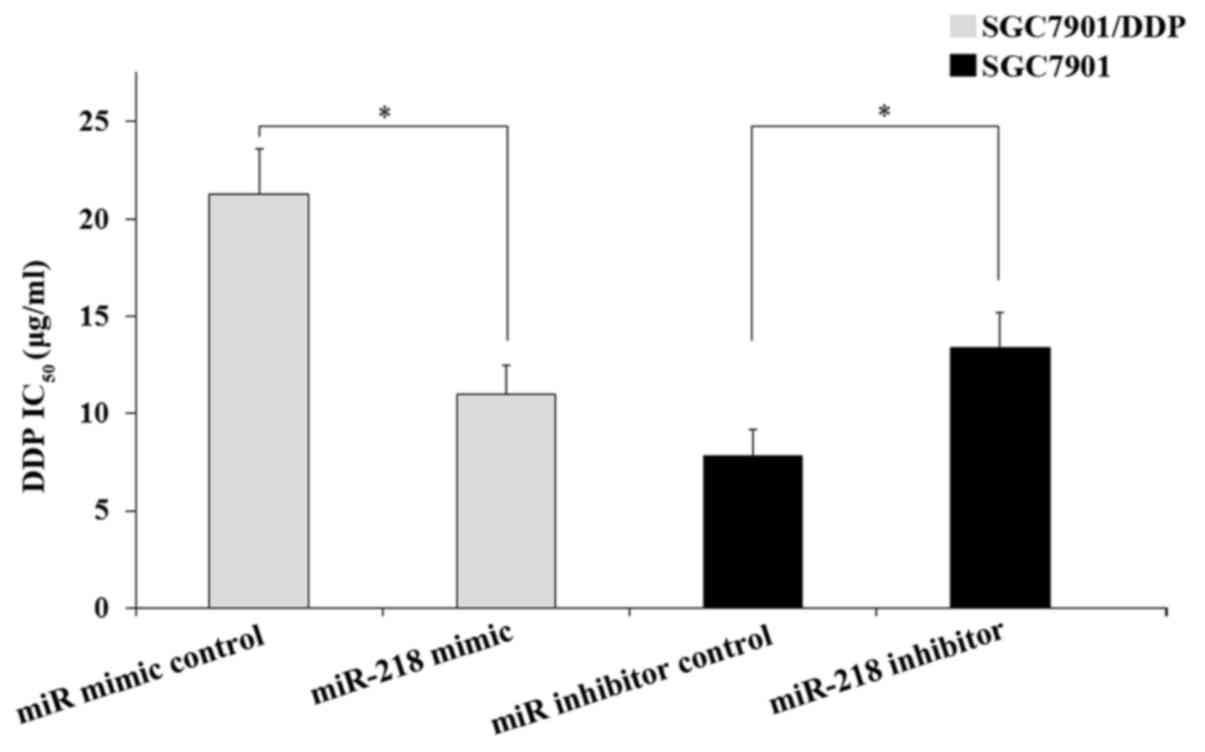
miR-218 modulates DDP resistance in
SGC7901/DDP cells
To evaluate the effect of miR-218 on DDP resistance
in GC cells, an MTT assay was performed. As presented in Fig. 2, the transfection of miR-218 mimics
into SGC7901/DDP cells induced a significant decrease in the
IC50 of DDP compared with those cells transfected with
the miRNA mimic control (P<0.05). However, transfection of the
miR-218 inhibitor into SGC7901 cells significantly increased the
IC50 of DDP compared with those cells transfected with
the miRNA inhibitor control (P<0.05). Therefore, the results
demonstrated that miR-218 modulates the DDP resistance of GC
cells.
Survivin is the target gene of
miR-218
To identify the target gene of miR-218, a search was
performed using the microRNA.org
database (www.microrna.org). The results indicated
that survivin was a potential target gene of miR-218 (Fig. 3A). To confirm this, a dual luciferase
assay was performed on SGC7901/DDP cells. WT and Mut survivin
3′-UTR was fused directly downstream of the firefly luciferase gene
(Fig. 3A) and then co-transfected
with the miR-218 mimic and various luciferase 3′-UTR constructs (WT
or Mut) into SGC7901/DDP cells. Transfected cells were then
cultured and luciferase activity was measured. The dual luciferase
assay results indicated that the enhanced expression of miR-218
significantly decreased luciferase activity in the WT pLuc-Survivin
3′-UTR reporter (P<0.05). However, no significant difference was
observed between cells treated with the miR-218 mimic and cells
that received the miRNA mimic control in the Mut pLuc-Survivin
3′-UTR reporter group (Fig. 3B).
These results demonstrated that survivin is a direct target gene of
miR-218.
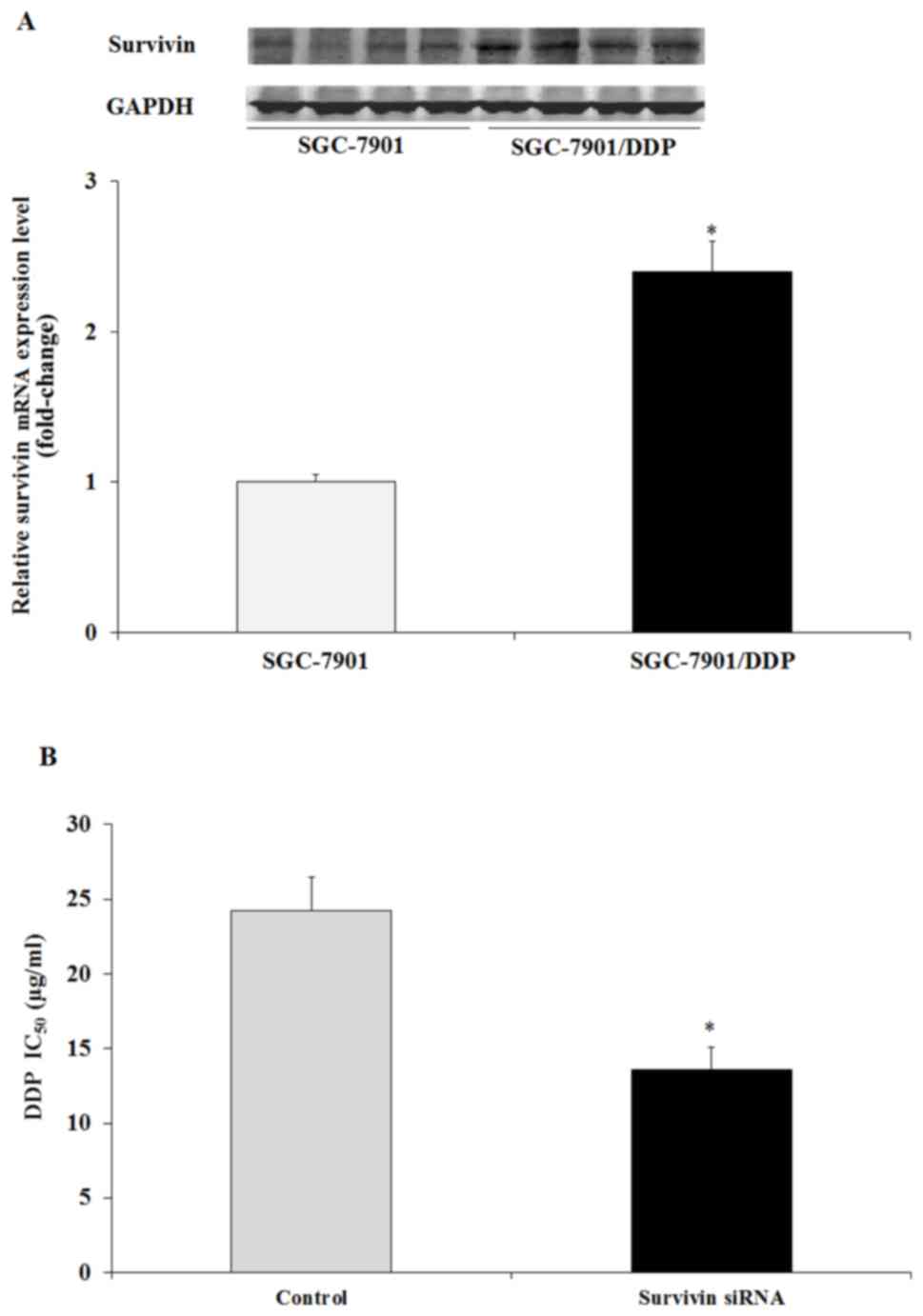
Survivin is overexpressed in resistant
GC cell lines and is associated with DDP resistance
To assess whether survivin gene expression was
associated with DDP resistance, survivin protein and mRNA
expression in SGC7901/DDP cells and SGC7901 cells was determined.
Western blotting and RT-qPCR results indicated that survivin
protein and mRNA expression was increased in SGC7901/DDP cells
compared with SGC7901 cells (P<0.05; Fig. 4A and B). It was further demonstrated
that the transfection of survivin siRNA resulted in a significant
decrease in DDP IC50 in SGC7901/DDP cells compared with
the untreated cells (P<0.05; Fig.
4B). The results indicated that the survivin gene was closely
associated with DDP resistance in GC cells.
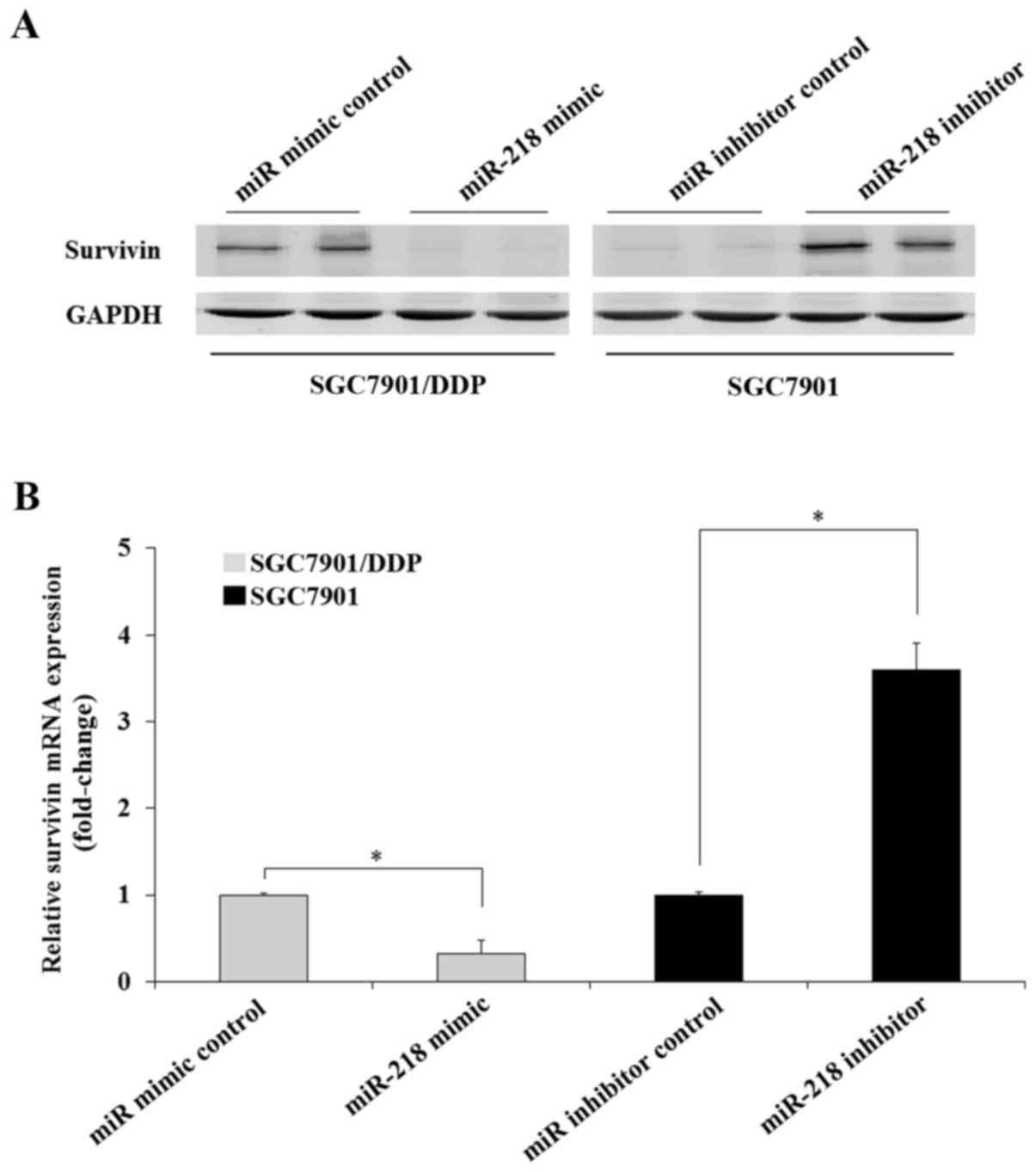
miR-218 modulates DDP resistance by
repressing survivin
In the present study it was demonstrated that
miR-218 and survivin expression was decreased and overexpressed,
respectively in SGC7901/DDP cells when compared with SGC7901 cells
(Figs. 1 and 4A). It was also revealed that survivin was
a direct target gene of miR-218 and that it was associated with DDP
resistance in GC cells. Therefore, it was hypothesized that miR-218
may regulate DDP resistance in GC cells by modulating the
expression of survivin. To confirm this, the expression of survivin
in GC cells transfected with miR-218 mimic or inhibitor were
assessed. The results indicated that transfection of the miR-218
mimic significantly decreased the expression of survivin in
SGC7901/DDP cells compared with those transfected with the miRNA
mimic control. Transfection of the miR-218 inhibitor increased the
expression of survivin protein and mRNA in SGC7901 cells compared
with those transfected with the miRNA inhibitor control (Fig. 5). The results indicated that miR-218
modulates DDP resistance in GC cells through the regulation of
survivin expression.
Transfected miR-218 sensitizes
SGC7901/DDP cells to DDP by inducing apoptosis
A previous study has indicated that drug resistance
in various types of cancer is closely associated with the
overexpression of anti-apoptotic proteins, including BCL2, MCL1 and
XIAP (25). Since survivin has been
demonstrated to be a direct target of miR-218 that is associated
with DDP resistance (26), the
present study hypothesized that miR-218 may modulate DDP resistance
in SGC7901/DDP cells by enhancing the DDP-induced apoptosis of
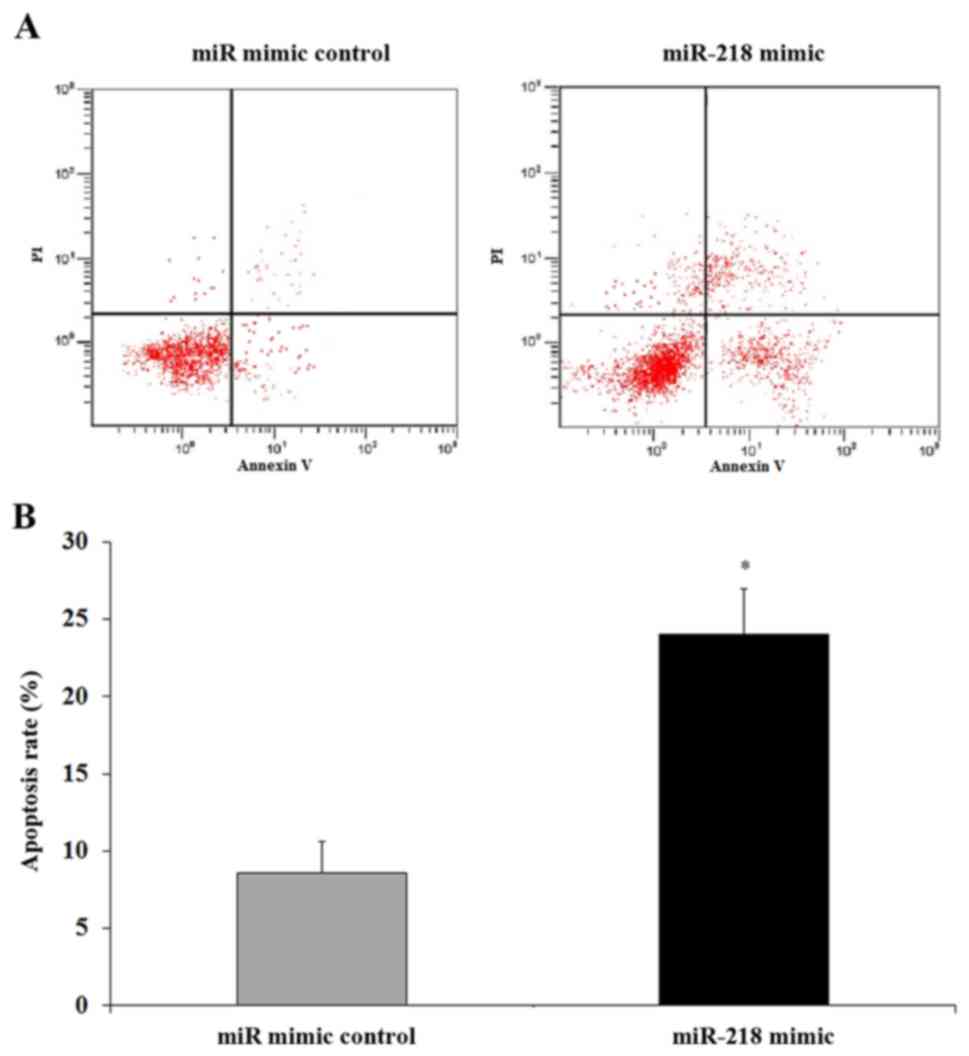
SGC7901/DDP cells. To confirm this, flow cytometry was performed.
The miR-218 mimic or miRNA mimic control was transfected into
SGC7901/DDP cells, which were subsequently treated with DDP and
cultured. Cells were then harvested and the apoptosis rate was
determined. The results indicated that there was a significant
induction of apoptosis in SGC7901/DDP cells transfected with the
miR-218 mimic compared with those transfected with the miRNA mimic
control (P<0.05; Fig. 6).
Therefore miR-218 enhanced the sensitivity of DDP resistance in GC
cells by inducing apoptosis, at least in part, by targeting
survivin.
Discussion
Surgery is the primary treatment for early stage
gastric cancer; however, the majority of patients are diagnosed at
an advanced stage and receive chemotherapy as a primary treatment
option (27). Furthermore, DDP-based
chemotherapy is the major strategy for the clinical treatment of GC
(16). However DDP resistance is an
obstacle for effective GC therapy (27). A previous study has demonstrated that
miRNA dysregulation is present in many different types of human
cancer (28). Other studies have
also indicated that certain miRNAs, including miR-128, miR-497 and
miR-200, may contribute to chemotherapeutic drug resistance
(29–31). A number of previous studies have
revealed that miR-218 was downregulated in various malignancies,
including colon cancer, cervical cancer and GC. Furthermore, these
studies also indicated that miR-218 serves an important biological
role in cancer proliferation and metastasis (32–35).
However, the functional and biological roles of miR-218 in GC cell
chemosensitivity are yet to be fully elucidated.
To investigate the role of miR-218 in GC cell
chemosensitivity, the present study assessed the expression of
miR-218 in DDP resistant GC SGC7901/DDP cells. The results of
RT-qPCR demonstrated that the expression of miR-218 was
significantly downregulated in SGC7901/DDP cells compared with
SGC7901 cells. Further assessment revealed that the upregulation of
miR-218 sensitized SGC7901/DDP cells to DDP via the inhibition of
cell viability and the induction of cell apoptosis.
Survivin is encoded by Baculoviral inhibitor of
apoptosis repeat containing 5 and is a member of the apoptosis
inhibitor family (31). Survivin may
therefore serve anti-apoptotic roles in cells due to its regulation
of caspase activity (36). It has
also been demonstrated that survivin exhibits an anti-apoptotic
role via the inhibition of caspase-9 activity, which is associated
with the hepatitis B X-interacting protein (37). These studies have also indicated that
survivin serves an important role in antitumor drug resistance and
may therefore be a promising biomarker of antitumor drug resistance
(37,38). A previous study has indicated that
dysregulated survivin is associated with many types of human
cancer, including breast and lung cancer, as well as prostate,
gastric, colon, bladder and esophageal carcinoma, osteosarcoma, and
lymphoma (39). A previous study has
also indicated that certain survivin-targeting miRNAs have been
identified in many types of cancer (40). The present study demonstrated that
survivin is a direct target gene of miR-218. It was also revealed
that survivin protein and mRNA expression was increased in DDP
resistant GC SGC7901/DDP cells and that decreased survivin
expression sensitized GC cells to DDP. Further experiments
demonstrated that the transfection of the miR-218 mimic inhibited
the expression of survivin in GC cells, whereas the transfection of
the miR-218 inhibitor led to the increase of survivin expression in
GC cells, which was associated with the induction of cell
apoptosis.
Taken together, the present findings demonstrated
that miR-218 may be associated with the development of DDP
resistance in human GC cells, at least in part, by targeting
survivin. The results indicated that the regulation of miR-218 in
combination with DDP chemotherapy may provide a novel treatment for
human GC in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ, YK and YH designed the present study; WY, BZ, FM
and HL performed the experiments; WY, BZ and FM analyzed the data;
BZ, FM and HL prepared the manuscript; and ZZ and FM reviewed the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Authors' information
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roder DM: The epidemiology of gastric
cancer. Gastric Cancer. 5 Suppl 1:S5–S11. 2002. View Article : Google Scholar
|
|
3
|
Torre LA, Siegel RL, Ward EM and Jemal A:
Global cancer incidence and mortality rates and Trends-an update.
Cancer Epidemiol Biomarkers Prev. 25:16–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mesner PW Jr, Budihardjo II and Kaufmann
SH: Chemotherapy-induced apoptosis. Adv Pharmacol. 41:461–499.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kaufmann SH and Earnshaw WC: Induction of
apoptosis by cancer chemotherapy. Exp Cell Res. 256:42–49. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hannun YA: Apoptosis and the dilemma of
cancer chemotherapy. Blood. 89:1845–1853. 1997.PubMed/NCBI
|
|
7
|
Sharma SV, Lee DY, Li B, Quinlan MP,
Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach
MA, et al: A chromatin-mediated reversible drug-tolerant state in
cancer cell subpopulations. Cell. 141:69–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rabik CA and Dolan ME: Molecular
mechanisms of resistance and toxicity associated with platinating
agents. Cancer Treat Rev. 33:9–23. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kostova I: Platinum complexes as
anticancer agents. Recent Pat Anticancer Drug Discov. 1:1–22. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma L, Feldstein Teruya J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of mir-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: Predictors and modifiers of chemo and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cunningham D, Allum WH, Stenning SP,
Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ,
Falk SJ, Iveson TJ, et al: Perioperative chemotherapy versus
surgery alone for resectable gastroesophageal cancer. N Engl J Med.
355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-218 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Petrocca F, Visone R, Onelli MR, Shah MH,
Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini
M, et al: E2F1-regulated microRNAs impair TGFbeta-dependent
cell-cycle arrest and apopto-sis in gastric cancer. Cancer Cell.
13:272–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Q, Yang Z, Chen M and Liu Y:
Downregulation of microRNA-196a enhances the sensitivity of
non-small cell lung cancer cells to cisplatin treatment. Int J Mol
Med. 37:1067–1074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moriyama M, Tsukamoto Y, Fujiwara M, Kondo
G, Nakada C, Baba T, Ishiguro N, Miyazaki A, Nakamura K, Hori N, et
al: Identification of a novelhuman ankyrin-repeated protein
homologous to CARP. Biochem Biophys Res Commun. 285:715–723. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang LL, Wang L, Wang XY, Shang D, Yin SJ,
Sun LL and Ji HB: MicroRNA-218 inhibits the proliferation,
migration, and invasion and promotes apoptosis of gastric cancer
cells by targeting LASP1. Tumour Biol. 37:15241–15252. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu W, Shan X, Wang TS, Shu YQ and Liu P:
miR-181b modulates multidrug resistance by targeting BCL2 in human
cancer cell lines. Int J Cancer. 127:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu Y, Xu K and Yagüe E: miR-218 targets
survivin and regulates resistance to chemotherapeutics in breast
cancer. Breast Cancer Res Treat. 151:269–280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang SM, Huang C, Li XF, Yu MZ, He Y and
Li J: miR-21 confers cisplatin resistance in gastric cancer cells
by regulating PTEN. Toxicology. 306:162–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao
H, Gong C, Chen J, Su F, Zhang Y and Song E: Reduced miR-128 in
breast tumor-initiating cells induces chemotherapeutic resistance
via Bmi-1 and ABCC5. Clin Cancer Res. 17:7105–7115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Creevey L, Ryan J, Harvey H, Bray IM,
Meehan M, Khan AR and Stallings RL: MicroRNA-497 increases
apoptosis in MYCN amplified neuroblastoma cells by targeting the
key cell cycle regulator WEE1. Mol Cancer. 12:232013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen DQ, Pan BZ, Huang JY, Zhang K, Cui
SY, De W, Wang R and Chen LB: HDAC 1/4-mediated silencing of
microRNA-200b promotes chemoresistance in human lung adenocarcinoma
cells. Oncotarget. 5:3333–3349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He X, Dong Y, Wu CW, Zhao Z, Ng SS, Chan
FK, Sung JJ and Yu J: MicroRNA-218 inhibits cell cycle progression
and promotes apoptosis in colon cancer by downregulating BMI1
poly-comb ring finger oncogene. Mol Med. 18:1491–1498.
2013.PubMed/NCBI
|
|
33
|
Li J, Ping Z and Ning H: MiR-218 impairs
tumor growth andincreases chemo-sensitivity to cisplatin in
cervical cancer. Int J Mol Sci. 13:16053–16064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Uesugi A, Kozaki K, Tsuruta T, Furuta M,
Morita K, Imoto I, Omura K and Inazawa J: The tumor suppressive
microRNAmiR-218 targets the mTOR component Rictor and inhibits
AKTphosphorylation in oral cancer. Cancer Res. 71:5765–5778. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xin SY, Feng XS, Zhou LQ, Sun JJ, Gao XL
and Yao GL: Reduced expression of circulating microRNA-218 in
gastric cancer and correlation with tumor invasion and prognosis.
World J Gastroenterol. 20:6906–6911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li F, Ambrosini G, Chu EY, Plescia J,
Tognin S, Marchisio PC and Altieri DC: Control of apoptosis and
mitotic spindle checkpoint by survivin. Nature. 396:580–584. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Marusawa H, Matsuzawa S, Welsh K, Zou H,
Armstrong R, Tamm I and Reed JC: HBXIP functions as a cofactor of
survivin in apoptosis suppression. EMBO J. 22:2729–2740. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Singh N, Krishnakumar S, Kanwar RK, Cheung
CH and Kanwar JR: Clinical aspects for survivin: A crucial molecule
for targeting drug-resistant cancers. Drug Discov Today.
20:578–587. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Waligórska-Stachura J, Jankowska A, Waśko
R, Liebert W, Biczysko M, Czarnywojtek A, Baszko-Błaszyk D, Shimek
V and Ruchała M: Survivin-prognostic tumor biomarker in human
neoplasms-review. Ginekol Pol. 83:537–540. 2012.PubMed/NCBI
|
|
40
|
Huang J, Lyu H, Wang J and Liu B: MicroRNA
regulation and therapeutic targeting of survivin in cancer. Am J
Cancer Res. 5:20–31. 2014.PubMed/NCBI
|