Introduction
Colorectal cancer (CRC) is one of the most common
human cancer types worldwide. According to the 2017 cancer
statistics, the incidence rate of CRC ranks third in the US
(1). Non-specific clinical symptoms,
rapid advancement and metachronous metastasis have become the most
common causes of colon adenocarcinoma (COAD)-associated death. A
previous report indicated that >1 million novel cases of colon
cancer are diagnosed and >0.5 million cancer-associated deaths
occur annually (2). Although the
definition of CRC patient subsets, drug development, combined
application of targeted therapy and immunotherapy, and other newer
technologies have allowed for great optimism for the future for
patients with CRC (3), patients
still have a poor outcome and there is a lack of effective
prognostic markers (4). Effective
diagnostic and prognostic markers may not only provide useful
prognostic information but also help to guide treatment.
COAD is the most common type of CRC. Thus far this
year, a large amount of research has been performed on diagnostic
and prognostic biomarkers for COAD. For instance, the adenomatous
polyposis coli (APC) gene encodes a multifunctional protein which
negatively regulates the Wnt signaling pathway and that have
various important roles, including intercellular adhesion, cell
cycle regulation and apoptosis (5).
Various reports have verified that mutations in the APC gene are
responsible for COAD (6–8). Furthermore, a high expression of
cellular communication network factor 1 was identified in CRC, and
its expression was associated with a poor prognosis for CRC
patients (9). Recently, an
increasing number of microRNAs (miRs) have been reported to hold
significant promise for the diagnosis and prognosis of CRC,
including miR-25 (10), miR-21
(11), let-7 (12) and miR-126 (13). However, most of the research on
diagnostic and prognostic markers for CRC is at the primary
experimental exploration stage and has limited value for clinical
applications. Thus, there is an enormous demand for additional
valuable markers that may help predict the prognosis and guide
treatments for CRC.
In the present study, an analysis of data from the
UALCAN database was performed, which indicated that cadherin 3
(CDH3) was significantly upregulated in COAD tissues, and COAD
patients with a high CDH3 level generally had a good prognosis. The
clinical data of the present study also support the above
conclusions. Furthermore, gene ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) analyses determined found that CDH3 is
mainly involved in regulating cell-cell adhesion, which may affect
the metastasis and progression of COAD. To the best of our
knowledge, the present study was the first to report on the value
of CDH3 in predicting the survival of COAD patients and its
potential mechanistic involvement in regulating the malignant
phenotype of tumors.
Materials and methods
Sample collection and patient
follow-up
A total of 48 paired COAD resection tissues and
adjacent tissues were obtained from patients undergoing CRC surgery
at HwaMei Hospital, University of the Chinese Academy of Sciences
(Ningbo, China) between January 2010 and April 2011, and stored in
liquid nitrogen. The cohort of 48 COAD patients comprised 27 males
and 21 females with an average age of 38.53±4.84 years (age range,
33–45), and all of them were diagnosed with primary COAD by
histopathology, while cases of recurrent and metastatic COAD were
excluded. These tissues were used to perform reverse
transcription-quantitative (RT-q)PCR analysis. The survival
information of 40 of the COAD patients was obtained by telephone
follow-up.
UALCAN database analysis
The differentially expressed genes from the COAD and
normal tissues were analyzed by using the UALCAN online database
(http://ualcan.path.uab.edu/index.html), according to
the website's instructions (14). In
the UALCAN database, COAD was selected, and the heatmap and
differentially expressed genes were downloaded. Next, according to
the average expression level of CDH3, the COAD patients from The
Cancer Genome Atlas (TCGA; TCGA and UALCAN data were for the same
cohort) (14) database were divided
into a CDH3 high expression group and a CDH3 medium/low expression
group, and the Kaplan-Meier method was used to analyze the overall
survival (OS) of the COAD patients.
RT-qPCR analysis
The sequences of 20 genes (CST1, MMP7, KRT23, KRT80,
CA9, FOXQ1, CDH3, KIAA1199, CLDN2, LY6G6D, ETV4, KLK10, CLDN1,
GRIN2D, NKD1, C6orf223, MMP3, MMP11, TRIM29 and TESC) were acquired
from GenBank (https://www.ncbi.nlm.nih.gov/nucleotide/). Primers
were designed using premier 5.0 software (http://www.premierbiosoft.com/primerdesign/;
Table I) and the synthesis was
performed by Invitrogen (Thermo Fisher Scientific, Inc.).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene symbol | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| CST1 |
ACTTGGACACCTGTGCCTTC |
TCACCAGGGACCTTCTGTTC |
| MMP7 |
AAGCCAAACTCAAGGAGATGC |
ATGTCAGCAGTTCCCCATACA |
| KRT23 |
ACCACATTTGACAGCCCATG |
CTTTCATCCCAGCACACCTC |
| KRT80 |
TCGGGCATCTCTATGAGGAATA |
AAGGTGAACTCCATGTCTGTG |
| CA9 |
GAAAACAGTGCCTATGAGCAGTTG |
TGCTTAAGCACTCAGCATCAC |
| FOXQ1 |
TGACAACTACTGGATGCTCATTCAAGAGATGAGCATCCAGTAGTTGTCTTTTTTC |
TCGAGGAAAAAAGACAACTACTGGATGCTCATCTCTTGAATGAGCATCCAGTAGTTGTCA |
| CDH3 |
AAACTTGGGGACAGCAACATCAG |
TCTTTTGGTTTGCAGAGACAGGG |
| KIAA1199 |
GGCTGTGGCCTATGCAGTCA |
TGTGACAAGGTTCCCACTGCTTAC |
| CLDN2 |
CTGCCAACACAGTCTCCTCA |
GGATTTGTTGCCTAGGGTGA |
| LY6G6D |
ATGAAACCCCAGTTTGTTGGG |
CTATCCGCTCCACAGTCCTGG |
| ETV4 |
CACCCGTCCTGCCCCTTCACCTT |
GGCTTCCCACGTGCGCAGCAGGA |
| KLK10 |
CTCTGGCGAAGCTGCTG |
ATAGGCTTCGGGGTCCAA |
| CLDN1 |
GGCAGATCCAGTGCAAAGTC |
TCTTCTGCACCTCATCGTCTT |
| GRIN2D |
CCTTCTTTGCCGTCATCTTTCTTGC |
AAACTTCAGGGGTGGGTATTGCTCC |
| NKD1 |
TCGCCGGGATAGAAAACTACA |
CAGTTCTGACTTCTGGGCCAC |
| C6orf223 |
ATCGCTTCGCCTCCTACAACA |
GACAGCGGCGGTGGGTAAT |
| MMP3 |
CGGTGGCTTCAGTACCTTTC |
ACCTCCTCCCAGACCTTCA |
| MMP11 |
TGGCTGTACGACGGTGAAAA |
CATGGGTCTCTAGCCCTGATA |
| TRIM29 |
GACATCATACCAGCCCTCGT |
ATCCCGTTGCCTTTGTTGAC |
| TESC |
AATTGTTCGTGCCTTCTTCG |
TCCGAGTCGTACATGTGGAA |
| GAPDH |
CTTCACCACCATGGAGAAGGC |
GGCATGGACTGTGGTCATGAG |
Total RNA was isolated from the COAD and normal
tissues with TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The total RNA was used for complementary (c)DNA synthesis
with a High Capacity cDNA Reverse Transcription Kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Next, Power SYBR Green
PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.)
was used to perform qPCR analysis. The thermocycling conditions for
PCR were as follows: 95°C for 10 min; followed by 40 cycles of 95°C
for 10 sec, 60°C for 20 sec and 72°C for 10 sec. All experiments
were performed according to the manufacturer's protocols and GAPDH
was used as a control. The fold change was determined relative to
the control after normalizing to a housekeeping gene using the
2−ΔΔCq method (15). All
experiments were independently replicated three times.
Analysis of other databases
The expression of CDH3 in colon cancer tissues was
assessed using the Human Protein Atlas (HPA) online database
(www.proteinatlas.org). In this database,
COAD data were selected, ‘CDH3’ was entered as a search term, the
IHC images were viewed and downloaded, and the representative
diagram of CDH3 expression levels in COAD and normal tissues was
displayed. Image-Pro Plus software (version 6.0; Media Cybernetics,
Inc.) was used to calculate the mean integrated optical density
(IOD) of these IHC images, and the sum IOD represented the relative
expression level of CDH3 in COAD and normal tissues.
GO and KEGG analyses were performed by using the
DAVID (https://david.ncifcrf.gov/; version 6.8)
and STRING (https://string-db.org/cgi/input.pl; version 11.0)
database, respectively.
Statistical methods
SPSS 21.0 software (IBM Corp.) was used for
statistical analysis. RT-qPCR data were analyzed by unpaired
Student's t-tests, and the IHC data were analyzed using
independent-samples t-tests. The Kaplan-Meier method and log-rank
test were employed to estimate OS. P<0.05 was considered to
indicate statistical significance.
Results
CDH3 mRNA is significantly upregulated
in COAD tissues
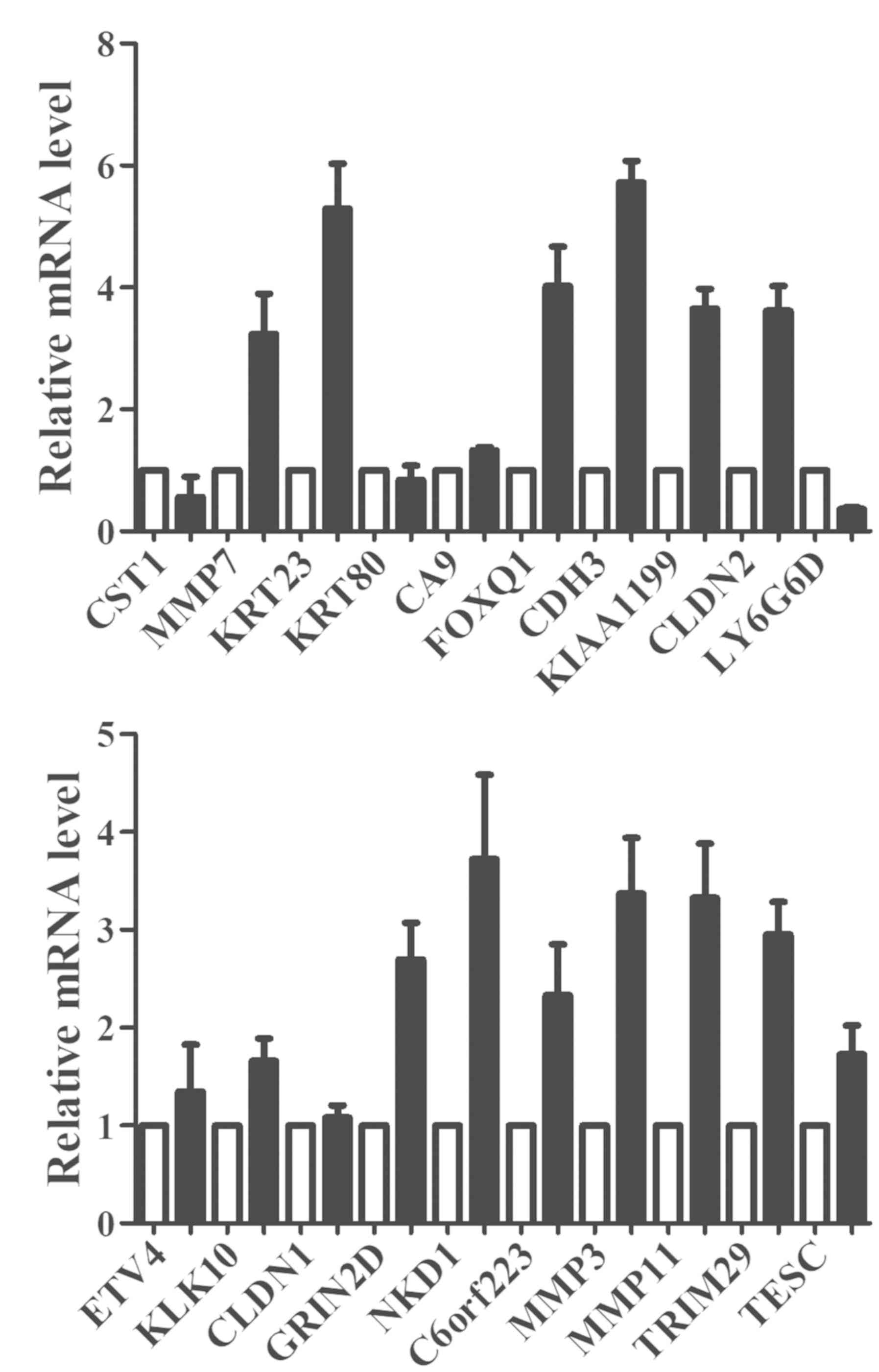
The top 20 significantly upregulated genes according
to the results of the UALCAN database analysis are presented in
Fig. 1; they were CST1, MMP7, KRT23,
KRT80, CA9, FOXQ1, CDH3, KIAA1199, CLDN2, LY6G6D, ETV4, KLK10,
CLDN1, GRIN2D, NKD1, C6orf223, MMP3, MMP11, TRIM29 and TESC. To
further confirm these differentially expressed mRNAs, the relative
levels of these 20 genes were detected by RT-qPCR analysis of
tissues from 48 paired COAD tissues. In the present study, MMP7,
KRT23, FOXQ1, CDH3, KIAA1199, CLDN2, GRIN2D, NKD1, C6orf223, MMP3,
MMP11 and TRIM29 were confirmed to be continuously upregulated in
the 48 COAD tissues compared with those in the normal colon tissues
(Fig. 2A and B). Among these
dysregulated genes, CDH3 was noted to have been examined by only
few studies with regard to its expression levels and prognostic
value in colon cancer. Hence, CDH3 was further investigated.
CDH3 protein is significantly
upregulated in colon cancer tissues
In order to further evaluate the expression levels
of CDH3 in colon cancer tissues, an analysis of IHC samples from
the HPA web portal database was performed. In the colon cancer
tissues and normal tissues, CDH3 was observed to be mainly
expressed in the cytoplasm and not in the nucleus. According to the
sum IOD value, the expression level of CDH3 in the colon cancer
tissues (n=11) was higher than that in the normal tissues (n=3;
P=0.0245). The above results suggested that the protein levels of
CDH3 were significantly upregulated in colon cancer tissues
(Fig. 3).
High CDH3 expression is associated
with a high survival rate for patients with COAD
The association between the expression of CDH3 and
other clinical characteristics was analyzed in TCGA database. The
results suggested that there was no significant association between
the expression level of CDH3 and age, gender, ethnicity, tumor
stage and tumor type (adenocarcinoma vs. mucinous-adenocarcinoma;
data not shown; P<0.05).
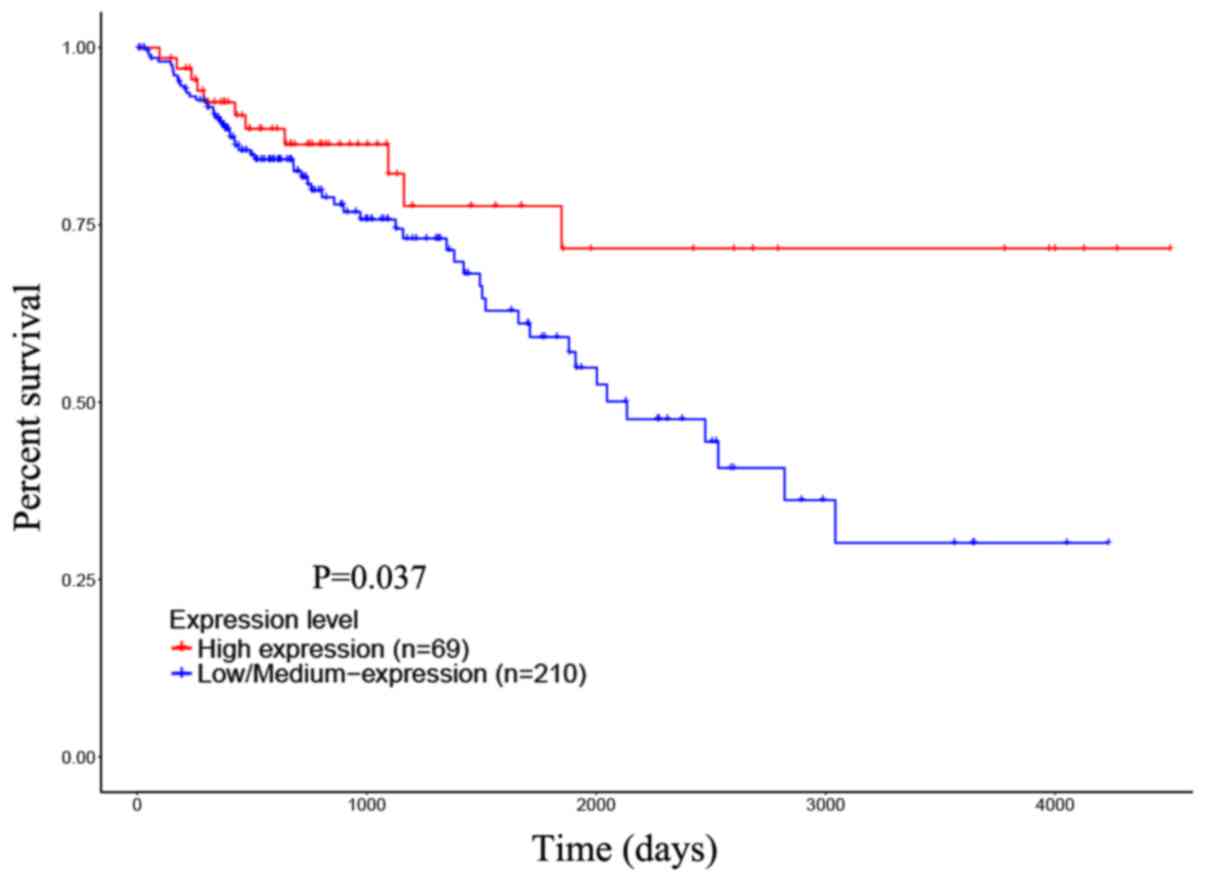
Next, to evaluate the association between
upregulated CDH3 and prognosis, a survival analysis of COAD
patients with different CDH3 mRNA levels was performed. First,
according to the CDH3 mRNA levels, the COAD patients from the TCGA
database were divided into a high expression group (n=69) and a
medium/low expression group (n=210). The prognostic implication of
CDH3 was investigated with the UALCAN online database. The
Kaplan-Meier curves indicated that the COAD patients with a high
CDH3 level had a better OS compared with that of the CDH3
medium/low expression group (P=0.037; Fig. 4). Next, follow-up was performed for
48 COAD patients who received surgery at our hospital and the
survival information of 40 patients was obtained. According to the
median CDH3 expression level, the 40 colon cancer patients were
divided into a high expression group (n=20) and a low expression
group (n=20), and Kaplan-Meier analysis was again performed. The
results indicated that the median survival time for the COAD
patients with a high or low CDH3 level was 55.50 or 43.50 months,
respectively. There was an obvious difference in the survival rate
between the two groups (P=0.0078; Fig.
5). Based on the above results, it was confirmed that a high
CDH3 level indicates a better prognosis for COAD patients.
CDH3 is associated with cell-cell
adherens junction
To explore the potential function of CDH3 in the
occurrence and development of tumors, GO and KEGG analyses were
performed. As presented in Table
II, the GO terms in the biological process category for genes
interacting with CDH3 mainly included adherens junction
organization and cell-cell adhesion. The GO terms in the category
molecular function mainly included calcium ion binding, cadherin
binding and cell adhesion molecular binding, while the terms in the
category cellular component mainly included cell-cell adherens
junction and catenin complexing. As presented in Table III, the KEGG analysis results
confirmed that CDH3 is mainly involved in adherens junctions,
leukocyte transendothelial migration, cell adhesion molecules and
pathways involved in cancer.
 | Table II.Gene Ontology analysis of genes
interacting with cadherin 3. |
Table II.
Gene Ontology analysis of genes
interacting with cadherin 3.
| A, Category cellular
component |
|---|
|
|---|
| Rank | Pathway
description | FDR |
|---|
| 1 | Cell-cell adherens
junction |
5.29×10−6 |
| 2 | Catenin complex |
5.29×10−6 |
| 3 | Plasma membrane |
1.30×10−5 |
| 4 | Cell periphery |
1.30×10−5 |
| 5 | Apical junction
complex | 0.000118 |
| 6 | Adherens
junction | 0.000364 |
| 7 | Zonula adherens | 0.0015 |
| 8 | Cell-cell
junction | 0.00353 |
| 9 | Focal adhesion | 0.00353 |
| 10 | Membrane part | 0.00353 |
| 11 | Extrinsic component
of plasma membrane | 0.00405 |
| 12 | Cell junction | 0.0118 |
| 13 | Intercalated
disc | 0.0277 |
|
| B, Category
molecular function |
|
| Rank | Pathway
description | FDR |
|
| 1 | Calcium ion
binding | 4.49E-07 |
| 2 | Cadherin
binding | 4.49E-07 |
| 3 | Cell adhesion
molecule binding | 5.45E-06 |
| 4 | Beta-catenin
binding | 0.00275 |
| 5 | Gamma-catenin
binding | 0.0119 |
|
| C, Category
biological process |
|
| Rank | Pathway
description | FDR |
|
| 1 | Adherens junction
organization | 1.70E-20 |
| 2 | Cell junction
assembly | 1.22E-14 |
| 3 | Cell-cell junction
organization | 1.22E-14 |
| 4 | Cell-cell
adhesion | 1.22E-14 |
| 5 | Homophilic cell
adhesion via plasma membrane adhesion molecules | 1.47E-12 |
| 6 | Cellular response
to indole-3-methanol | 1.77E-06 |
| 7 | Cellular component
assembly | 0.000299 |
| 8 | Vascular
endothelial growth factor receptor signaling pathway | 0.000307 |
| 9 | Cellular component
biogenesis | 0.000496 |
| 10 | Single organismal
cell-cell adhesion | 0.00137 |
| 11 | Cell adhesion | 0.00227 |
| 12 | Epithelial
cell-cell adhesion | 0.0157 |
| 13 | Cellular component
organization | 0.0181 |
| 14 | Regulation of cell
proliferation | 0.019 |
 | Table III.Kyoto Encyclopedia of Genes and
Genomes analysis of genes interacting with cadherin 3. |
Table III.
Kyoto Encyclopedia of Genes and
Genomes analysis of genes interacting with cadherin 3.
| Rank | Pathway
description | FDR |
|---|
| 1 | Adherens
junction | 0.00178 |
| 2 | Leukocyte
transendothelial migration | 0.00376 |
| 3 | Cell adhesion
molecules | 0.00436 |
| 4 | Endometrial
cancer | 0.0228 |
| 5 | Bacterial invasion
of epithelial cells | 0.0277 |
| 6 | Pathways in
cancer | 0.0277 |
| 7 | Arrhythmogenic
right ventricular cardiomyopathy | 0.0277 |
The above comprehensive results confirmed that CDH3
is closely associated with cell-cell adherens junctions. Hence,
CDH3 may be involved in the progression and metastasis of colon
cancer by regulating cell adhesion.
Discussion
The present study first examined the expression
level and prognostic value of CDH3 in COAD with the TCGA database.
It was revealed that CDH3 is a significantly upregulated gene in
COAD tissues compared with normal tissues. Furthermore, COAD
patients with a high CDH3 level in their tumor tissues usually had
a longer survival time compared with that of patients with
medium/low levels. Based on the results from the database analysis,
the relative expression level of CDH3 was further verified in 48
COAD tissues and the association between CDH3 and survival time was
assessed 4 in 0 COAD patients. Of note, the results were consistent
with those of the database analysis. The GO and KEGG analyses
further indicated that CDH3 is mainly involved in regulating
cell-cell adhesion, which may be one of the reasons for affecting
the prognosis of COAD patients.
With the development of proteomics and genomics,
high-throughput technology has been applied to explore biomarkers
associated with diagnosis and prognosis. In colon cancer, an
increasing number of prognostic markers have been identified, and
these biomarkers have contributed to the determination of the
prognosis and provide a theoretical basis for individualized
treatment (8). These biomarkers are
contained in various body fluids and include multiple proteins,
mRNA, non-coding RNA and exosomes. For instance, a high level of
SOX9 has recently been reported to be associated with a good
prognosis for stage II CRC (16).
Furthermore, in colon cancer tissues, various lncRNAs were
downregulated in COAD tissues and contributed to a poor prognosis
for colon cancer; they were CASC2, CTD903, GASS, MEG3, RPI-13P20.6
and TUSC7 (4). Exosomal CD151
expression has been investigated in CRC tissues and elevated
expression was observed in patients with advanced disease; higher
levels of expression were retrospectively identified to be
associated with a poorer prognosis (17). However, only few studies have
reported on the expression of CDH3 in colon cancer and its
association with prognosis. A previous study confirmed that CDH3
expression was elevated in COAD tissues compared with that in
normal tissues (18). However, the
association between the expression levels of CDH3 and patient
prognosis was not previously described. To the best of our
knowledge, the present study revealed that CDH3 was significantly
upregulated in COAD tissues and predicted a good prognosis for COAD
patients.
An increasing number of studies have confirmed that
CDH3 was more frequently demethylated in advanced colorectal
carcinomas (19), and this
phenomenon was also observed in advanced gastric carcinomas
(20). Hence, CDH3 is a potential
diagnostic and prognostic marker for various tumor types. The mRNA
expression levels of CDH3 were reported to be able to distinguish
between malignant and benign biliary strictures in brush cytology
specimens (21). However, CDH3
expression was closely associated with a clinicopathological
features and poor prognosis in gallbladder cancer (22). However, the potential mechanism by
which CDH3 affects the prognosis of COAD patients remains
elusive.
In the present study, it was revealed that CDH3 is
highly expressed in colon cancers, but patients with a higher CDH3
expression usually had a better prognosis. As an explanation for
this contradiction, it is possible that high CDH3 expression may
have a protective function in colon cancer. A previous, similar
study also reported that high expression of Beclin-1 was associated
with a better OS and disease-free survival in CRC, which is
possible due to the promotion of the formation of the autophagic
vesicle (23). A previous study
confirmed that the mRNA levels of several genes encoding adherens
junction proteins were dysregulated in 26 CRC, 42 adenoma and 24
normal mucosa samples. Among these genes, CDH3 was significantly
upregulated in CRC tissues (24). In
the present study, the GO and KEGG analyses indicated that CDH3 was
closely associated with a cell adhesion function, which may be the
major reason for its influence on the prognosis of COAD patients.
However, the direct empirical evidence provided by the present
study is sparse. Furthermore, owing to the lack of cohort clinical
characteristic determination and an analysis assessing its
association with CDH3 levels, the present study is limited.
In conclusion, the present study indicated that CDH3
was significantly upregulated in COAD tissues, and a high CDH3
level was predictive of a good prognosis. In addition,
Bioinformatics revealed that CDH3 is mainly involved in regulating
cell-cell adhesion. To the best of our knowledge, the present study
was the first to report on the role of CDH3 in the prognosis of
COAD and provided novel insight into the mechanism by which CDH3
affects the prognosis of COAD patients. However, whether CDH3 is a
possible candidate marker for guiding targeted or individualized
treatment still requires to be verified with a larger number of
samples.
Acknowledgements
Not applicable.
Funding
This study was supported by the Zhejiang Medical and
Health Science and Technology Planning Project (grant no.
2017KY593), and the HwaMei Science Project (grant no.
2019HMKY68).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX performed the experiments and collected and
analyzed the data. JZ designed the study, coordinated the
experiments and acquired the data. XD, MD and YX designed the
present study, collected and analyzed the, data, and wrote the
manuscript. All authors read and approved the final manuscript.
Ethical approval and consent to
participate
The present study was approved by the Ethics
Committee of the HwaMei Hospital, University of the Chinese Academy
of Sciences (Ningbo, China), and all of the patients provided
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. 67:7–30. 2017.
|
|
2
|
Wu K, Xu K, Liu K, Huang J, Chen J, Zhang
J and Zhang N: Long noncoding RNA BC200 regulates cell growth and
invasion in colon cancer. Int J Biochem Cell Biol. 99:219–225.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mody K and Bekaii-Saab T: Clinical trials
and progress in metastatic colon cancer. Surg Oncol Clin N Am.
27:349–365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deng H, Wang JM, Li M, Tang R, Tang K, Su
Y, Hou Y and Zhang J: Long non-coding RNAs: New biomarkers for
prognosis and diagnosis of colon cancer. Tumour Biol.
39:10104283177063322017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hankey W, Frankel WL and Groden J:
Functions of the APC tumor suppressor protein dependent and
independent of canonical WNT signaling: Implications for
therapeutic targeting. Cancer Metastasis Rev. 37:159–172. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raskov H, Pommergaard HC, Burcharth J and
Rosenberg J: cColorectal carcinogenesis-update and perspectives.
World J Gastroenterol. 20:18151–18164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malhotra P, Anwar M, Nanda N, Kochhar R,
Wig JD, Vaiphei K and Mahmood S: Alterations in K-ras, APC and
p53-multiple genetic pathway in colorectal cancer among Indians.
Tumour Biol. 34:1901–1911. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hammoudi A, Song F, Reed KR, Jenkins RE,
Meniel VS, Watson AJ, Pritchard DM, Clarke AR and Jenkins JR:
Proteomic profiling of a mouse model of acute intestinal Apc
deletion leads to identification of potential novel biomarkers of
human colorectal cancer (CRC). Biochem Biophys Res Commun.
440:364–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeong D, Heo S, Sung Ahn T, Lee S, Park S,
Kim H, Park D, Byung Bae S, Lee SS, Soo Lee M, et al: Cyr61
expression is associated with prognosis in patients with colorectal
cancer. BMC Cancer. 14:1642014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Yang C, Wang X, Zhang J, Zhang R and
Liu R: The expression of miR-25 is increased in colorectal cancer
and is associated with patient prognosis. Med Oncol. 31:7812014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin PL, Wu DW, Huang CC, He TY, Chou MC,
Sheu GT and Lee H: MicroRNA-21 promotes tumour malignancy via
increased nuclear translocation of β-catenin and predicts poor
outcome in APC-mutated but not in APC-wild-type colorectal cancer.
Carcinogenesis. 35:2175–2182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hansen TF, Christensen Rd, Andersen RF,
Sørensen FB, Johnsson A and Jakobsen A: MicroRNA-126 and epidermal
growth factor-like domain 7-an angiogenic couple of importance in
metastatic colorectal cancer. Results from the Nordic ACT trial. Br
J Cancer. 109:1243–1251. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B and Varambally
S: UALCAN: A portal for facilitating tumor subgroup gene expression
and survival analyses. Neoplasia. 19:649–658. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Prevostel C and Blache P: The
dose-dependent effect of SOX9 and its incidence in colorectal
cancer. Eur J Cancer. 86:150–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malla RR, Pandrangi S, Kumari S, Gavara MM
and Badana AK: Exosomal tetraspanins as regulators of cancer
progression and metastasis and novel diagnostic markers. Asia Pac J
Clin Oncol. 14:383–391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kumara HMCS, Bellini GA, Caballero OL,
Herath SAC, Su T, Ahmed A, Njoh L, Cekic V and Whelan RL:
P-Cadherin (CDH3) is overexpressed in colorectal tumors and has
potential as a serum marker for colorectal cancer monitoring.
Oncoscience. 4:139–147. 2017.PubMed/NCBI
|
|
19
|
Hibi K, Goto T, Mizukami H, Kitamura YH,
Sakuraba K, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H and
Sanada Y: Demethylation of the CDH3 gene is frequently detected in
advanced colorectal cancer. Anticancer Res. 29:2215–2217.
2009.PubMed/NCBI
|
|
20
|
Hibi K, Kitamura YH, Mizukami H, Goto T,
Sakuraba K, Sakata M, Saito M, Ishibashi K, Kigawa G, Nemoto H and
Sanada Y: Frequent CDH3 demethylation in advanced gastric
carcinoma. Anticancer Res. 29:3945–3947. 2009.PubMed/NCBI
|
|
21
|
Kim TH, Chang JH, Lee HJ, Kim JA, Lim YS,
Kim CW and Han SW: mRNA expression of CDH3, IGF2BP3 and BIRC5 in
biliary brush cytology specimens is a useful adjunctive tool of
cytology for the diagnosis of malignant biliary stricture. Medicine
(Baltimore). 95:e41322016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yi S, Yang ZL, Miao X, Zou Q, Li J, Liang
L, Zeng G and Chen S: N-cadherin and P-cadherin are biomarkers for
invasion, metastasis and poor prognosis of gallbladder carcinomas.
Pathol Res Pract. 210:363–368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Z, Ghoorun RA, Fan X, Wu P, Bai Y, Li
J, Chen H, Wang L and Wang J: High expression of Beclin-1 predicts
favorable prognosis for patients with colorectal cancer. Clin Res
Hepatol Gastroenterol. 39:98–106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bujko M, Kober P, Mikula M, Ligaj M,
Ostrowski J and Siedlecki JA: Expression changes of cell-cell
adhesion-related genes in colorectal tumors. Oncol Lett.
9:2463–2470. 2015. View Article : Google Scholar : PubMed/NCBI
|