Introduction
Endometrial cancer is one of the most common
invasive malignancies of the female genital tract. The incidence of
endometrial cancer has gradually increased, approaching or even
surpassing that of cervical cancer, posing a serious threat to
women's health (1). Surgery is the
standard treatment for endometrial cancer, followed by
chemotherapy, radiotherapy or hormone (progesterone) therapy
(1). However, the prognosis of
patients with advanced endometrial cancer remains poor following
standard treatment and is often associated with significant side
effects, including myelosuppression, liver damage, radiation
enteritis and radiation cystitis (1). In recent years, gene therapy has become
increasingly common in clinical cancer research (2). Gene therapy has several advantages,
which include high selectivity and efficacy on metastases in
advanced cancer, making it a promising therapeutic strategy for the
treatment of patients with advanced cancer and a possible
alternative to surgery, radiotherapy and chemotherapy (2).
S-phase kinase-associated protein 2 (Skp2) is a
substrate recognition component of the E3 ubiquitin-protein ligase
complex SCF (Skpl-Cullin-F-box) that recognizes specific
phosphorylated substrates and mediates the ubiquitination and
subsequent degradation of cellular regulatory factors involved in
regulating cell cycle progression (3), signal transduction and transcription.
Several cell cycle regulators are substrates of the ubiquitin
proteasome pathway, which include cyclin D1 and p27 (4). Skp2 is involved in cell signal
transduction and transcriptional regulation and is closely
associated with several oncogenes and tumor suppressor genes,
indicating that Skp2 possesses oncogenic potential (5). Skp2 serves a regulatory role in several
processes including cell cycle regulation, growth, differentiation,
proliferation, metastasis and apoptosis. Skp2 has also been
reported to serve a role in drug resistance and is closely
associated with the development of tumors, demonstrating a
prognostic value in cancer (6–8). Skp2 is
overexpressed in several types of human cancer, and it therefore
targeting Skp2 may be a promising strategy for cancer treatment
(9–11). Several studies have demonstrated that
Skp2 expression is significantly increased in endometrial cancer
compared with normal endometrium tissues (9–11). Skp2
is first produced in the G1/S phase of the cell cycle, increases in
the S/G2 phase and decreases rapidly in the M phase (12,13).
Skp2 thus exhibits time-dependent expression in the cell (12,13).
Therefore, targeted therapy may increase the sensitivity of
chemotherapy and radiotherapy at specific cell cycle phases.
In the current study, Skp2 was selected as an RNAi
target gene. Specific RNAi lentiviral expression vectors were
constructed to inhibit Skp2 expression at the mRNA level to
determine the effect of Skp2 inhibition on the characteristics of
the endometrial cancer cell line, HEC-1-A, including the cell
cycle, apoptosis and proliferation. The aim of the current study
was to investigate the underlying mechanism of Skp2 in endometrial
cancer progression as a potential for targeted gene therapy.
Materials and methods
Materials
HEC-1-A and 293T cell lines were obtained from Cell
Bank of the Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). The lentiviral vector system [which includes the
lentiviral vector pLV-green fluorescent protein (GFP), the
lentivirus plasmid and two auxiliary packaging plasmids] was
obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China). T4 DNA
ligase and restriction endonucleases were purchased from New
England BioLabs, Inc., Ipswitch, MA, USA. A QIAGEN®
Plasmid Extraction kit was purchased from Qiagen GmbH (Hilden,
Germany). The PrimeScript™ RT reagent kit and SYBR®
Premix Ex Taq™ were purchased from Takara Biotechnology Co., Ltd.
(Dalian, China). PCR primer synthesis was performed by Shanghai
Shenggong Biology Engineering Technology Service, Ltd. (Shanghai,
China). Mouse anti-human Skp2 monoclonal antibodies were purchased
from Invitrogen; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Mouse anti-human p27 monoclonal antibodies were purchased from
Abcam (Cambridge, UK). Mouse anti-human cyclin D1 antibodies was
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Mouse anti-human caspase-3 antibodies were purchased from Santa
Cruz Biotechnology, Inc. (Dallas, TX, USA). A bicinchoninic acid
(BCA) protein assay kit, radioimmunoprecipitation (RIPA) assay
buffer, tetramethylethylenediamine, phenylmethylsulfonyl fluoride,
a BeyoECL Plus kit, mouse anti-human β-actin antibodies, and
horseradish peroxidase (HRP)-labeled goat anti-mouse IgG secondary
antibodies were all purchased from Beyotime Institute of
Biotechnology (Haimen, China). The Cell Cycle Assay kit [which
includes propidium iodide and RNaseA] was purchased from Nanjing
KeyGen Biotech Co., Ltd (Nanjing, China; cat. no. KGA512). The Cell
Counting Kit-8 (CCK-8) assay was purchased from Tongren Chemical
Institute (Kyushu, Japan).
Recombinant lentiviral vector
construction
Two Skp2 transcript variants (ACCESSION NM_032637
and NM_005983) were identified in GenBank (https://www.ncbi.nlm.nih.gov/gene/6502) and the target
was designed according to the homologous region. According to the
design principle of RNA interference sequences, a pair of negative
control nonsense sequences and four pairs of shRNA sequences were
designed using BLOCK-iT™ RNAi Designer software from Ambion (Thermo
Fisher Scientific, Inc.; http://rnaidesigner.thermofisher.com/rnaiexpress/).
The constructs were named accordingly: Skp2-NC, Skp2-1, Skp2-2,
Skp2-3 and Skp2-4. The siRNA target sequence was: Skp2-NC,
5′-TTCTCCGAACGTGTCACGT-3′; Skp2-1, 5′-CCTTAGACCTCACAGGTAA-3′;
Skp2-2, 5′-CCAACCATTGGCTGAACAT-3′; Skp2-3,
5′-CAGAAAGAATCTCCAGAAA-3′ and Skp2-4, 5′-GTGTCATGCTAAAGAATGA-3′.
BLASTN was used to align the sequences with the corresponding human
genome database, and homologous sequences of other coding sequences
were excluded. The double-strand DNA oligonucleotide containing the
interference sequence was synthesized by Shanghai GeneChem Co.,
Ltd. and cloned into the AgeI and EcoRI digested lentiviral vector
pLV-GFP (part of the aforementioned lentiviral vector system). Upon
transformation into E. coli competent cells, the positive
clones were identified by PCR using the following upstream and
downstream primers: 5′-CCTATTTCCCATGATTCCTTCATA-3′ and
5′-GTAATACGGTTATCCACGCG-3′. A stock solution containing 10× buffer,
0.5 mM MgCl2, 2.5 mM dNTPs (Shanghai GeneChem Co., Ltd.,
Shanghai, China), 0.2 U/µl, Taq DNA polymerase (Takara
Biotechnology Co., Ltd., Dalian, China) and 0.4 µM primers
(Shanghai GeneChem Co., Ltd., Shanghai, China). A total of 20 µl of
PCR stock solution was added to each tube and the following
thermocycling conditions were applied: 94°C for 2 min, followed by
30 cycles of denaturation at 94°C for 30 sec, annealing at 60°C for
30 sec, elongation at 72°C for 30 sec and final extension at 72°C
for 7 min. Agarose gel electrophoresis was then used to analyze the
result. PCR primers were designed to reside within 2 separate DNA
fragments so that a positive PCR band of expected size reflected
the correct joining of 2 DNA fragments. For effective colony PCR
and subsequent analysis by agarose gel, PCR product sizes of ~343
bp were found to be optimal. The positive clones were selected and
sent to Shanghai Meiji Biotechnology Co., Ltd., (Shanghai, China)
for sequencing analysis.
Lentivirus packaging
The recombinant lentivirus plasmid and two auxiliary
packaging plasmids (provided by the aforementioned lentiviral
vector system) were purified using the QIAGEN Plasmid Extraction
kit, according to the manufacturer's protocol. Recombinant
lentiviral particles were generated using 293T cells which were
co-transfected using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Following 48 h transfection, 293T cell supernatants rich
in lentivirus particles were collected. After the supernatant was
obtained, virus titers were determined in 293T cells using an
stepwise dilution method (14).
Cell culture and transfection of
HEC-1-A cells
HEC-1-A cells were cultured in high glucose
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 100 U/ml
penicillin-streptomycin and 10% newborn bovine serum (Zhejiang
Tianhang Biotechnology Co., Ltd., Zhejiang, China), and cells were
maintained at 37°C in a 5% CO2-humidified incubator. The
following groups were included: The negative control group
(LV-Skp2-NC) and the experimental groups (LV-Skp2-1, LV-Skp2-2,
LV-Skp2-3 and LV-Skp2-4). Cells were seeded into six-well plates at
a density of 1×105 cells/well. Once cells reached 30%
confluence, the recombinant lentiviruses LV-Skp2-1, LV-SKP-2,
LV-Skp2-3 or LV-SKP-4 were used to transfect HEC-1-A cells (5 ·g/ml
polybrene; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany);
multiplicity of infection, 10). Transfection efficiency was
determined by observing GFP expression in HEC-1-A cells using an
inverted fluorescence microscope (magnification, ×40) under six
fields of view.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from HEC-1-A cells on day 4
or 5 following recombinant lentiviral transfection using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (2.5 ·g) was reversed transcribed into cDNA using
the PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.),
according to the manufacturer's protocol. Subsequently, qPCR was
performed using the SYBR® Premix Ex Taq™ (Takara
Biotechnology Co., Ltd.), according to the manufacturer's protocol.
PRIMER3 5.0 software (PREMIER Biosoft International, Inc., Palo
Alto, CA, USA) was used to design specific primers for Skp2, p27,
Cyclin D1, and β-actin. The following primer pairs were used for
qPCR: Skp2 forward, 5′-CCAGGAGATTCCAGACCTGAGT-3′ and reverse,
5′-TGTCACTCCCTTTGCTCTTCAG-3′ (212 bp); p27 forward,
5′-GGGGTATGAAGAGCTTGCTTTG-3′ and reverse,
5′-GGGCAGTGAGGATAGGTTTCTG-3′ (308 bp); cyclin D1 forward,
5′-TCAAATGTGTGCAGAAGGAGGT-3′ and reverse,
5′-ATGGAGTTGTCGGTGTAGATGC-3′ (262 bp); β-actin forward,
5′-TCGTGCGTGACATTAAGGAG-3′ and reverse, 5′-AAGGTAGTTTCGTGGATGCC-3′
(214 bp). RT-qPCR uses a two-step reaction according to the
protocol of SYBR® PremixEx Taq™. The following
thermocycling conditions were utilized: Initial denaturation at
95°C for 15 sec; 40 cycles of 95°C for 5 sec and 60°C for 31 sec.
The relative mRNA expression of Skp2, p27 and cyclin D1 were
quantified using the 2−∆∆Cq method (15) and normalized to the internal
reference gene, β-actin.
Western blot analysis
Total protein was extracted from HEC-1-A cells on
day 7 following recombinant lentiviral transfection using RIPA
lysis buffer and low-temperature centrifugation at 10,000 × g for
10 min at 4°C. Total protein was quantified using a BCA assay and
50 ·g protein/lane was separated via SDS-PAGE on a 12% gel at 110
V. The separated proteins were transferred onto polyvinylidene
difluoride membranes and blocked with 10% skim milk solution at
room temperature for 2–3 h. The membranes were then incubated with
primary antibodies against Skp2 (1:500; cat. no. 32-3300), p27
(1:500; cat. no. ab54563), cyclin D1 (1:1,000; cat. no. 2926),
caspase-3 (1:200; cat. no. sc-396225) or β-actin (1:1,000; cat. no.
AA128) overnight at 4°C. The membranes were washed with
Tris-buffered saline containing 0.04% Tween-20 (TBST), followed by
incubation with HRP-labeled secondary antibodies (1:2,000; cat. no.
A0216) for 2 h at room temperature. The membranes were washed with
TBST prior to chemiluminescence imaging. Protein bands were
visualized using the BeyoECL Plus (Beyotime Institute of
Biotechnology) and protein expression was quantified using the
ImageScanner Imaging System (Amersham; GE Healthcare Life Sciences,
Little Chalfont, UK) to process the film, Imagequant TL 7.0 image
analysis software (Amersham; GE Healthcare Life Sciences, Little
Chalfont, UK) was utilized to analyze the molecular weight and net
optical density of the protein band.
Flow cytometry
Cell cycle and apoptosis assays were performed using
a Beckman Coulter EPICS XL flow cytometer (Beckman Coulter, Inc.,
Brea, CA., USA). HEC-1-A cells were seeded into 6-well plates at
density of 1×105 cells/well and upon reaching 30%
confluence, the negative control virus LV-Skp2-NC and recombinant
lentiviruses LV-Skp2-3 and LV-Skp2-4 were transfected into HEC-1-A
cells. A total of 6 days following infection, EDTA-free trypsin was
added and the cells were harvested. Following centrifugation at
1,000 × g for 5 min, cell pellets were washed twice with pre-cooled
PBS and fixed with 70% pre-cooled ethanol at −20°C for >12 h.
Following further centrifugation at 1,000 × g for 5 min, ethanol
was removed and cells were washed twice with PBS. Cells were
resuspended with PBS, then propidium iodide and RNaseA were added
at a final concentration of 20 and 50 µg/ml, respectively,
according to the instructions of the Cell Cycle Assay kit. Samples
were then incubated for 30 min at 37°C in the dark. Analysis of the
flow cytometric data were performed using System IITM 3.0 software
(Beckman Coulter, Inc.). All experiments were performed in
triplicate.
Cell proliferation assay
Cell proliferation was analyzed using a CCK-8 assay
(Tongren Chemical Institute). HEC-1-A cells were seeded into
96-well plates at density of 5×103 cells/well in high
glucose DMEM supplemented with 100 U/ml penicillin-streptomycin and
10% newborn bovine serum and once the cells reached 30% confluence,
the negative control virus LV-Skp2-NC and recombinant lentiviruses
LV-Skp2-3 and LV-Skp2-4 were transfected into HEC-1-A cells. The
medium was replaced with DMEM supplemented with 10% newborn bovine
serum (90 ·l/well) and 10 ·l CCK-8 reagent at 12, 36, 60, 84, 108,
132, and 156 h time points following infection and cells were
incubated at 37°C in a 5% CO2-humidified incubator for
30 min. The absorbance was measured at a wavelength of 450/630 nm
with a microplate reader (Bio-Rad Model 550; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Three biological and three technical
replicates were performed.
Bioinformatics analysis
Skp2 co-expression gene data was analyzed using
Oncomine (www.oncomine.org) from published data
obtained from obtained from Wu et al (16). The heatmap and the association
between Skp2 and co-expressed genes in the cohort of patients with
endometrial cancer in the TGCA database were analyzed using UCSC
Xena (xena.ucsc.edu), and the expression data
for each gene was used for further analysis.
Statistical analysis
Data presented as the mean ± standard
deviation. All statistical analyses were performed using SPSS
statistical software (version 17.0; SPSS, Inc., Chicago, IL, USA).
One-way analysis of variance followed by a Dunnett's post hoc test
was used to analyze differences among multiple groups. A Student's
t-test was used to analyze differences between the expression level
of specific genes in the Skp2-high and Skp2-low endometrial cancer
samples. P<0.05 was considered to indicate a statistically
significant difference.
Results
Transfection efficiency of recombinant
lentivirus
Transfection efficiency was examined in HEC-1-A
cells on day 7 following transfection with recombinant lentiviral
vectors LV-Skp2-1, LV-Skp2-2, LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC.
The percentage of cells expressing GFP (the transfection
efficiency) observed under a fluorescent microscope was >70%
compared with cells observed under light microscopy (data not
shown).
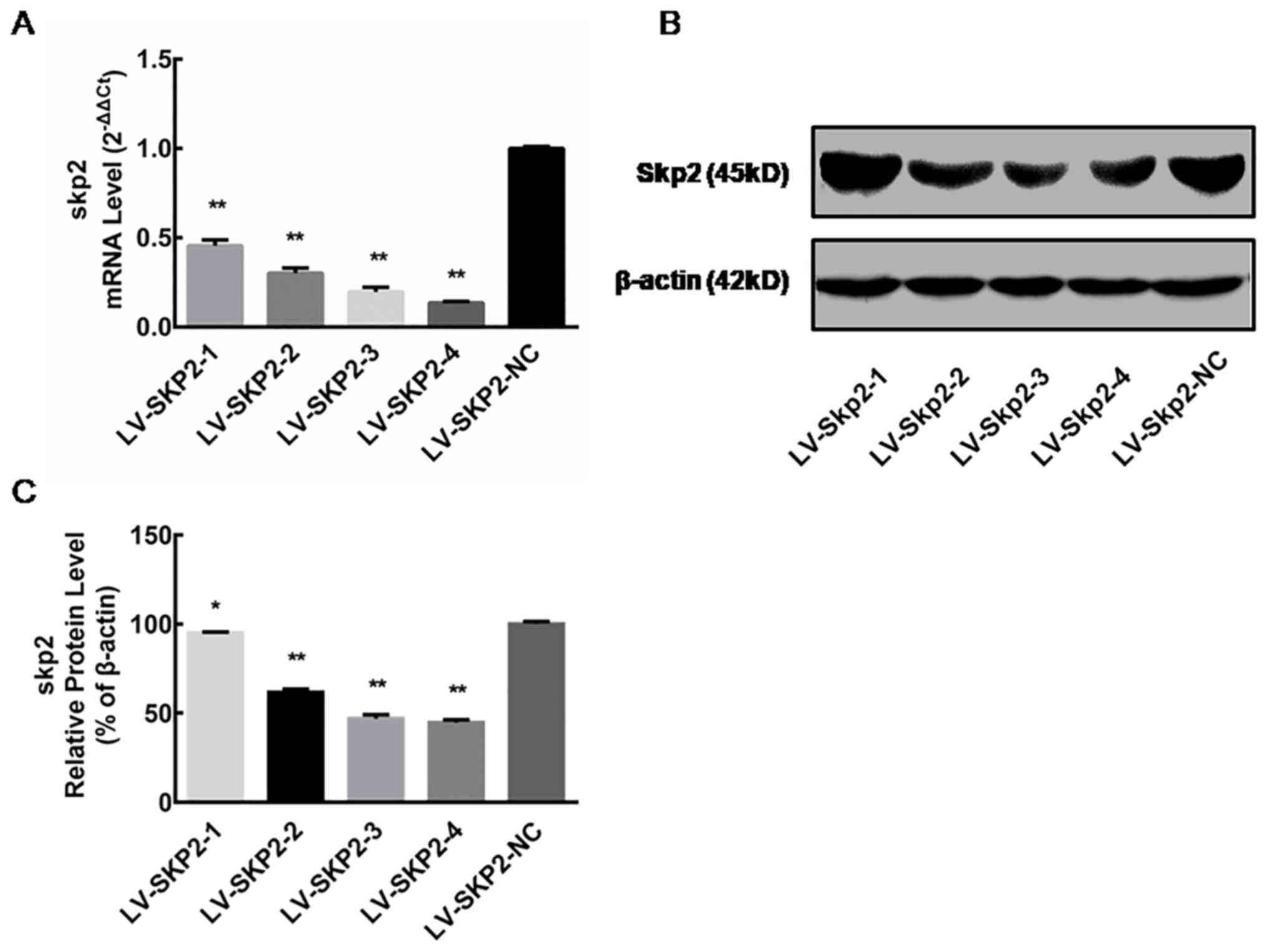
Skp2 expression following lentiviral
transfection
The relative Skp2 mRNA expression level was
determined via RT-qPCR in HEC-1-A cells on day 5 following
transfection with recombinant lentiviral vectors (LV-Skp2-1,
LV-Skp2-2, LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC). The mRNA expression
of Skp2 was significantly decreased in cells transfected with
LV-Skp2-3 or LV-Skp2-4 compared with the negative control,
LV-Skp2-NC (P<0.01; Fig. 1A).
These results indicate that recombinant lentiviruses can
specifically inhibit the expression of Skp2 mRNA, with inhibition
rates of 54.39±3.19, 69.87±2.88, 80.36±2.61 and 86.46±0.77%,
respectively (data not shown). The relative protein expression of
Skp2 was determined by western blot analysis in HEC-1-A cells on
day 7 following transfection with recombinant lentiviral vectors.
The protein expression of Skp2 was significantly decreased in cells
transfected with all four lentiviral vectors compared with cells
transfected with the negative control (P<0.01; Fig. 1B and C). These results suggest that
all four recombinant lentiviruses can influence Skp2 protein
inhibition, with inhibition rates of 5.11±0.68, 38.16±0.85,
53.04±1.51 and 55.34±0.97%, respectively (data not shown).
Furthermore, the inhibitory effect of LV-Skp2-3 and LV-Skp2-4 was
greater than LV-Skp2-1 or LV-Skp2-2 (Fig. 1C). The two recombinant lentiviral
vectors LV-Skp2-3 and LV-Skp2-4 could therefore inhibit the
expression of Skp2 at the mRNA and protein level, achieving the
greatest effect on Skp2 gene silencing.
Effect of Skp2 inhibition on cell
cycle and apoptosis
Flow cytometry was used to detect the effect of RNA
interference of (RNAi)-induced Skp2 inhibition on the cell cycle
and apoptosis of HEC-1-A cells following transfection with
recombinant lentiviral vectors LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC.
The proportion of cells in the G0/G1 phase
was significantly decreased, while the proportion of cells in the
G2/M phase was significantly increased in cells transfected with
LV-Skp2-3 or LV-Skp2-4 compared with LV-Skp2-NC (P<0.05;
Table I). In addition, there were no
significant differences observed in the rate of apoptosis in cells
transfected with LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC, 2.59±0.10,
2.68±0.15 and 2.54±0.25%, respectively (Fig. 2A). Furthermore, there were no
significant differences in the protein expression of caspase-3 in
cells transfected with LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC (Fig. 2B and C).
 | Table I.Effect of recombinant lentivirus
interference on the cell cycle, as detected via flow cytometry. |
Table I.
Effect of recombinant lentivirus
interference on the cell cycle, as detected via flow cytometry.
| Group |
G0/G1 | S |
G2/M |
|---|
| LV-Skp2-NC | 40.00±4.90 | 25.77±2.55 | 34.23±2.35 |
| LV-Skp2-3 |
32.63±0.83a | 25.23±0.65 |
42.13±0.25a |
| LV-Skp2-4 |
30.26±1.55a | 26.40±0.80 |
43.33±0.75a |
Effect of RNAi-induced Skp2 inhibition
on cell proliferation
Cell proliferation was assessed in HEC-1-A cells
following transfection with the recombinant lentiviral vectors
LV-Skp2-3 and LV-Skp2-4. Although there were no significant
differences at 12, 36 and 60 h, cell proliferation at 132 and 156 h
was significantly decreased in cells transfected with LV-Skp2-3 or
LV-Skp2-4 compared with LV-Skp2-NC (P<0.01; Fig. 3A). In addition, cell proliferation
significantly decreased at 84 and 108 h in cells transfected with
LV-Skp2-4 compared with LV-Skp2-NC (P<0.05; Fig. 3A). The relative mRNA expression of
cyclin D1 was determined via RT-qPCR in HEC-1-A cells on day 4 and
5 following transfection with recombinant lentiviral vectors
LV-Skp2-3, LV-Skp2-4 or LV-Skp2-NC. The results revealed that there
was no significant change in the mRNA expression of cyclin D1
(Fig. 3B). The relative protein
expression of cyclin D1 was detected via western blotting in
HEC-1-A cells on day 7 following transfection with recombinant
lentiviral vectors (Fig. 3C). The
protein expression of cyclin D1 was significantly decreased in
cells transfected with LV-Skp2-4 compared with LV-Skp2-NC
(P<0.01; Fig. 3D).
Changes in p27 expression following
letiviral transfection
The relative mRNA expression of p27 was determined
via RT-qPCR in HEC-1-A cells on day 4 and 5 following transfection
with recombinant lentiviral vectors LV-Skp2-3, LV-Skp2-4 or
LV-Skp2-NC. The results revealed that there was no significant
change in the mRNA expression of p27 (Fig. 4A). The relative protein expression of
p27 was detected via western blotting in HEC-1-A cells on day 7
following transfection with recombinant lentiviral vectors
(Fig. 4B). The protein expression of
p27 was significantly increased in cells transfected with LV-Skp2-3
or LV-Skp2-4 compared with LV-Skp2-NC transfected cells (P<0.01;
Fig. 4C).
Skp2 co-expressed genes
To further determine the underlying regulatory
mechanism of Skp2 in endometrial cancer, data mining was performed
using the Oncomine and TCGA databases. The co-expression data
obtained from Wu et al (16)
was analyzed using Oncomine. The results demonstrated that the
genes involved in the insulin-like growth factor 1 receptor (IGFR)
signaling pathway were enriched in the Skp2 co-expression gene
dataset (data not shown). Heatmaps of these genes were generated
and analyzed using UCSC Xena browser (Fig. 5A). The results demonstrated that the
expression of Skp2 was associated with insulin-like growth factor 2
receptor (IGF2R), insulin-like growth factor 2 mRNA binding protein
3 (IGF2BP3), insulin-like growth factor 2 mRNA binding protein 1
(IGFBP1) and cyclin F (CCNF), while no association was observed
with insulin-like growth factor 2 (IGF2), insulin-like growth
factor binding protein 6 (IGFBP6), insulin-like growth factor
binding protein 7 (IGFBP7) and insulin-like growth factor binding
protein 3 (IGFBP3; data not shown). In addition, there was no
association observed between Skp2 and insulin like growth factor 1,
IGFR and insulin-like growth factor 2 mRNA binding protein 2 (data
not shown). The gene expression data for the cohort of patients
(n=201) with endometrial cancer in the TGCA database were analyzed.
The 201 TCGA-EC samples were divided into two groups: Skp2-High
(n=100) and Skp2-low (n=101), according to the expression of Skp2
and the expression of each co-expressed gene. In the Skp2-High
group, the expression of IGF2R (P<0.001), IGF2BP3 (P<0.001),
IGF2BP1 (P<0.001) and CCNF (P<0.001) were significantly
increased, while the expression of IGF2 (P<0.05), IGFBP6
(P<0.001), IGFBP7 (P<0.001) and IGFBP3 (P<0.05) were
significantly decreased compared with the Skp2-low group (Fig. 5B). Taken together, these results
suggest that Skp2 may be regulated by the IGFR signaling
pathway.
 | Figure 5.Skp2 co-expressed with genes in the
IGFR signaling pathway. (A) Heatmap identification of gene
co-expression patterns for Skp2, IGF2R, IGF2BP3, IGF2BP1, CCNF,
IGF2, IGFBP6, IGFBP7 and IGFBP3 in the TCGA-EC dataset. (B) The
relative gene expression of these genes in Skp2-High and Skp2-Low
endometrial cancer samples. *P<0.05; ***P<0.001 as indicated.
Skp2, S-phase kinase associated protein 2; IGFR, insulin-like
growth factor 1 receptor; IGF2R, insulin-like growth factor 2
receptor; IGF2BP3, insulin-like growth factor 2 mRNA binding
protein 3; IGF2BP1, insulin-like growth factor 2 mRNA binding
protein 1; CCNF, Cyclin F; IGF2, insulin-like growth factor 2;
IGFBP6, insulin-like growth factor binding protein 6; IGFBP7,
insulin-like growth factor binding protein 7; IGFBP3, insulin-like
growth factor binding protein 3. |
Discussion
The ubiquitin-proteasome system is an important
mechanism for the regulation of protein content in eukaryotes via
protein degradation, which involves three sequential reactions
catalyzed by a ubiquitin-activating enzyme, a ubiquitin-conjugating
enzyme (E2) and a ubiquitin-protein ligase (E3) (10). In eukaryotic cells, protein
degradation is a highly selective and efficient ATP-dependent
process, which occurs in the cytoplasm and nucleus (17). Substrate proteins are ubiquitinated,
which specifically target the substrate protein for degradation by
the 26S proteasome (18). Protein
degradation regulates several cellular processes including cell
cycle progression. SCF (Skpl-Cullin-F-box) is a multisubunit E3
ubiquitin-protein ligase complex, consisting of F-box proteins,
Skp1, Cullins, Rbxl, Nedd8 and Skp2 (13,18).
Although RNAi is a powerful gene-silencing process, there are
relatively few studies that have assessed the application of RNAi
technology towards Skp2 gene regulation in endometrial cancer. The
aim of the current study was to investigate the expression of Skp2
in endometrial cancer and examine the effect of RNAi-induced Skp2
inhibition on the cell cycle, apoptosis and proliferation of the
endometrium cancer cell line HEC-1-A.
Cyclins are a family of regulatory proteins that
control cell cycle progression. Cyclins bind to and activate
cyclin-dependent kinases (Cdk), which control cell cycle processes
via the phosphorylation of its target substrate, consequently
driving the cell through G1 to S and G2 to M phase (19). If the G1-S or G2-M cell cycle
checkpoints are blocked, cell proliferation may be inhibited,
resulting in slow cell growth (19).
In normal cell cycle progression, cyclin D1 forms a complex with
Cdk4. Cyclin D1:Cdk4 phosphorylates and inactivates the
retinoblastoma (RB) protein during early G1 phase to release E2F
transcription factors, which initiate DNA synthesis, enabling cell
cycle progression from the G1 to S phase during cell proliferation
(20). Cyclin D1 overexpression
occurs in several types of malignant tumor and dysregulated cyclin
D1 expression can lead to the abnormal regulation of RB by Cdk4
(21). In addition, cyclin D1 can
induce the expression of cyclin E and the activation of the Cyclin
E-Cdk-2 complex in resting cells, causing the abnormal initiation
of the cell cycle, which leads to the persistent proliferation of
malignant cells (19). Several
studies have demonstrated that cyclin D1 is closely associated with
the occurrence, development, metastasis, prognosis and recurrence
of several types of tumor (22–24).
p27 specifically binds to various cyclin-Cdk
complexes and their monomers, blocking RB phosphorylation and
inhibiting cell proliferation (25).
In addition, p27 is also involved in cell-cell adhesion, promoting
cell differentiation and inducing apoptosis (26,27).
Several studies have demonstrated that the low expression of p27
occurs in several types of malignant tumor and is closely
associated with tumor invasion, metastasis, staging and prognosis
(28–30). In tumors, p27 is associated with a
reduction or loss of protein expression, while gene mutations are
rare (31). The low expression of
p27 also attenuates the inhibition of positive regulatory cell
cycle proteins, resulting in excessive cell proliferation and tumor
development (32).
It is generally considered that Skp2 expression is
negatively associated with p27 expression (33,34);
however, Kim et al (35)
reported that in 332 untreated patients with cervical cancer, there
was no correlation between Skp2 and p27. Additionally, Handra-Luca
et al (36) and Fagan-Solis
et al (25) reported that
there was no correlation between Skp2 and p27 in salivary gland
epidermoid carcinoma and breast cancer, respectively. Several
studies have also demonstrated that low expression levels of p27 in
malignant tumors are associated with high expression levels of
cyclin D1 (37–39), although the correlation between p27
and cyclin D1 remains unclear. A previous study revealed that p27
and cyclin D1 are negatively correlated (37); however, Jonason et al
(40) reported that cyclin D1 has a
specific function in the post-transcriptional regulation of p27.
There may therefore be a feedback pathway between p27 and cyclin
D1, to maintain a dynamic balance between the positive and negative
regulators of the cell cycle. However, this feedback pathway may
not be a unidirectional regulatory pathway, but rather, an
intricate regulatory system distributed among various members. An
abnormality in one or more of these members may be involved in
tumor development and progression, however this remains to be
confirmed (41).
In the current study, the application of
lentivirus-mediated RNAi technology was used to inhibit Skp2 gene
expression. RT-qPCR results revealed that the inhibition of Skp2
expression did not affect the mRNA expression of p27 and cyclin D1.
However, downregulation of Skp2 increased the expression levels of
p27 and decreased the expression levels of Cychin D1. Flow
cytometry demonstrated that RNAi-induced Skp2 inhibition exerts an
anti-proliferative effect by inducing cell cycle arrest in the G2/M
phase, with no evidence of apoptosis. The inactive 35 kDa subunit
(but not the cleaved, activated 17 kDa subunit) of caspase-3 was
detected. However, there was no difference in the protein
expression of caspase-3 following RNAi-induced Skp2 inhibition
compared with the control group. These results indicate that the
inhibition of Skp2 expression does not promote apoptosis of HEC-1-A
cells. Therefore, RNAi-induced Skp2 inhibition does not affect the
mRNA expression level of p27, but it does upregulate its protein
expression by reducing the ubiquitin degradation of P27
protein.
Tsvetkov et al (42) revealed that SCF-Skp2 binds to
phosphorylated Thr187 in p27 to specifically target p27 for
degradation via a ubiquitin-dependent process during cell-cycle
progression. In the current study, the inhibition of Skp2
expression did not increase the protein expression of cyclin D1,
but instead downregulated cyclin D1 expression, which indicated
that cyclin D1 may not be a substrate of the SCF complex, but a
protein closely associated with Skp2. A study by Carrano et
al (43) revealed that no direct
biochemical data obtained using a reconstituted in vitro
ubiquitination system, was available to verify whether cyclin D1,
p21 and E2F-1 were true Skp2 substrates. However, It was likely,
that Skp2 targets other substrates, including p27, for
ubiquitination (43). Furthermore,
the previous study revealed that two different Skp2 antisense
oligonucleotides decrease Skp2 expression and concomitantly
increase the levels of endogenous p27 (43). The study reveals that the phosphatase
and tensin homolog/phosphoinositide 3-kinase signaling pathway and
Cyclin D1 control a novel pathway that regulates the assembly of
the SCF-Skp2 complex by modulating cullin neddylation and cullin
associated and neddylation dissociated 1 binding at the G1/S cell
cycle transition (37). Cyclin D1
regulates cyclin-dependent kinase inhibitor 1B (CDKN1B) abundance
at a post-translational level, inhibiting the Skp2 promoter, Skp2
abundance and inducing CDKN1B phosphorylation at Ser10 (41). Therefore, it was hypothesized that
cyclin D1 downregulation may be due to the reduced stability of
other factors in the regulatory network affected by Skp2
expression. In the current study, inhibition of Skp2 led to the
upregulation of p27 and G2/M arrest, which in turn inhibited the
proliferation of HEC-1-A cells. These results correlated with other
previous studies (44,45). Nakayama et al (44) demonstrated that Skp2 regulates the
stability of p27, which participates in the regulation of G2/M
phase. Pagano et al (45)
also revealed that p27 inhibited Cdk1 activity at the G2/M phase,
which is an important regulatory factor for G2-M checkpoints. In
addition to p27, several cell cycle regulatory proteins also serve
as substrates for SCF complexes, including cyclin A, cyclin B, p21
and p53, which are degraded via this pathway and are regulators of
the G2-M checkpoint (45). Wu et
al (46) identified small
molecule inhibitors specific to SCF-Skp2 activity using in
silico screens targeted at the binding interface for p27. These
compounds selectively inhibited Skp2-mediated p27 degradation by
reducing p27 binding through key compound-receptor contacts
(46). In cancer cells, these
compounds induce p27 accumulation in a Skp2-dependent manner and
promote cell-type specific blocks in the G1 or G2/M phases
(46). Furthermore, although the
expression of p27 was upregulated in the current study, cell
apoptosis was not affected. This may be due to the inhibition of
Skp2 affecting the stability of unknown factors, offsetting the
pro-apoptotic effect of p27. RNAi-induced Skp2 inhibition affects
the stability of downstream factors, but may also affect the
stability of upstream factors via feedback regulation, thereby
causing changes in multiple factors along the entire cell
regulatory network.
The results of the current study demonstrated that
lentivirus-mediated RNAi technology can effectively inhibit the
expression of Skp2, thereby upregulating the protein expression of
p27 and inhibiting the proliferation of human endometrial cancer
HEC-1-A cells. The current study verified the use of in
vitro experiments to assess the mechanism underlying
endometrial cell growth via Skp2 inhibition. Therefore, these
methods may be used to further investigate the role of Skp2 in
tumorigenesis in future studies. Furthermore, Skp2 is thought to be
a potential target for gene therapy in patients with endometrial
cancer (47). However, multi-level
and multi-factor interactions involved in cell proliferation,
apoptosis and cycle regulation need to be examined further and
considered in an integrative manner. In addition, as there may be
several factors underlying disease etiology, the effect of Skp2 on
the expression levels of cyclin D1 and p27 as well as on cell
proliferation, apoptosis and cycle regulation, and the association
between Skp2 expression and IGFR signaling require further
study.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Science Foundation of Fujian (grant nos. 2016J01428 and
2016J01491) and the Joint Funds for the Innovation of Science &
Technology (grant no. 2017Y9062).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL, XZ and PS designed the study. HL and XZ
collected the data. HL, GR, XS and XC analyzed the data. HL and GR
prepared the manuscript and PS revised the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vanderstichele A, Neven P and Vergote I:
Combined modality adjuvant therapy for high-risk endometrial
cancer. Lancet Oncol. 17:1029–1030. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wirth T, Parker N and Ylä-Herttuala S:
History of gene therapy. Gene. 525:162–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ezoe S, Matsumura I, Nakata S, Gale K,
Ishihara K, Minegishi N, Machii T, Kitamura T, Yamamoto M, Enver T
and Kanakura Y: GATA-2/estrogen receptor chimera regulates
cytokine-dependent growth of hematopoietic cells through
accumulation of p21(WAF1) and p27(Kip1) proteins. Blood.
100:3512–3520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bochis OV, Irimie A, Pichler M and
Berindan-Neagoe I: The role of Skp2 and its substrate CDKN1B (p27)
in colorectal cancer. J Gastrointestin Liver Dis. 24:225–234.
2015.PubMed/NCBI
|
|
5
|
Wang Z, Inuzuka H, Zhong J, Liu P, Sarkar
FH, Sun Y and Wei W: Identification of acetylationdependent
regulatory mechanisms that govern the oncogenic functions of Skp2.
Oncotarget. 3:1294–1300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu SY, Wang F, Wei G, Wang B, Yang JY,
Huang YZ, Zhang L, Zheng F, Guo LY, Wang JN and Tang JM: S-phase
kinase-associated protein 2 knockdown blocks colorectal cancer
growth via regulation of both p27 and p16 expression. Cancer Gene
Ther. 20:690–694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chung YK, Chi-Hung Or R, Lu CH, Ouyang WT,
Yang SY and Chang CC: Sulforaphane down-regulates SKP2 to stabilize
p27(KIP1) for inducing antiproliferation in human colon
adenocarcinoma cells. J Biosci Bioeng. 119:35–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Masumoto K and Kitagawa M: E3 ubiquitin
ligases as molecular targets in human oral cancers. Curr Cancer
Drug Targets. 16:130–135. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pratheeshkumar P, Siraj AK, Divya SP,
Parvathareddy SK, Begum R, Melosantos R, Al-Sobhi SS, Al-Dawish M,
Al-Dayel F and Al-Kuraya KS: Downregulation of SKP2 in papillary
thyroid cancer acts synergistically with TRAIL on inducing
apoptosis via ROS. J Clin Endocrinol Metab. 103:1530–1544. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang W, Cao L, Sun Z, Xu J, Tang L, Chen
W, Luo J, Yang F, Wang Y and Guan X: Skp2 is over-expressed in
breast cancer and promotes breast cancer cell proliferation. Cell
Cycle. 15:1344–1351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamada S, Yanamoto S, Naruse T, Matsushita
Y, Takahashi H, Umeda M, Nemoto TK and Kurita H: Skp2 Regulates the
expression of MMP-2 and MMP-9, and enhances the invasion potential
of oral squamous cell carcinoma. Pathol Oncol Res. 22:625–632.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Kobayashi R, Galaktionov K and
Beach D: p19Skp1 and p45Skp2 are essential elements of the cyclin
A-CDK2 S phase kinase. Cell. 82:915–925. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wirbelauer C, Sutterlüty H, Blondel M,
Gstaiger M, Peter M, Reymond F and Krek W: The F-box protein Skp2
is a ubiquitylation target of a Cul1-based core ubiquitin ligase
complex: Evidence for a role of Cul1 in the suppression of Skp2
expression in quiescent fibroblasts. EMBO J. 19:5362–5375. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tiscornia G, Singer O and Verma IM:
Production and purification of lentiviral vectors. Nat Protoc.
1:241–245. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang
Y, Wang D, Li R, Yi X, Zhang H, et al: Hypomethylation-linked
activation of PAX2 mediates tamoxifen-stimulated endometrial
carcinogenesis. Nature. 438:981–987. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peth A, Uchiki T and Goldberg AL:
ATP-dependent steps in the binding of ubiquitin conjugates to the
26S proteasome that commit to degradation. Mol Cell. 40:671–681.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bornstein G, Ganoth D and Hershko A:
Regulation of neddylation and deneddylation of cullin1 in SCFSkp2
ubiquitin ligase by F-box protein and substrate. Proc Natl Acad Sci
USA. 103:11515–11520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stacey DW: Cyclin D1 serves as a cell
cycle regulatory switch in actively proliferating cells. Curr Opin
Cell Biol. 15:158–163. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiao B, Spencer J, Clements A, Ali-Khan N,
Mittnacht S, Broceño C, Burghammer M, Perrakis A, Marmorstein R and
Gamblin SJ: Crystal structure of the retinoblastoma tumor
suppressor protein bound to E2F and the molecular basis of its
regulation. Proc Natl Acad Sci USA. 100:2363–2368. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ru Y, Chen XJ, Zhao ZW, Zhang PF, Feng SH,
Gao Q, Gao SG and Feng XS: Cyclin D1 and p57 as biomarkers in
differentiation, metastasis and prognosis of gastric cardia
adenocarcinoma. Oncotarget. 8:73860–73870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Georgiadou D, Sergentanis TN, Sakellariou
S, Filippakis GM, Zagouri F, Vlachodimitropoulos D, Psaltopoulou T,
Lazaris AC, Patsouris E and Zografos GC: Cyclin D1, p16(INK) (4A)
and p27(Kip1) in pancreatic adenocarcinoma: Assessing prognostic
implications through quantitative image analysis. APMIS.
122:1230–1239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoo J, Jung JH, Lee MA, Seo KJ, Shim BY,
Kim SH, Cho DG, Ahn MI, Kim CH, Cho KD, et al: Immunohistochemical
analysis of non-small cell lung cancer: Correlation witll clinical
parameters and prognosis. J Korean Med Sci. 22:318–325. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu ZJ, Ma ZY, Wang QR, Lei DP, Wang R, Liu
CX and Pan XL: Overexpression of CyclinD1 and underexpression of
p16 correlate with lymph node metastases in laryngeal squamous cell
carcinoma in Chinese patients. Clin Exp Metastasis. 25:887–892.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fagan-Solis KD, Pentecost BT, Gozgit JM,
Bentley BA, Marconi SM, Otis CN, Anderton DL, Schneider SS and
Arcaro KF: SKP2 overexpression is associated with increased serine
10 phosphorylation of p27 (pSer10p27) in triple-negative breast
cancer. J Cell Physiol. 229:1160–1169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang H, Chen H, Zhou H, Yu W and Lu Z:
Cyclin-dependent kinase inhibitor 3 promotes cancer cell
proliferation and tumorigenesis in nasopharyngeal carcinoma by
targeting p27. Oncol Res. 25:1431–1440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Biçer A, Orlando S, Islam ABMMK,
Gallastegui E, Besson A, Aligué R, Bachs O and Pujol MJ: ChIP-Seq
analysis identifies p27(Kip1)-target genes involved in cell
adhesion and cell signalling in mouse embryonic fibroblasts. PLoS
One. 12:e01878912017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Diersch S, Wenzel P, Szameitat M, Eser P,
Paul MC, Seidler B, Eser S, Messer M, Reichert M, Pagel P, et al:
Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer
cells towards a dual PI3K/mTOR inhibitor in preclinical models.
Oncotarget. 4:277–288. 2013.PubMed/NCBI
|
|
29
|
Santala S, Talvensaari-Mattila A, Soini Y,
Kuvaja P and Santala M: Cyclins A, B, E and p27 in endometrial
endometrioid adenocarcinoma. Anticancer Res. 36:6467–6473. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu L, Chiao CY, Enzer KG, Stankiewicz AJ,
Faller DV and Dai Y: SIRT1 inactivation evokes antitumor activities
in NSCLC through the tumor suppressor p27. Mol Cancer Res.
13:41–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lindberg D, Akerström G and Westin G:
Mutational analysis of p27 (CDKN1B) and p18 (CDKN2C) in sporadic
pancreatic endocrine tumors argues against tumor-suppressor
function. Neoplasia. 9:533–535. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhu CQ, Shih W, Ling CH and Tsao MS:
Immunohistochemical markers of prognosis in non-small cell lung
cancer: A review and proposal for a multiphase approach to marker
evaluation. J Clin Pathol. 59:790–800. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jung D, Khurana A, Roy D, Kalogera E,
Bakkum-Gamez J, Chien J and Shridhar V: Quinacrine upregulates
p21/p27 independent of p53 through autophagy-mediated
downregulation of p62-Skp2 axis in ovarian cancer. Sci Rep.
8:24872018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang ST, Ho HJ, Lin JT, Shieh JJ and Wu
CY: Simvastatin-induced cell cycle arrest through inhibition of
STAT3/SKP2 axis and activation of AMPK to promote p27 and p21
accumulation in hepatocellular carcinoma cells. Cell Death Dis.
8:e26262017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim JY, Lim SJ, Kim HJ, Shin E, Park K and
Lee CM: Clinical significance of p27 and Skp2 protein expression in
uterine cervical neoplasm. Int J Gynecol Pathol. 26:242–247. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Handra-Luca A, Ruhin B, Lesty C and Fouret
P: P27, SKP2, and extra-cellular signal-related kinase signalling
in human salivary gland mucoepidermoid carcinoma. Oral Oncol.
42:1005–1010. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guan G, Bakr MM, Firth N and Love RM:
Expression of cyclin D1 correlates with p27KIP1 and
regulates the degree of oral dysplasia and squamous cell carcinoma
differentiation. Oral Surg Oral Med Oral Pathol Oral Radiol.
126:174–183. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee KH, Lee HE, Cho SJ, Cho YJ, Lee HS,
Kim JH, Nam SY, Chang MS, Kim WH and Lee BL: Immunohistochemical
analysis of cell cycle-related molecules in gastric carcinoma:
Prognostic significance, correlation with clinicopathological
parameters, proliferation and apoptosis. Pathobiology. 75:364–372.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pesutić-Pisac V, Punda A, Gluncić I,
Bedeković V, Pranić-Kragić A and Kunac N: Cyclin D1 and p27
expression as prognostic factor in papillary carcinoma of thyroid:
Association with clinicopathological parameters. Croat Med J.
49:643–649. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jonason JH, Gavrilova N, Wu M, Zhang H and
Sun H: Regulation of SCF(SKP2) ubiquitin E3 ligase assembly and
p27(KIP1) proteolysis by the PTEN pathway and cyclin D1. Cell
Cycle. 6:951–961. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L,
Lisanti MP, Katiyar S and Pestell RG: Cyclin D1 induction of
cellular migration requires p27(KIP1). Cancer Res. 66:9986–9994.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tsvetkov LM, Yeh KH, Lee SJ, Sun H and
Zhang H: p27(Kip1) ubiquitination and degradation is regulated by
the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr
Biol. 9:661–664. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carrano AC, Eytan E, Hershko A and Pagano
M: SKP2 is required for ubiquitin-mediated degradation of the CDK
inhibitor p27. Nat Cell Biol. 1:193–199. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nakayama K, Nagahama H, Minamishima YA,
Miyake S, Ishida N, Hatakeyama S, Kitagawa M, Iemura S, Natsume T
and Nakayama KI: Skp2-mediated degradation of p27 regulates
progression into mitosis. Dev Cell. 6:661–672. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pagano M: Control of DNA synthesis and
mitosis by the Skp2-p27-Cdk1/2 axis. Mol Cell. 14:414–416. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu L, Grigoryan AV, Li Y, Hao B, Pagano M
and Cardozo TJ: Specific small molecule inhibitors of Skp2-mediated
p27 degradation. Chem Biol. 19:1515–1524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pavlides SC, Huang KT, Reid DA, Wu L,
Blank SV, Mittal K, Guo L, Rothenberg E, Rueda B, Cardozo T and
Gold LI: Inhibitors of SCF-Skp2/Cks1 E3 ligase block
estrogeninduced growth stimulation and degradation of nuclear
p27kip1: Therapeutic potential for endometrial cancer.
Endocrinology. 154:4030–4545. 2013. View Article : Google Scholar : PubMed/NCBI
|