Introduction
Chronic rhinosinusitis (CRS), a frequently seen
disease in the rhinology department, involves inflammation of the
nose and paranasal sinuses mucosa lasting >12 weeks (1). Chronic rhinosinusitis without nasal
polyposis (CRSsNP) accounts for about 60% of chronic rhinosinusitis
and is a common refractory disease in rhinology (2). CRS includes symptoms of nasal
discharge, nasal obstruction, facial pain, and a decreased sense of
smell. It severely affects the quality of life of patients.
CRSsNP often originates from bacterial infection and
develops into chronic inflammation in uncorrected treatment.
Gram-positive bacteria such as coagulase-negative staphylococci
(such as Staphylococcus aureus and Streptococcus
viridans) and Gram-negative enteric rods are known to exist in
the sinuses of adults with CRS. Bacterial infection causes mucosal
inflammation with infiltration of neutrophils and monocytes,
reconstruction of the extracellular matrix, goblet cell
hyperplasia, and tissue necrosis (3,4).
Several mediators and cytokines are involved in this
pathologic process, with increased expression levels of interleukin
(IL)-1 and 6, tumor necrosis factor (TNF-α), eosinophil cationic
protein, collagen, and urokinase plasminogen activator (Plau).
Increased expression levels of tissue remodeling genes such as
matrix metalloproteinase (MMPs), fibroblast growth factor (FGF),
bone morphogenetic proteins (BMP), and transforming growth factor
(TGF-β1) have also been found in bacterial chronic rhinosinusitis
(5). However, the genomic expression
of coagulation cascade-related cytokines, which might serve a
crucial role in host defense, inflammation and tissue remodeling,
has not been found.
The present study was carried out to investigate the
changes in the gene profile in bacterial CRS using a gene array
test. Bacterial chronic rhinosinusitis, simulating CRSsNP, was
induced in a mouse model, the pathway analyzed and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) used
to verify the key pathway in this pathogenesis.
Materials and methods
Animals and experiment groups
All experimental procedures were approved by the
Institutional Animal Care and Use Committee of Shanghai Jiao Tong
University. Adult male specific pathogen-free C57BL/6J mice (n=24)
were obtained from Shanghai Laboratory Animal Research Center
(Shanghai, China). Mice were aged between 10–12 weeks and weighed
24±3 g. All mice were housed at room temperature (20–23°C) in a 12
h light/ dark cycle with soft food and tap water provided
continuously ad libitum. Two groups were created according
to a randomization schedule (n=12 each): 1) bacterial chronic
rhinosinusitis (CRS) group: The right nasal cavity was occluded for
3 months with Merocel nasal pack (Medtronic, Minneapolis, MN, USA)
inoculated with 20 µl overnight cultured Staphylococcus
aureus (cat. no. ATCC25293; American Type Culture Collection,
Manassas, VA, USA), 2) Sham operation group (control group): The
nasal cavity was opened but no Merocel was inserted.
The bacterial CRS mouse model was built according to
the method described by Jacob et al (6), with certain modifications. A peritoneal
injection of 1% pentobarbital sodium (0.6 ml/100 g; Sinopharm
Chemical Reagent Co., Ltd, Shanghai, China) was given to
anesthetize the animals. An incision was then made over the right
snout up to the nasal dorsum to expose the right bony anterior
naris. A Merocel nasal pack of about 5 mm length soaked in 20 µl of
a Staphylococcus aureus solution was inserted in the right
nasal cavity and the incision was closed. Following the operation,
the mice were returned to the mouse care facility where they were
housed for the next 3 months.
Histological analysis
For histological studies, the mice were anesthetized
with chloral hydrate (35 mg/100 g; Sinopharm Chemical Reagent Co.,
Ltd.) and sacrificed to harvest the nose. The tissue samples were
fixed in 4% paraformaldehyde solution, decalcified in EDTA
solution, embedded in paraffin blocks, sectioned, and then stained
with hematoxylin staining solution for 5 min and eosin staining
solution for 1 min at room temperature. Sections were imaged with a
Leica light microscope system (Leica Microsystems GmbH, Wetzlar,
Germany).
ELISA analysis
The mouse nasal mucosa was harvested and the total
protein content quantified using Bicinchoninic Acid (BCA) kit
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). The test was
processed following the protocol on the IL-1β (Cat. no. MU30369,
Bio-Swamp; www.bio-swamp.com/) and TNF-α ELISA
(Cat. no. MU30030, Bio-Swamp) kit. The extracted protein samples of
bacterial CRS and control nasal mucosa were added to the pre-coated
96-well standard strip plate. Measurements of the optical density
were performed at 450 nm. The IL-1β and TNF-α expression levels
were determined using the standard curve prepared for each
assay.
cDNA microarray analysis
The nasal mucosal membrane was collected using
angled tweezers and washed with PBS solution. The Takara RNA
isolation kit (Takara Bio, Inc., Otsu, Japan) was used to extract
the total RNA of each sample following the operating manual. The
RNA integrity number of the inspected RNA samples was assessed
using the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The sample was further purified using the
RNeasy Micro Kit (Qiagen GmbH, Hilden, Germany). According to the
manufacturer's protocol, the total RNA was amplified and labeled
using a low input quick amp labeling kit after purification and
hybridized using the gene expression hybridization kit (Agilent
Technologies, Inc.). The microarrays were scanned using an Agilent
microarray scanner system (G2565CA). Data were extracted from
images using Feature Extraction software v10.7 (Agilent
Technologies, Inc.) with default settings. Raw data were normalized
by the quantile algorithm in Gene Spring v12.6.1 software (Agilent
Technologies, Inc.).
Gene regulation in bacterial CRS
The significantly differential expressed genes
(P<0.05) between the bacterial CRS and control groups were
selected. David v6.7 online software (david.abcc.ncifcrf.gov/) was used to perform molecular
function and pathway analysis.
Real time-quantitative polymerase
chain reaction (RT-qPCR)
The selected genes were tested on the 7900 HT
Sequenced Detection System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the iScript cDNA synthesis kit (Bio-Rad
Laboratories, Hercules, CA, USA) and ABI Power SYBR®
Green PCR Master Mix Kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The procedure was: 50°C for 2 min incubation;
95°C for 10 min; 95°C for 15 sec, and 60°C for 1 min for 40 cycles.
The primers are presented in Table
I. The relative quantification of the target genes were
normalized to the GAPDH gene expression level. Relative
quantification of the gene expression level was presented using the
2−ΔΔCq method (7).
 | Table I.Reverse transcription-quantitative
polymerase chain reaction primers used. |
Table I.
Reverse transcription-quantitative
polymerase chain reaction primers used.
| Primer Name | Sequence (5′ to
3′) |
|---|
| CF XII-F |
ACAAGCCCGGAGTCTACACA |
| CF XII-R |
GGGAAGGATAAAGCCTGGTT |
| Plau-F |
CCTGAGCAAAAGCTGGTGTA |
| Plau-R |
GGAGCATCACATGGAAGACC |
| Plaur-F |
GTGCCCTCGTGTTGTCTTCT |
| Plaur-R |
CAGCCCTTGTTCCAATTCTC |
| Col1a1-F |
GCCAAGAAGACATCCCTGAA |
| Col1a1-R |
TCAAGCATACCTCGGGTTTC |
| Col5a1-F |
TGGCATCCGAGGTCTGAAG |
| Col5a1-R |
CCCCCGGTCACCCTTTATT |
| GAPDH-F |
CGTGTTCCTACCCCCAATGT |
| GAPDH-R |
TGTCATCATACTTGGCAGGTTTCT |
Statistical analysis
The quantile algorithm in the Gene Spring v12.6.1
software (Agilent Technologies, Inc.) was used to normalize the
gene expression microarray raw data. Differential expression
>2-fold and a two-tailed probability value P<0.05 were
considered statistically significant. The Student's t-test and SPSS
v17.0 (SPSS, Inc., Chicago, IL, USA) were used for the statistical
analysis. Data were presented as the mean ± standard deviation and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Histological structure of the nasal
mucosa in the bacterial CRS mouse model
Histological structures of the nasal mucosa was
observed in the bacterial CRS mouse model and control group. In the
bacterial CRS group, hematoxylin and eosin staining showed damaged
cilia of nasal epithelium, increased lymphocytes and macrophages
infiltration, obvious lamina propria edema and gland hyperplasia
(Fig. 1).
IL-1β and TNF-α in bacterial CRS
murine model
The expression level of IL-1β and TNF-α in nasal
mucosa of both bacterial CRS and control group were evaluated using
ELISA. As shown in Fig. 2, the
levels of IL-1β (P<0.01) and TNF-α (P<0.01) were
significantly increased in bacterial CRS compared with the
control.
Gene expression in the nasal mucosa in
bacterial CRS and control group
Gene microarray showed there were 6,018 genes
expressed 2-fold differentially in the bacterial CRS group compared
with the control. Among them, 2,865 genes were upregulated 2-fold,
and 3,153 genes were downregulated 2-fold in bacterial CRS group as
shown by the heat map (Fig. 3).
Gene Ontology (GO) analysis of
differentially expressed genes
Out of the total number of 6,018 genes which were
differentially expressed, 3,945 genes underwent GO analysis in
DAVID v6.7 to determine molecular function. The important GO terms.
demonstrated to have the higher fold enrichment GO socres, were:
Dopamine receptor binding, 2-acylglycerol O-acyltransferase
activity, eicosatetraenoic acid binding, intracellular calcium
activated chloride channel activity, arachidonic acid binding,
diacylglycerol O-acyltransferase activity, eicosanoid binding,
6-phosphofructokinase activity, anaphylatoxin receptor activity,
prostaglandin-D synthase activity, C-X-C chemokine receptor
activity and C-X-C chemokine binding, folic acid transporter
activity, ligase activity, carbon-carbon bonding and
phosphofructokinase activity (Fig.
4).
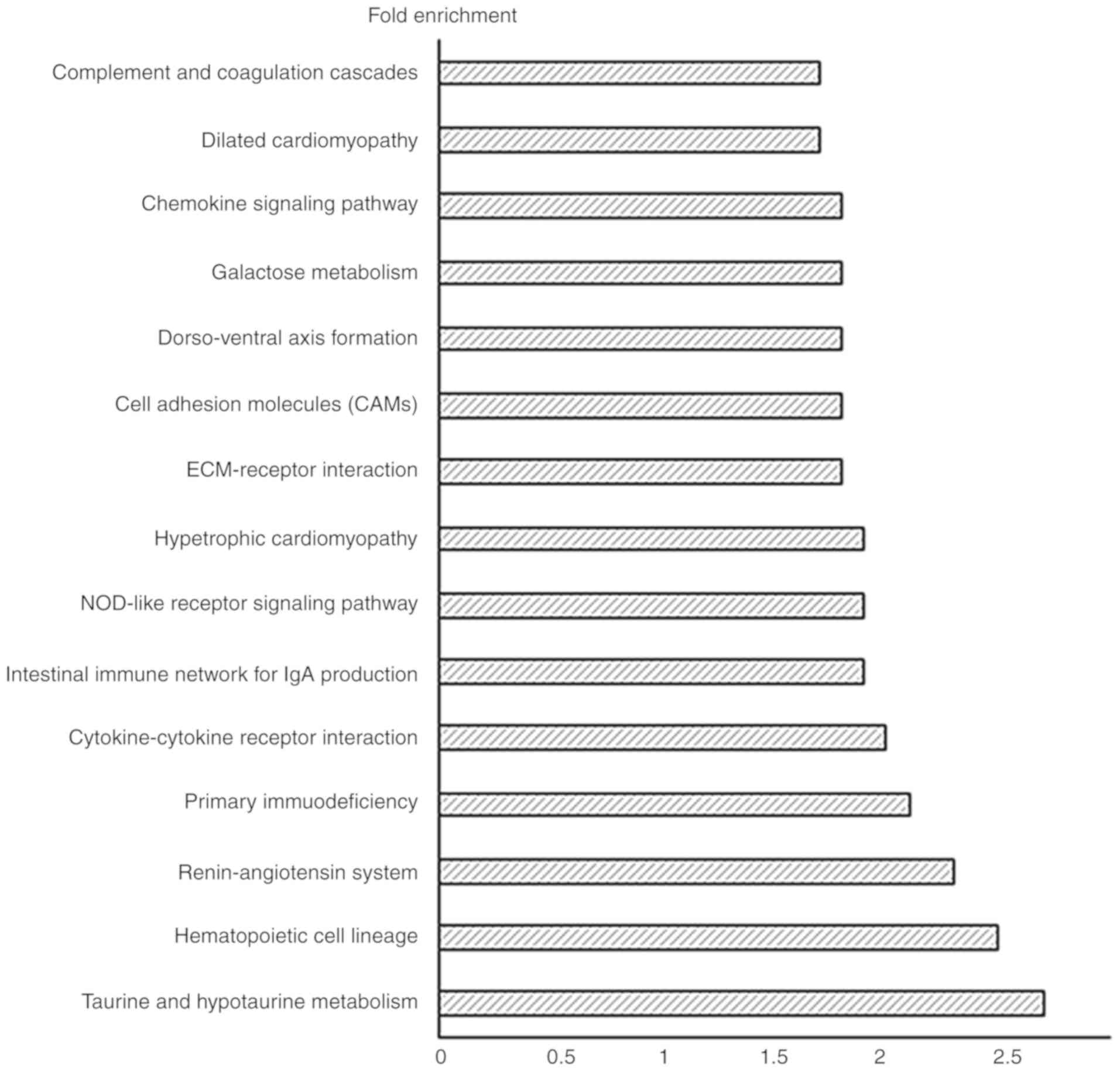
Pathway analysis of the differentially
expressed genes
The differentially expressed genes in bacterial CRS
underwent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis using DAVID v6.7. The important pathways in the
bacterial CRS mouse model were: Taurine and hypotaurine metabolism,
hematopoietic cell lineage, renin-angiotensin system, primary
immunodeficiency, cytokine-cytokine receptor interaction,
intestinal immune network for Immunoglobulin (Ig)A production,
nucleotide-binding oligomerization domain-like (NOD-like) receptor
signaling pathway, hypertrophic cardiomyopathy, ECM-receptor
interaction, cell adhesion molecules, dorso-ventral axis formation,
galactose metabolism, chemokine signaling pathway, dilated
cardiomyopathy, and complement and coagulation cascades (Fig. 5).
Differentially expressed gene analysis
in the coagulation cascade pathway
Twenty-eight genes were expressed differentially
(Table II) as detected in the KEGG
coagulation cascade pathway; of these, 24 were upregulated and four
genes were downregulated: Alpha-2-macroglobulin, coagulation factor
VIII, complement component 9 and Mannan-binding lectin (protein C);
RT-qPCR verified Coagulation factor XII (CF XII), urokinase
plasminogen activator (Plau), collagen type V alpha 1 chain
(Col5a1), urokinase receptor plasminogen activator (Plaur), and
collagen type I alpha 1 (Col1a1) genes which may serve an important
role in the activation of the coagulation cascade pathway. All
verified genes showed similar alterations in the gene microarray
test (P<0.05; Fig. 6).
 | Figure 6.Reverse transcription-quantitative
polymerase chain reaction test on selectively upregulated genes (A)
CFXII, Plau, Plaur, Col1a1 and Col5a1 (n=3 for each group). (B)
Gene array test of CFXII, Plau, Plaur, Col1a1 and Col5a1, in
bacterial CRS and control groups (n=3 for each group). *P<0.05
vs. control. CFXII, coagulation factor XII; Col1a1, collagen type I
alpha 1; Col5a1, collagen type V alpha 1 chain; CRS, chronic
rhinosinusitis; Plau, urokinase plasminogen activator; Plaur,
urokinase receptor plasminogen activator; B-CRS, bacterial chronic
rhinosinusitis. |
 | Table II.Genes with fold change >2 or
<0.5 (differential expression of genes between bacterial CRS
group and control group; P<0.05). |
Table II.
Genes with fold change >2 or
<0.5 (differential expression of genes between bacterial CRS
group and control group; P<0.05).
| NM_No | Gene Symbol | Fold change | P-values |
|---|
| NM_010776 | Mbl2 |
0.061 |
0.041 |
| NM_013485 | C9 |
0.124 |
0.008 |
| NM_007977 | F8 |
0.248 |
0.001 |
| NM_181858 | Cd59b |
0.448 |
0.001 |
| NM_028066 | F11 |
2.056 |
0.030 |
| NM_019775 | Cpb2 |
2.109 |
0.031 |
| NM_009779 | C3ar1 |
2.166 |
0.016 |
| NM_009776 | Serping1 |
2.272 |
0.002 |
| NM_010406 | Hc |
2.458 |
0.019 |
| NM_008873 | Plau |
2.773 |
0.020 |
| NM_144938 | C1s1 |
2.804 |
0.005 |
| NM_011576 | Tfpi |
2.893 |
0.001 |
| NM_023143 | C1ra |
3.217 |
0.001 |
| NM_028784 | F13a1 |
3.637 |
0.007 |
| NM_175628 | A2m |
4.251 |
0.016 |
| D16492 | Masp1 |
4.373 |
0.003 |
| NM_008223 | Serpind1 |
4.634 |
0.001 |
| NM_010172 | F7 |
4.875 |
0.001 |
| NM_021489 | F12 |
6.707 | <0.001 |
| NM_009778 | C3 | 12.681 | <0.001 |
| NM_008198 | Cfb | 19.482 | <0.001 |
| NM_011113 | Plaur | 20.906 |
0.006 |
| NM_007972 | F10 | 25.379 |
0.001 |
| NM_007577 | C5ar1 | 30.771 |
0.010 |
| NM_009245 | Serpina1c | 35.437 | <0.001 |
| BC037008 | Serpina1b | 78.520 | <0.001 |
| NM_009246 | Serpina1d | 83.682 | <0.001 |
Discussion
Chronic rhinosinusitis (CRS) has a prevalence of 8%
in China and 10.9% in Europe (1,8). CRSsNP
is the most common type of CRS (2).
Previous studies suggested that genes responsible for antigen
presentation, innate and adaptive immune responses, tissue
remodeling, and arachidonic acid metabolism are involved in the
pathological process of CRS (9).
However, no reports have shown comprehensive gene expression
patterns in bacterial CRS.
The present study utilized a murine model to profile
gene expression in bacterial CRS to prove that bacterial CRS causes
significant tissue inflammation by the alteration of multiple
genes. Some of these are the T-cell receptor complex,
immunoglobulin complex, integrin complex, ciliary rootlet and
collagen cell component, which may be relevant to cytokine-cytokine
receptor interaction, chemokine signaling, cell adhesion molecules,
and the complement and coagulation cascade pathway in bacterial
CRS.
To the best of our knowledge, this finding of gene
expression profile being changed in bacterial CRS has not been
reported before. This provides a basis for further identification
of pivotal cell pathways and key target molecules in bacterial CRS,
and has potential importance for the diagnosis and treatment of
bacterial CRS.
Activation of the coagulation cascade and deposition
of fibrin as a consequence of inflammation is well known, and is
thought to serve a critical role in host defense and in containing
microbial or toxic agents (10).
Previous studies have proved that in CRSsNP, excessive
extracellular collagen deposition occurs in the sinus cavities and
pseudocysts are absent in the thickened submucosa (11). In addition, dysregulation of the
coagulation cascade may serve an etiological role in many other
diseases, including rheumatoid arthritis, severe asthma
glomerulonephritis, and delayed-type hypersensitivity through
excessive fibrin deposition (12–14).
Infiltration of chronic inflammatory cells such as
neutrophils and mononuclear cells occurs in the nasal mucosa
(15). In this pathological process,
genes involved in antigen presentation, the innate and adaptive
immune response and tissue remodeling are regulated (9). Many studies have focused on the gene
expression in chronic rhinosinusitis with nasal polyposis using DNA
microarray (16,17) but none have studied the gene
expression profile in CRSsNP. In the present study, bacterial CRS
in the mice were studied using gene expression profile.
Consistent with previous studies in CRSsNP patients
using nasal membrane samples (18),
ciliary damage of the nasal epithelium, lamina propria edema, and
infiltration of inflammatory cells were found in the sinus
membranes of mice used in this study. A total of 6,018 genes were
found to have a 2-fold differential expression in bacterial CRS.
Among them, immunoglobulin-associated genes such as the
immunoglobulin (Ig) heavy chain variable, Ig light chain, and Ig
kappa chain variable were increased.
As reported in previous studies (19,20),
MMP-7, MMP-9, TGF-β1, Plau (uPA) and Plaur (uPAR) also showed
increased expression in this study. In the study by Sejima et
al uPA/uPAR expression was found to be upregulated in bacterial
CRS (20). Both Plau and Plaur are
believed to activate inflammatory cells such as neutrophils and
monocytes, releasing many kinds of inflammatory and chemotactic
mediators, with direct cytolytic effects on several bacterial
species (21,22). Additionally, uPA/uPAR could modulate
the extracellular matrix by activating plasmin and MMPs and affect
leukocyte migration (23). There are
many other unreported genes associated with bacterial CRS, further
bioinformatics and experimental studies are required to identify
the crucial genes.
Coagulation and inflammation are considered two
distinct pathologies, but they closely interact with each other at
multiple levels, for example, coagulation factor Xa, coagulation
factor XIa and plasmin activate both complement component 5 and
complement component 3 to complement component 5a and complement
component 3a, respectively, which are involved in the inflammatory
response (24). In the present
study, GO term molecular function analysis and pathway analysis
were used to study the differentially expressed genes. Apart from
the immune-related pathways, 28 genes involved in the complement
and coagulation cascade pathways were expressed differentially.
RT-qPCR revealed that the levels of coagulation factor XII (CF
XII), Plau, Col5a1, Plaur, and Col1a1, which serve a key role in
the complement and coagulation pathways (22,25),
were found to be upregulated in bacterial CRS. Of these, Col1a1
(important components of collagen I) is significantly elevated in
chronic sinusitis (26), and the
expression of Plau and Plaur are elevated in sinusitis (27).
The CFXII-driven plasma contact system provided a
link between procoagulant and proinflammatory reactions and
contributes to host defense mechanisms as a component of the innate
immune system. It was also found to promote inflammatory reactions
by activating the kallikrein-kinin system (28), increasing the formation of bradykinin
which caused vascular permeability and vasodilation (29) and enhanced the inflammatory
reaction.
The present study demonstrated that CFXII is an
important factor in the coagulation system and inflammatory
reaction, which requires further study. The major coagulation
factors CFXII, Plau and Plaur serve a vital role in inflammation
and immunity, they may also be crucial to the pathogenesis of
bacterial CRS (Fig. 7).
In summary, the gene expression profile in the nasal
mucosa of mice in bacterial CRS is presented in the present study,
to the best of our knowledge, for the first time. This study
demonstrates that genes responsible for the complement and
coagulation cascade pathways have an important immunomodulatory
function. Further studies are required to better understand the
pathogenesis of bacterial CRS.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant no. 81400445).
Availability of the data and materials
All data generated or analyzed during the present
study is included in this published article.
Authors' contributions
LG and SW performed the bacterial chronic
rhinosinusitis animal model, mucosa tissue collection, and the
histological and ELISA assays. YZ and HH designed the present
study, including analysis and interpretation of the cDNA microarray
data. All authors read and approved the final manuscript.
Ethical approval and consent to
participate
The animal experiments and procedures were performed
in accordance with the guidelines for the use and care of
laboratory animals in Shanghai Jiao Tong University, the present
study was approved by the Institutional Animal Care and Use
Committee of Shanghai Jiao Tong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fokkens WJ, Lund VJ, Mullol J, Bachert C,
Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, et
al: European position paper on rhinosinusitis and nasal polyps
2012. Rhinol Suppl. 23:3, p preceding table of contents, 1–298.
2012.PubMed/NCBI
|
|
2
|
Dykewicz MS and Hamilos DL: Rhinitis and
sinusitis. J Allergy Clin Immunol. 125 (Suppl 2):S103–S115. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meltzer EO, Hamilos DL, Hadley JA, Lanza
DC, Marple BF, Nicklas RA, Bachert C, Baraniuk J, Baroody FM,
Benninger MS, et al: Rhinosinusitis: Establishing definitions for
clinical research and patient care. J Allergy Clin Immunol. 114
(Suppl 6):S155–S212. 2004. View Article : Google Scholar
|
|
4
|
Malekzadeh S, Hamburger MD, Whelan PJ,
Biedlingmaier JF and Baraniuk JN: Density of middle turbinate
subepithelial mucous glands in patients with chronic
rhinosinusitis. Otolaryngol Head Neck Surg. 127:190–195. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takabayashi T, Kato A, Peters AT, Hulse
KE, Suh LA, Carter R, Norton J, Grammer LC, Tan BK, Chandra RK, et
al: Increased expression of factor XIII-A in patients with chronic
rhinosinusitis with nasal polyps. J Allergy Clin Immunol.
132:584–592.e4. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jacob A, Faddis BT and Chole RA: Chronic
bacterial rhinosinusitis: Description of a mouse model. Arch
Otolaryngol Head Neck Surg. 127:657–664. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi JB, Fu QL, Zhang H, Cheng L, Wang YJ,
Zhu DD, Lv W, Liu SX, Li PZ, Ou CQ and Xu G: Epidemiology of
chronic rhinosinusitis: Results from a cross-sectional survey in
seven Chinese cities. Allergy. 70:533–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hsu J, Avila PC, Kern RC, Hayes MG,
Schleimer RP and Pinto JM: Genetics of chronic rhinosinusitis:
State of the field and directions forward. J Allergy Clin Immunol.
131:977–993, 993.e1-e5. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jennewein C, Tran N, Paulus P, Ellinghaus
P, Eble JA and Zacharowski K: Novel aspects of fibrin(ogen)
fragments during inflammation. Mol Med. 17:568–573. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Bruaene N, Derycke L, Perez-Novo CA,
Gevaert P, Holtappels G, De Ruyck N, Cuvelier C, Van Cauwenberge P
and Bachert C: TGF-beta signaling and collagen deposition in
chronic rhinosinusitis. J Allergy Clin Immunol. 124:253–259.e1-e2.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Boer JD, Majoor CJ, van't Veer C, Bel
EH and van der Poll T: Asthma and coagulation. Blood.
119:3236–3244. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gabazza EC, Osamu T, Yamakami T, Ibata H,
Sato T, Sato Y and Shima T: Correlation between clotting and
collagen metabolism markers in rheumatoid arthritis. Thromb
Haemost. 71:199–202. 1994.PubMed/NCBI
|
|
14
|
Neale TJ, Tipping PG, Carson SD and
Holdsworth SR: Participation of cell-mediated immunity in
deposition of fibrin in glomerulonephritis. Lancet. 2:421–424.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Bruaene N and Bachert C: Tissue
remodeling in chronic rhinosinusitis. Curr Opin Allergy Clin
Immunol. 11:8–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Platt M, Metson R and Stankovic K:
Gene-expression signatures of nasal polyps associated with chronic
rhinosinusitis and aspirin-sensitive asthma. Curr Opin Allergy Clin
Immunol. 9:23–28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Payne SC, Han JK, Huyett P, Negri J, Kropf
EZ, Borish L and Steinke JW: Microarray analysis of distinct gene
transcription profiles in non-eosinophilic chronic sinusitis with
nasal polyps. Am J Rhinol. 22:568–581. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Van Bruaene N, C PN, Van Crombruggen K, De
Ruyck N, Holtappels G, Van Cauwenberge P, Gevaert P and Bachert C:
Inflammation and remodelling patterns in early stage chronic
rhinosinusitis. Clin Exp Allergy. 42:883–890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Meng J, Qiao X, Liu Y, Liu F, Zhang
N, Zhang J, Holtappels G, Luo B, Zhou P, et al: Expression of TGF,
matrix metalloproteinases, and tissue inhibitors in Chinese chronic
rhinosinusitis. J Allergy Clin Immunol. 125:1061–1068. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sejima T, Holtappels G and Bachert C: The
expression of fibrinolytic components in chronic paranasal sinus
disease. Am J Rhinol Allergy. 25:1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mondino A and Blasi F: uPA and uPAR in
fibrinolysis, immunity and pathology. Trends Immunol. 25:450–455.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Del Rosso M, Margheri F, Serratì S, Chillà
A, Laurenzana A and Fibbi G: The urokinase receptor system, a key
regulator at the intersection between inflammation, immunity, and
coagulation. Curr Pharm Des. 17:1924–1943. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gong Y, Hart E, Shchurin A and Hoover-Plow
J: Inflammatory macrophage migration requires MMP-9 activation by
plasminogen in mice. J Clin Invest. 118:3012–3024. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dahlbäck B: Coagulation and
inflammation-close allies in health and disease. Semin
Immunopathol. 34:1–3. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gorbet MB and Sefton MV:
Biomaterial-associated thrombosis: Roles of coagulation factors,
complement, platelets and leukocytes. Biomaterials. 25:5681–5703.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kim DK, Kang SI, Kong IG, Cho YH, Song SK,
Hyun SJ, Cho SD, Han SY, Cho SH and Kim DW: Two-track medical
treatment strategy according to the clinical scoring system for
chronic rhinosinusitis. Allergy Asthma Immunol Res. 10:490–502.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li X, Zhan Z, Sun J, Zeng B, Xue Y and
Wang S: Nasal mucosa remodeling in chronic rhinosinusitis without
nasal polyps. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi.
27:1110–1113, 1117, 2013 (In Chinese). PubMed/NCBI
|
|
28
|
Kenne E, Nickel KF, Long AT, Fuchs TA,
Stavrou EX, Stahl FR and Renné T: Factor XII: A novel target for
safe prevention of thrombosis and inflammation. J Intern Med.
278:571–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Björkqvist J, Jämsä A and Renné T: Plasma
kallikrein: The bradykinin-producing enzyme. Thromb Haemost.
110:399–407. 2013. View Article : Google Scholar : PubMed/NCBI
|