Introduction
Osteosarcoma is the most common primary malignant
bone tumor with the highest incidence among patients aged 10–20
years (1,2). The annual incidence of osteosarcoma is
five per million. Since, in 80% of patients with osteosarcoma,
metastasis or micrometastasis has occurred at the time of
diagnosis, almost all patients receive multiple doses of
chemotherapy in addition to surgery (3). Recent improvements in surgical methods
resulted in decreased numbers of amputations among patients with
osteosarcoma (4). The understanding
of molecular mechanisms of osteosarcoma may contribute to the
development of targeted therapy for this disease (5). Development of novel treatment
strategies and improvement of the currently available methods may
increase the survival rate of patients with osteosarcoma.
In recent years, numerous studies have demonstrated
that microRNAs (miRNAs) serve important regulatory roles in tumor
formation and progression (6–9). A
number of oncogenes and tumor suppressor genes are regulated by
miRNAs (10). miRNAs are
single-stranded, non-coding RNAs that recognize specific target
mRNAs and regulate the expression of target genes at the
post-transcriptional level (11–13).
miRNAs promote the degradation of mRNA and/or inhibit the
translation process, and regulate the translation of ~30% of the
protein-coding mRNA in the human genome (11–13).
Previous studies indicated that the expression of miRNAs is
abnormal in numerous types of tumors (6–9). miRNAs
may serve carcinogenic or tumor suppressive roles by regulating
downstream target genes and influencing the biological behavior of
tumor cells, including proliferation, apoptosis, invasion,
metastasis and angiogenesis (14).
The majority of tumor suppressor miRNAs are downregulated in
malignant cells, while oncogenic miRNAs are upregulated and affect
tumor pathology through multiple mechanisms (15,16).
miR-708-5p is a miRNA expressed in a number of
diseases (17). miR-708-5p was first
identified in normal tissues and tumor samples from patients with
cervical cancer, and exhibits a high degree of sequence similarity
with miR-28 (18,19). The passenger strand of miR-708
(miR-708-3p) shows potential biological function and is
incorporated into the RNA-induced silencing complex (20–25).
miR-708-5p is involved in numerous diseases, including cancer,
neurodegenerative diseases and cardiovascular diseases (26–30).
The current study aimed to determine the role of
miR-708-5p in the occurrence and development of osteosarcoma. This
miRNA may become a clinical marker for the diagnosis of this type
of cancer and the results of the current study may provide
theoretical basis for diagnosis and treatment.
Materials and methods
Clinical specimens
Osteosarcoma tissue and adjacent normal tissue
samples were obtained from 60 patients (age, 14–65 years; 32 males
and 28 females; 9 cases without tumor metastasis; 23 cases with
lymph node metastasis without distant metastasis; 28 cases with
distant metastasis) diagnosed with osteosarcoma by pathological
examination between May 2011 and May 2016 at the First Affiliated
Hospital of Anhui Medical University and The First Affiliated
Hospital of University of Science and Technology of China (Hefei,
China). Patients were included in the present study if they did not
receive radiotherapy or chemotherapy, and exhibited normal
cardiopulmonary, liver and kidney function. Patients with chronic
conditions including high blood pressure, chronic heart disease and
kidney failure were excluded. All experiments involving human
tissues were reviewed and approved by the Ethics Committee of the
First Affiliated Hospital of Anhui Medical University and by the
Ethics Committee of the First Affiliated Hospital of University of
Science and Technology of China. All patients provided written
informed consent for the use of their tissues.
Cell culture and treatment
Human osteosarcoma cell line SaOS-2 and normal
osteoblast cell line hFOB1.19 were purchased from American Type
Culture Collection (Manassas, VA, USA) and cultured in the First
Affiliated Hospital of Anhui Medical University. Cells were grown
in RPMI 1640 medium containing 10% fetal bovine serum (FBS; both
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 1%
penicillin-streptomycin solution (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), and incubated at 37°C with 5% CO2.
Cells were passaged every 2–3 days.
SaOS-2 cells (3×104 cells/well) were
transiently transfected with 100 nM miRNA-708-5p mimics
(AAGGAGCUUACAAUCUAGCUGGG and CAGCUAGAUUGUAAGCUCCUUUU), negative
control mimics (UUCUCCGAACGUGUCACGUTT and ACGUGACACGUUCGGAGAATT),
miR-708-5p inhibitors (CCCAGCUAGAUUGUAAGCUCCUU) or negative control
inhibitors (CAGUACUUUUGUGUAGUACAA) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The miR-708-5p mimics, miR-708-5p
inhibitor and negative controls were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). After 48 h, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to determine transfection efficiency, as described below.
Cells without any treatment were used as the control group.
Western blot analysis
Following treatment, total cellular proteins from
SaOS-2 cells were extracted using radioimmunoprecipitation assay
buffer (Hunan Auragene Biotech Co., Ltd, Changsha, China).
Bicinchoninic acid protein quantification kit (Thermo Fisher
Scientific, Inc.) was used to measure the protein concentration in
samples. Equal amounts of protein (30 µg/lane) were resolved by
SDS-PAGE on a 12% gel and transferred onto polyvinylidene fluoride
membranes. The membranes were blocked with 5% non-fat milk at room
temperature for 1 h, followed by incubation with primary
antibodies, including anti-up-regulator of cell proliferation
(URGCP; cat. no. ab103323; 1:1,000; Abcam, Cambridge, MA, USA),
anti-nuclear factor (NF)-κB (cat no. 8242) and anti-GAPDH (cat. no.
8884; both 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA) at 4°C overnight. Subsequently, the membranes were incubated
with an anti-rabbit immunoglobulin G horseradish
peroxidase-conjugated secondary antibody (cat. no. 7074; 1:2,000;
Cell Signaling Technology, Inc.) at room temperature for 2 h. To
visualize the protein blots, the enhanced chemiluminescence Western
Blotting Detection kit (Applygen Technologies, Inc., Beijing,
China) was used according the manufacturer's protocol.
RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from cells and
tissues. GAPDH or U6 were used as internal controls for mRNA or
miRNA expression, respectively. cDNA was synthesized using
PrimeScript™ RT reagent kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's protocol. SYBR® Premix
Ex Taq™ (Takara Bio, Inc.) was used for qPCR according to the
manufacturer's protocol. The following primer sequences were used
for the qPCR: miR-708-5p, forward 5′-GGCGCGCAAGGAGCTTACAATC-3′ and
reverse 5′-GTGCAGGGTCCGAGGTAT-3′; URGCP, forward:
5′-CTTCATCCTGAGTCCCTACCG-3′ and reverse: 5′-GCCGTTCTGCTGCATTCG-3′;
NF-κB, forward: 5′-AACACTGCCGACCTCAAGAT-3′ and reverse:
5′-CATCGGCTTGAGAAAAGGAG-3′; U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse
5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH, forward
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse
5′-GTAGAGGCAGGGATGATGTTCT-3′. Relative expression of each gene was
calculated using the 2−ΔΔCq method (31).
Cell migration and invasion
assays
An in vitro invasion assay was performed
using transwell plates (BD Biosciences, Franklin Lakes, NJ, USA)
with 8 µm pores. Chamber inserts were coated with 200 mg/ml BD
Matrigel™ matrix (BD Biosciences) at room temperature
overnight. The SaOS-2 cells (1×104 cells) in RPMI 1640
medium were added to the upper chamber of the transwell plates.
RPMI 1640 medium containing 20% FBS as a chemoattractant was added
to the lower chamber. After a 48-h incubation, cells were removed
from the upper surface using cotton swabs and the invasive cells
were fixed with methanol and stained with 0.5% crystal violet at
room temperature for 30 min. Images were captured and the cells
were counted using a light photomicroscope (Olympus Corporation,
Tokyo, Japan) at a magnification of ×200.
For the wound healing assay, confluent monolayers of
SaOS-2 cells cultured in 24-well plates were scratched using a
10-µl pipette tip. The wells were washed to remove cellular debris
and the cells were allowed to migrate for 48 h. Representative
images were captured under an light inverted microscope (Olympus
Corporation; magnification, ×100). The experiments were repeated at
least three times.
Cell Counting Kit-8 (CCK-8) assay
SaOS-2 cells were seeded into a 96-well plate
(2×105 cells/well) and cultured in RPMI-1640 medium at
37°C for 24, 48 and 72 h respectively. Cell viability was detected
using the CCK-8 kit according to the manufacturer's protocol. The
absorbance was measured at a wavelength of 450 nm using an
iMark® microplate absorbance reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The experiments were
repeated at least 3 times.
Cell apoptosis detection
Following transfection, SaOS-2 cells in the
logarithmic growth phase were collected and washed at least three
times with cold PBS. Annexin V-FITC Early Apoptosis Detection kit
(cat. no. 6592; Cell Signaling Technology, Inc.) was used for cell
apoptosis analysis. In brief, SaOS-2 cells (1×106) from
different groups were re-suspended in binding buffer, labeled with
1 µl Annexin V-fluorescein isothiocyanate (FITC) and 12.5 µl
propidium iodide (PI) and then incubated for 10 min on ice in the
dark. A flow cytometer (FACSCalibur™; BD Biosciences, Franklin
Lakes, NJ, USA) was used to analyze cell apoptosis. Data were
analyzed using WinMDI software (version 2.5; Purdue University
Cytometry Laboratories, West Lafayette, IN, USA). The experiments
were repeated at least 3 times.
Bioinformatics prediction and
dual-luciferase reporter assay
Targetscan (version 7.1; www.targetscan.org/vert_71) was used to predict the
putative target genes of miR-708-5p. To confirm whether miR-708-5p
directly targets URGCP, a luciferase reporter assay was performed
using a pEZX-MT01 target reporter plasmid containing the URGCP
3′-untranslated region (UTR; GeneCopoeia, Inc., Rockville, MD,
USA). Additionally, a mutant (MUT) URGCP 3′-UTR reporter construct
was generated by site-directed mutagenesis in the putative target
site of miR-708-5p in the wild-type (WT) URGCP 3′-UTR using the
QuikChange XL site-directed mutagenesis kit (Agilent Technologies,
Inc., Santa Clara, CA, USA). The reporter plasmids were
co-transfected into SaOS-2 cells with miR-708-5p mimics or the NC
using Lipofectamine 3000® (Invitrogen; Thermo Fisher
Scientific, Inc.) in 24-well plates. A total of 48 h following
transfection, dual-luciferase reporter assay system (Promega
Corporation, Madison, WI, USA) was used to measure luciferase
activity according to the manufacturer's protocol. Relative
luciferase activity was normalized to the Renilla luciferase
activity. The results were obtained from three independent
experiments.
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS software (version 17.0; SPSS, Inc., Chicago, IL,
USA) was used for statistical analyses. Comparisons between groups
were performed using Student's t-test or one-way analysis of
variance followed by Tukey's test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of miR-708-5p in
osteosarcoma
The expression levels of miR-708-5p were detected in
osteosarcoma tissues, adjacent normal tissues, human osteosarcoma
cell line SaOS-2 and normal osteoblast cell line hFOB1.19 using
RT-qPCR. The results indicated that compared with the adjacent
normal tissues, the level of miR-708-5p in osteosarcoma tissues
significantly decreased (Fig. 1A).
Furthermore, the level of miR-708-5p in SaOS-2 cells was
significantly lower compared with the hFOB1.19 cells (Fig. 1B).
miR-708-5p suppresses SaOS-2 cell
viability and induces cell apoptosis in vitro
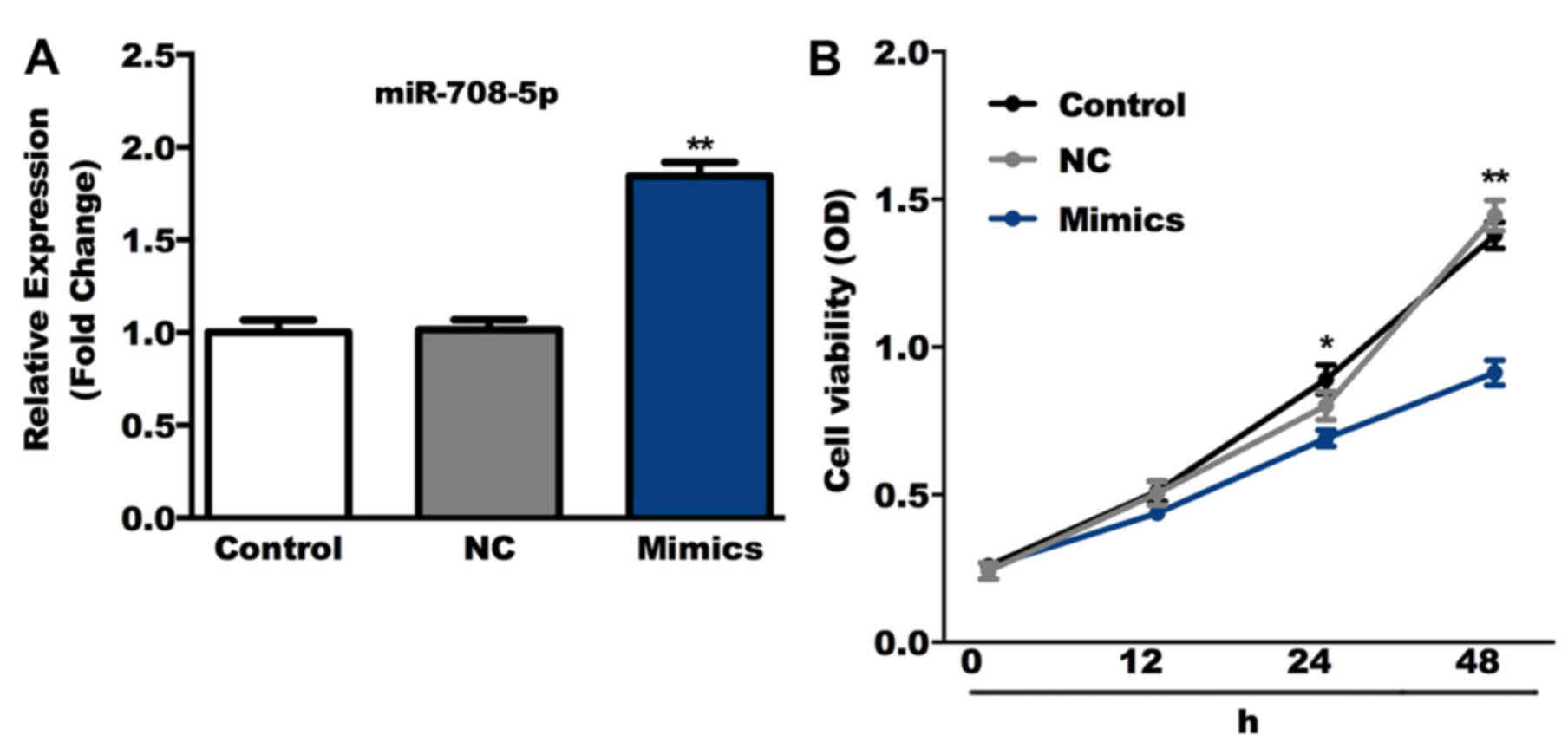
The role of miR-708-5p overexpression was determined
in SaOS-2 cells in the present study. Cells were transiently
transfected with miR-708-5p mimics or NC. RT-qPCR results confirmed
that compared with the control group, the level of miR-708-5p was
significantly increased in the miR-708-5p mimics group (Fig. 2A). Cell viability was subsequently
detected using the CCK-8 assay and the results indicated that,
compared with the control group, miR-708-5p mimics significantly
suppressed SaOS-2 cell viability (Fig.
2B). Furthermore, SaOS-2 cell apoptosis was detected 48 h
following transfection with miR-708-5p mimics or NC. Flow cytometry
analysis indicated that cell apoptosis significantly increased in
SaOS-2 cells transfected with miR-708-5p mimics compared with the
cells from the control group (Fig.
3).
miR-708-5p suppresses SaOS-2 cell
migration and invasion
Cell migration and invasion were measured in the
present study. A total of 48 h after cell transfection, wound
healing and transwell assays were used to detect cell migration and
invasion respectively. The results demonstrated that both migration
and invasion of SaOS-2 cells in the miR-708-5p mimics group were
significantly inhibited compared with the control group (Fig. 4).
miR-708-5p directly targets URGCP
Subsequent experiments were performed to determine
the underlying mechanism of the effect of miR-708-5p on SaOS-2
cells. The potential targets of miR-708-5p were predicted using
TargetScan. URGCP was identified as a potential target gene of
miR-708-5p. To verify the binding site, the 3′-UTR of URGCP
containing a WT or MUT sequence was cloned into SaOS-2 cells for a
subsequent firefly luciferase reporter assay. The results indicated
that URGCP was a direct target gene of miR-708-5p (Fig. 5A and B). To determine the effect of
miR-708-5p on the URGCP signaling pathway, cells were transfected
with miR-708-5p mimics and miR-708-5p inhibitors, and expression
levels of URGCP and NK-κB were analyzed at the mRNA and protein
levels. The results indicated that compared with the control group,
miR-708-5p mimics and miR-708-5p inhibitors significantly decreased
and increased the mRNA and protein levels of URGCP and NK-κB in
SaOS-2 cells, respectively (Fig.
5C-I).
 | Figure 5.miR-708-5p directly targets URGCP.
(A) TargetScan was used to predict the interactions between
miR-708-5p and the WT 3′-UTR of URGCP. (B) Relative luciferase
activity was detected by a dual-luciferase assay. **P<0.01 vs.
NC. After a 48 h transfection with NC and miR-708-5p mimics, (C)
the protein level of URGCP and NK-κB were detected using a western
blot assay, and the mRNA levels of (D) URGCP and (E) NK-κB were
detected using RT-qPCR. (F) miR-708-5p levels were detected after a
48 h transfection with NC and a miR-708-5p inhibitor using RT-qPCR.
After a 48 h transfection with NC and a miR-708-5p inhibitor, (G)
the protein level of URGCP and NK-κB were detected using a western
blot assay, and the mRNA levels of (H) URGCP and (I) NK-κB were
detected using RT-qPCR. Data are presented as the mean ± standard
deviation. **P<0.01 vs. the control group. miR, microRNA; URGCP,
up-regulator of cell proliferation; UTR, untranslated region; WT,
wild type; NC, negative control; NK-κB, nuclear factor-κB. |
Discussion
Osteosarcoma is the most common primary malignant
bone tumor with the highest incidence among individuals aged 10–20
years (2). However, the therapeutic
effect of osteosarcoma is still unsatisfactory (32,33).
Therefore, identification of novel effective treatment methods for
patients with osteosarcoma is required. miRNAs serve important
roles in the occurrence and development of a number of diseases
(34–36). Furthermore, previous studies
indicated that miRNAs participate in the occurrence and development
of osteosarcoma (37,38). The role of miR-708-5p was previously
studied in several types of cancer including cervical (18,19),
lung (39) and prostate (40) cancer. However, to the best of our
knowledge, the role of miR-708-5p in osteosarcoma has not been
previously studied.
The present study aimed to investigate the potential
role of miR-708-5p in the development and progression of
osteosarcoma in vitro, and that miR-708-5p may be a
potential marker for the diagnosis of osteosarcoma. The present
study detected the level of miR-708-5p in osteosarcoma tissues,
adjacent normal tissues, human osteosarcoma cell line SaOS-2 and
normal osteoblast hFOB1.19 cell line. miR-708-5p was significantly
downregulated in osteosarcoma tissues and cells, indicating that
miR-708-5p may be involved in the occurrence and development of
this disease. The effects of miR-708-5p overexpression on SaOS-2
cells were studied using miR-708-5p mimics. Transfection with
miR-708-5p mimics significantly inhibited cell growth, induced cell
apoptosis, and inhibited cell invasion and migration in
vitro. The present study also demonstrated that URGCP was a
direct target of miR-708-5p.
URGCP is an oncogene that contributes to
carcinogenesis, cell cycle regulation and cell proliferation in
cells (41). URGCP is involved in
the development of various types of cancer (42–44), and
promotes the malignant behavior of cancer cells by regulating the
NF-κB signaling pathway (45–47). In
the present study, miR-708-5p overexpression inhibited the
expression of URGCP and NF-κB in SaOS-2 cells, while miR-708-5p
downregulation enhanced the expression of URGCP and NF-κB. The data
indicated that miR-708-5p may inhibit osteosarcoma cell viability
and invasion by regulating the URGCP/NF-κB signaling pathway.
In conclusion, to the best of our knowledge, the
present study is the first to indicate that miR-708-5p was
significantly downregulated in osteosarcoma tissues and cells.
miR-708-5p overexpression inhibited osteosarcoma cell viability,
migration and invasion, and induced cell apoptosis. Furthermore,
the results of the present study indicated that URGCP was a direct
target gene of miR-708-5p. The current study provides novel
insights into osteosarcoma research and targeted therapies.
According to the present preliminary study, miR-708-5p may be a
novel and promising therapeutic target for the treatment of
osteosarcoma. However, the tumor suppressor role of miR-708-5p in
osteosarcoma requires further investigation. Future studies should
determine whether the overexpression of URGCP or NF-kB could
reverse the effects of miR-708-5p on SaOS-2 cells. Furthermore,
potential association between patient prognosis and miR-708-5p
expression may be studied in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CS and DL collaborated to design the study. CS, DL
and YH were responsible for data access and analysis. LZ
interpreted the results. All authors collaborated to develop the
manuscript.
Ethics approval and consent to
participate
All experiments involving human tissues were
reviewed and approved by the Ethics Committee of the First
Affiliated Hospital of Anhui Medical University and by the Ethics
Committee of the First Affiliated Hospital of University of Science
and Technology of China. All patients provided written informed
consent for the use of their tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li X, Lu Q, Xie W, Wang Y and Wang G:
Anti-tumor effects of triptolide on angiogenesis and cell apoptosis
in osteosarcoma cells by inducing autophagy via repressing
Wnt/β-Catenin signaling. Biochem Biophys Res Commun. 496:443–449.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lo YC, Lin YC, Huang YF, Hsieh CP, Wu CC,
Chang IL, Chen CL, Cheng CH and Chen HY: Carnosol-induced ROS
inhibits cell viability of human osteosarcoma by apoptosis and
autophagy. Am J Chin Med. 45:1761–1772. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu ZR, Sun LZ, Jia TH and Jia DF:
β-Aescin shows potent antiproliferative activity in osteosarcoma
cells by inducing autophagy, ROS generation and mitochondrial
membrane potential loss. J BUON. 22:1582–1586. 2017.PubMed/NCBI
|
|
4
|
Xu HY, Fang W, Huang ZW, Lu JC, Wang YQ,
Tang QL, Song GH, Kang Y, Zhu XJ, Zou CY, et al: Metformin reduces
SATB2-mediated osteosarcoma stem cell-like phenotype and tumor
growth via inhibition of N-cadherin/NF-kB signaling. Eur Rev Med
Pharmacol Sci. 21:4516–4528. 2017.PubMed/NCBI
|
|
5
|
Shaikh AB, Li F, Li M, He B, He X, Chen G,
Guo B, Li D, Jiang F, Dang L, et al: Present advances and future
perspectives of molecular targeted therapy for osteosarcoma. Int J
Mol Sci. 17:5062016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu L, Ai J, Long H, Liu W, Wang X, Zuo Y,
Li Y, Wu Q and Deng Y: Intergrative microRNA and gene profiling
data analysis reveals novel biomarkers and mechanisms for lung
cancer. Oncotarget. 7:8441–8454. 2016.PubMed/NCBI
|
|
7
|
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi
HC, Lin YH, Lu PH and Lin KH: Potential diagnostic, prognostic and
therapeutic targets of microRNAs in human gastric cancer. Int J Mol
Sci. 17:E9452016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao H, Li M, Li L, Yang X, Lan G and
Zhang Y: MiR-133b is down-regulated in human osteosarcoma and
inhibits osteosarcoma cells proliferation, migration and invasion,
and promotes apoptosis. PLoS One. 8:e835712013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kaukoniemi KM, Rauhala HE, Scaravilli M,
Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL and
Visakorpi T: Epigenetically altered miR-193b targets cyclin D1 in
prostate cancer. Cancer Med. 4:1417–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schwarzenbacher D, Balic M and Pichler M:
The role of microRNAs in breast cancer stem cells. Int J Mol Sci.
14:14712–14723. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Friedman JM and Jones PA: MicroRNAs:
Critical mediators of differentiation, development and disease.
Swiss Med Wkly. 139:466–472. 2009.PubMed/NCBI
|
|
14
|
Schneider MR: MicroRNAs as novel players
in skin development, homeostasis and disease. Br J Dermatol.
166:22–28. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Coghlin C and Murray GI: Current and
emerging concepts in tumour metastasis. J Pathol. 222:1–15. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Kim J, Mueller AC, Dey B, Yang Y,
Lee DH, Hachmann J, Finderle S, Park DM, Christensen J, et al:
Multiple receptor tyrosine kinases converge on microRNA-134 to
control KRAS, STAT5B, and glioblastoma. Cell Death Differ.
21:720–734. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garg M: Emerging role of microRNAs in
cancer stem cells: Implications in cancer therapy. World J Stem
Cells. 7:1078–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bicker S and Schratt G: microRNAs: Tiny
regulators of synapse function in development and disease. J Cell
Mol Med. 12:1466–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Condorelli G and Dimmeler S: MicroRNAs:
Components of an integrated system controlling cardiac development,
physiology, and disease pathogenesis. Cardiovasc Res. 79:551–552.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He X, Eberhart JK and Postlethwait JH:
MicroRNAs and micromanaging the skeleton in disease, development
and evolution. J Cell Mol Med. 13:606–618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chhabra R and Saini N: MicroRNAs in cancer
stem cells: Current status and future directions. Tumour Biol.
35:8395–8405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu M, Lu S, He W, Zhang L, Ma Y, Lv P, Ma
M, Yu W, Wang J, Zhang M, et al: ULK1-regulated autophagy: A
mechanism in cellular protection for ALDH2 against hyperglycemia.
Toxicol Lett. 283:106–115. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ji Q, Karnak D, Hao P, Wang R and Xu L: No
small matter: MicroRNAs - key regulators of cancer stem cells. Int
J Clin Exp Med. 3:84–87. 2010.PubMed/NCBI
|
|
25
|
Nimmo RA and Slack FJ: An elegant miRror:
MicroRNAs in stem cells, developmental timing and cancer.
Chromosoma. 118:405–418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Meng F, Glaser SS, Francis H, DeMorrow S,
Han Y, Passarini JD, Stokes A, Cleary JP, Liu X, Venter J, et al:
Functional analysis of microRNAs in human hepatocellular cancer
stem cells. J Cell Mol Med. 16:160–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xia H and Hui KM: MicroRNAs involved in
regulating epithelial-mesenchymal transition and cancer stem cells
as molecular targets for cancer therapeutics. Cancer Gene Ther.
19:723–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen Y, Pan Y, Xu L, Chen L, Liu L, Chen
H, Chen Z and Meng Z: Identifying microRNA-mRNA regulatory network
in gemcitabine-resistant cells derived from human pancreatic cancer
cells. Tumour Biol. 36:4525–4534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang YH, Yang YL, Huang FC, Tiao MM, Lin
YC, Tsai MH and Wang FS: MicroRNA-29a mitigation of endoplasmic
reticulum and autophagy aberrance counteracts in obstructive
jaundice-induced fibrosis in mice. Exp Boil Med (Maywood).
243:13–21. 2018. View Article : Google Scholar
|
|
30
|
Tian J, An X and Niu L: Role of microRNAs
in cardiac development and disease. Exp Ther Med. 13:3–8. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsuchiya H, Tomita K, Mori Y, Asada N,
Morinaga T, Kitano S and Yamamoto N: Caffeine-assisted chemotherapy
and minimized tumor excision for nonmetastatic osteosarcoma.
Anticancer Res. 18:657–666. 1998.PubMed/NCBI
|
|
34
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Magri F, Vanoli F and Corti S: miRNA in
spinal muscular atrophy pathogenesis and therapy. J Cell Mol Med.
22:755–767. 2018.PubMed/NCBI
|
|
36
|
Romakina VV, Zhirov IV, Nasonova SN,
Zaseeva AV, Kochetov AG, Liang OV and Tereshchenko SN: MicroRNAs as
biomarkers of cardiovascular diseases. Kardiologiia. 66–71.
2018.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tian K, Wang L, Di R, Xu J, Li G and Li Z:
Effect and mechanism of miRNA to osteosarcoma cell. Pak J Pharm
Sci. 27 (5 Suppl):1657–1660. 2014.PubMed/NCBI
|
|
38
|
Xie B, Li Y, Zhao R, Xu Y, Wu Y, Wang J,
Xia D, Han W and Chen D: Identification of key genes and miRNAs in
osteosarcoma patients with chemoresistance by bioinformatics
analysis. Biomed Res Int. 2018:47610642018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu X, Liu T, Fang O, Dong W, Zhang F,
Leach L, Hu X and Luo Z: MicroRNA-708-5p acts as a therapeutic
agent against metastatic lung cancer. Oncotarget. 7:2417–2432.
2016.PubMed/NCBI
|
|
40
|
Yang J, Wei J, Wu Y, Wang Z, Guo Y, Lee P
and Li X: Metformin induces ER stress-dependent apoptosis through
miR-708-5p/NNAT pathway in prostate cancer. Oncogenesis.
4:e1582015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dodurga Y, Seçme M and Lale
Şatıroğlu-Tufan N: A novel oncogene URG4/URGCP and its role in
cancer. Gene. 668:12–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yu G, Zhang T, Jing Y, Bao Q, Tang Q and
Zhang Y: miR-519 suppresses nasopharyngeal carcinoma cell
proliferation by targeting oncogene URG4/URGCP. Life Sci.
175:47–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tokay E and Kockar F: Identification of
intracellular pathways through which TGF-β1 upregulates URG-4/URGCP
gene expression in hepatoma cells. Life Sci. 144:121–128. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dodurga Y, Seçme M, Eroğlu C, Gündoğdu G,
Avcı ÇB, Bağcı G, Küçükatay V and Lale Şatıroğlu-Tufan N:
Investigation of the effects of a sulfite molecule on human
neuroblastoma cells via a novel oncogene URG4/URGCP. Life Sci.
143:27–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cai J, Li R, Xu X, Zhang L, Wu S, Yang T,
Fang L, Wu J, Zhu X, Li M and Huang Y: URGCP promotes non-small
cell lung cancer invasiveness by activating the NF-κB-MMP-9
pathway. Oncotarget. 6:36489–36504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu M, Chen J, Wang Y, Hu J, Liu C, Feng C
and Zeng X: URGCP/URG4 promotes apoptotic resistance in bladder
cancer cells by activating NF-κB signaling. Oncotarget.
6:30887–30901. 2015.PubMed/NCBI
|
|
47
|
Xing S, Zhang B, Hua R, Tai WC, Zeng Z,
Xie B, Huang C, Xue J, Xiong S, Yang J, et al: URG4/URGCP enhances
the angiogenic capacity of human hepatocellular carcinoma cells in
vitro via activation of the NF-κB signaling pathway. BMC Cancer.
15:3682015. View Article : Google Scholar : PubMed/NCBI
|