Introduction
Nephroblastoma is the most common pediatric kidney
tumor with a prevalence of 1 in 10,000 children (1). Nephroblastoma originates from the
embryo, develops in the renal parenchyma, undergoes a form
distortion during growth, and invades the surrounding kidney
tissue. Currently, combination therapy has improved the prognosis
of most patients, but there are still certain patients with poor
survival due to recurrence and metastasis (2–5).
Therefore, understanding the occurrence and development of
nephroblastoma in children and seeking for new targets and methods
for the treatment of nephroblastoma are essential.
MicroRNAs (miRNAs/miRs) are a group of highly
conserved endogenous small non-coding RNAs (21–23 nucleotides in
length) in eukaryotes (6). miRNAs do
not encode proteins, and can inhibit target gene expression by
binding to the 3 prime-untranslated region (3′-UTR) of the target
gene (7–9). There is ample evidence indicating that
miRNAs are involved in the regulation of multiple cellular events,
including cell proliferation, differentiation, and apoptosis
(10–12). In addition, abnormal expression of
miRNAs has been shown to be involved in the development and
progression of various types of tumors (9). miRNAs are promising biomarkers and
therapeutic targets for cancer treatment.
miR-130b functions as an oncogenic factor or tumor
suppressor miRNA, and shows dysregulation in various cancers
including gliomas, gastric cancers, and endometrial cancers
(13–15). Phosphatase and tensin homolog (PTEN)
is one of the most common tumor suppressors in many human cancers
and serves a vital role in the regulation of cell growth and
apoptosis (16). In addition, it has
been reported that PTEN is the target of various miRNAs. For
example, miR-106b induces radio-resistance in colorectal cancer
through the PTEN/Phophatidyl inositol 3′-OH kinase (PI3K)/Akt
pathway (17). However, there is
currently no report on the role of miR-130b-3p in nephroblastoma
and the association of miR-130b-3p and PTEN in nephroblastoma.
Therefore, the present study aimed to investigate
the role and underlying mechanism of miR-130b-3p/PTEN axis in
nephroblastoma in children. The results of the present study may
clarify the role and mechanism of miR-130b-3p in nephroblastoma and
provide novel strategies and a theoretical basis for the treatment
of nephroblastoma in children.
Materials and methods
Clinical samples
A total of 30 samples of nephroblastoma and normal
adjacent tissues were obtained from children with nephroblastoma
who had undergone surgery at the Department of Pediatric Surgery,
Zaozhuang City Hospital between December 2015 and December 2017.
All patients did not received chemotherapy or radiotherapy prior to
surgery. The present study was approved by the Ethical Committee of
the Zaozhuang City Hospital and informed consent was obtained from
each child and their guardian.
Cell culture and cell
transfection
Nephroblastoma cell line WiT49 was provided by The
Hospital for Sick Children (Toronto, Canada) and was subsequently
grown in Dulbecco's Modified Eagle's Medium/Nutrient Mixture F-12
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) and
1% penicillin streptomycin. These cells were cultured in a
humidified chamber at 37°C with 5% CO2. miR-130b-3p
inhibitor (miR-130b-3p antagomir; sequence:
5′-UGCCAACCUUGCAAGCCGAAG-3′) and inhibitor control
(antagomir-negative control; sequence: 5′-CAGUACUUUUGUGUAGUACAA-3′)
were purchased from GenePharma Co. Ltd. WiT49 cells were
transfected with either 100 nM inhibitor control, 100 nM
miR-130b-3p inhibitor, 2 µl control-siRNA (cat. no. Sc-36869; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA), 2 µl PTEN-siRNA (cat.
no. Sc-29459; Santa Cruz Biotechnology, Inc.), 100 nM miR-130b-3p
inhibitor + 2 µl control-siRNA or 100 nM miR-130b-3p inhibitor + 2
µl PTEN-siRNA by using the Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells without any treatment were
considered as the control group. The cells were subjected to the
following experiments 48 h after transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The transfection efficiency was detected 48 h after
cell transfection, using RT-qPCR and/or western blot assay. Total
RNA from tissues and cells were extracted using the TRIzol Reagent
(Takara Bio, Inc.) according to the manufacturer's protocol. The
RNA concentration was detected by NanoDrop spectrophotometer
(ND-2000; Nanodrop Technologies; Thermo Fisher Scientific, Inc.).
The total RNA was stored at −80°C until use. cDNAs were synthesized
by using PrimeScript RT Reagent Kit (Takara Biotechnology Co.,
Ltd.) according to the manufacturer's protocol and analyzed by
performing SYBR® Premix Ex Taq™ II (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. The following
primer sequences were used for qPCR: GAPDH forward,
5′-CGGGAAGCTTGTCATCAATGG-3′ and reverse,
5′-GGCAGTGATGGCATGGACTG-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; PTEN forward,
5′-GGAAAGGGACGAACTGGTGT-3′ and reverse-5′-CAGGTAACGGCTGAGGGAAC-3′;
miR-130b-3p forward, 5′-CTGGTAGGGTACAGTACTGTGATA-3′ and reverse,
5′-CTGGTGTCGTGGAGTCGGC-3′. The thermocycling conditions were as
follows: 95°C for 5 min, followed by 38 cycles of denaturation at
95°C for 15 sec and annealing/elongation at 60°C for 30 sec. GAPDH
(for mRNA) and U6 (for miRNA) were used as the internal controls.
Relative expression levels were calculated by the 2−ΔΔCq
method (18). All experiments were
performed in triplicate.
Western blot assay
Radioimmunoprecipitation assay (RIPA) buffer
(Beyotime Institute of Biotechnology) was used to extract proteins
from cells or tissues according to the manufacturer's protocol.
Bicinchoninic acid assay kit (Pierce Biotechnology; Thermo Fisher
Scientific, Inc.) was used to quantify the protein samples in line
with the manufacturer's protocol. The protein samples (25 µg/lane)
were separated on 10% SDS-PAGE gel and transferred to
polyvinylidene difluoride membranes. The membranes were blocked
with 5% non-fat milk for 1.5 h at room temperature, followed by
incubation with primary antibody against PTEN (cat. no. 9188;
1:5,000; Cell Signaling Technology Inc.), Akt (cat. no. 4691;
1:5,000; Cell Signaling Technology Inc.), phospho-Akt-B (p-Akt;
cat. no. 4060; 1:5,000; Cell Signaling Technology Inc.), nuclear
factor (NF)-κB-p65 subunit (p65; cat. no. 8242; 1:5,000; Cell
Signaling Technology Inc.), phospho-NF-κB (p-NF-κB or p-p65; cat.
no. 3033; 1:5,000; Cell Signaling Technology Inc.), survivin (cat.
no. 2808; 1:5,000; Cell Signaling Technology Inc.), or β-actin
(cat. no. 4970; 1:5,000; Cell Signaling Technology Inc.) overnight
at 4°C. The membranes were then washed with Tris buffered saline
with 0.1% Tween-20 (TBST) three times. Subsequently, the membranes
were incubated with the anti-rabbit IgG, horse radish
peroxidase-linked antibody (cat. no. 7074; 1:5,000; Cell Signaling
Technology Inc.) for 2 h at room temperature. Finally, enhanced
chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was used
to determine the immunoreactive bands. The band density was
quantified with Gel-Pro Analyzer densitometry software (Version
6.3, Media Cybernetics, Inc.).
Cell proliferation assay
Cell proliferation was analyzed using the Cell
Counting Kit-8 (CCK-8) assay (cat no. C0037; Beyotime Institute of
Biotechnology). Logarithmic phase cells were seeded in a 96-well
plate at a density of 1×104 cells per well and incubated
in the 37°C, 5% CO2 incubator for 12 h. Subsequently, 10
µl CCK-8 was added to each well, and the cells were incubated for a
further 2 h at 37°C with 5% CO2. The absorbance was
measured at a wavelength of 450 nm using a microplate reader.
Flow cytometry assay
WiT49 cells were transfected with either 100 nM
inhibitor control, 100 nM miR-130b-3p inhibitor, 100 nM miR-130b-3p
inhibitor + 2 µl control-siRNA or 100 nM miR-130b-3p inhibitor + 2
µl PTEN-siRNA for 48 h. Then, WiT49 cells were collected by 0.25%
Trypsin. Apoptotic cells were detected by using Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis
detection kit (cat. no. 70-AP101-100; MultiSciences) according to
the manufacturer's protocol. Briefly, cells (106) were
incubated with 5 µl Annexin V-FITC and 5 µl PI for 30 min in the
absence of light at room temperature. Finally, flow cytometry (BD
Biosciences) was used to detect cell apoptosis. The data were
analyzed using WinMDI software (version 2.5; Purdue University
Cytometry Laboratories; www.cyto.purdue.edu/flowcyt/software/Catalog.htm).
Dual-luciferase reporter analysis
TargetScan (www.targetscan.org) was used to predict the binding
sites of miR-130b-3p and PTEN. To confirm the association between
miR-130b-3p and PTEN, the dual-luciferase reporter vector
pmiR-RB-REPORT™ (Guangzhou RiboBio Co., Ltd.) that contains a
wild-type (WT) or mutant (MUT) 3′-UTR sequence of PTEN was
constructed. Cells seeded in 24-well plates were co-transfected
with miR-130b-3p mimics or mimic control and the MUT or WT 3′-UTR
of PTEN using Lipofectamine® 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). After transfection for 48 h, cells
were lysed with RIPA buffer. 48 h after transfection, the relative
luciferase activity was detected using the
Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, WI, USA) according to the manufacturer's
protocol. Firefly luciferase activity was normalized to that of
Renilla luciferase.
Statistical analyses
All experiments were performed in triplicates. All
data were shown as the mean ± standard deviation and SPSS v17.0
statistical software (SPSS, Inc.) was used for data analyses.
Comparison between groups were performed by Student's t-tests and
one-way analysis of variance followed by Tukey's test. P<0.05
was considered to indicate a statistically significant
difference.
Results
The expression of miR-130b-3p in
nephroblastoma tissues
To explore the role of miR-130b-3p in
nephroblastoma, RT-qPCR was performed to detect the relative
expression of miR-130b-3p in nephroblastoma (tumor tissues) of
children with nephroblastoma. The results revealed that compared
with the adjacent tissues, the expression of miR-130b-3p was
significantly upregulated in nephroblastoma tissues in children
with nephroblastoma (Fig. 1).
PTEN is a direct target gene of
miR-130b-3p
TargetScan analysis was performed to identify
functional targets of miR-130b-3p. Hundreds of target genes were
identified to be the potential target genes of miR-130b-3p and as
shown in Fig. 2A, PTEN was one such
target. Based on the literature analysis, it was identified that
PTEN is one of the most common tumor suppressor in many human
cancers and serves a vital role in the regulation of cell growth
and apoptosis (16,19). Therefore, PTEN was selected for
further investigations in the present study. Thus, to further
examine whether miR-130b-3p directly targets PTEN, LucPTEN
−3′UTR-WT and its 3′UTR-MUT plasmids were constructed. Luciferase
reporter assay revealed that miR-130b-3p mimics clearly suppressed
the luciferase activity of the WT-PTEN-3′UTR (Fig. 2B). Therefore, this provides evidence
that PTEN is a direct target of miR-130b-3p.
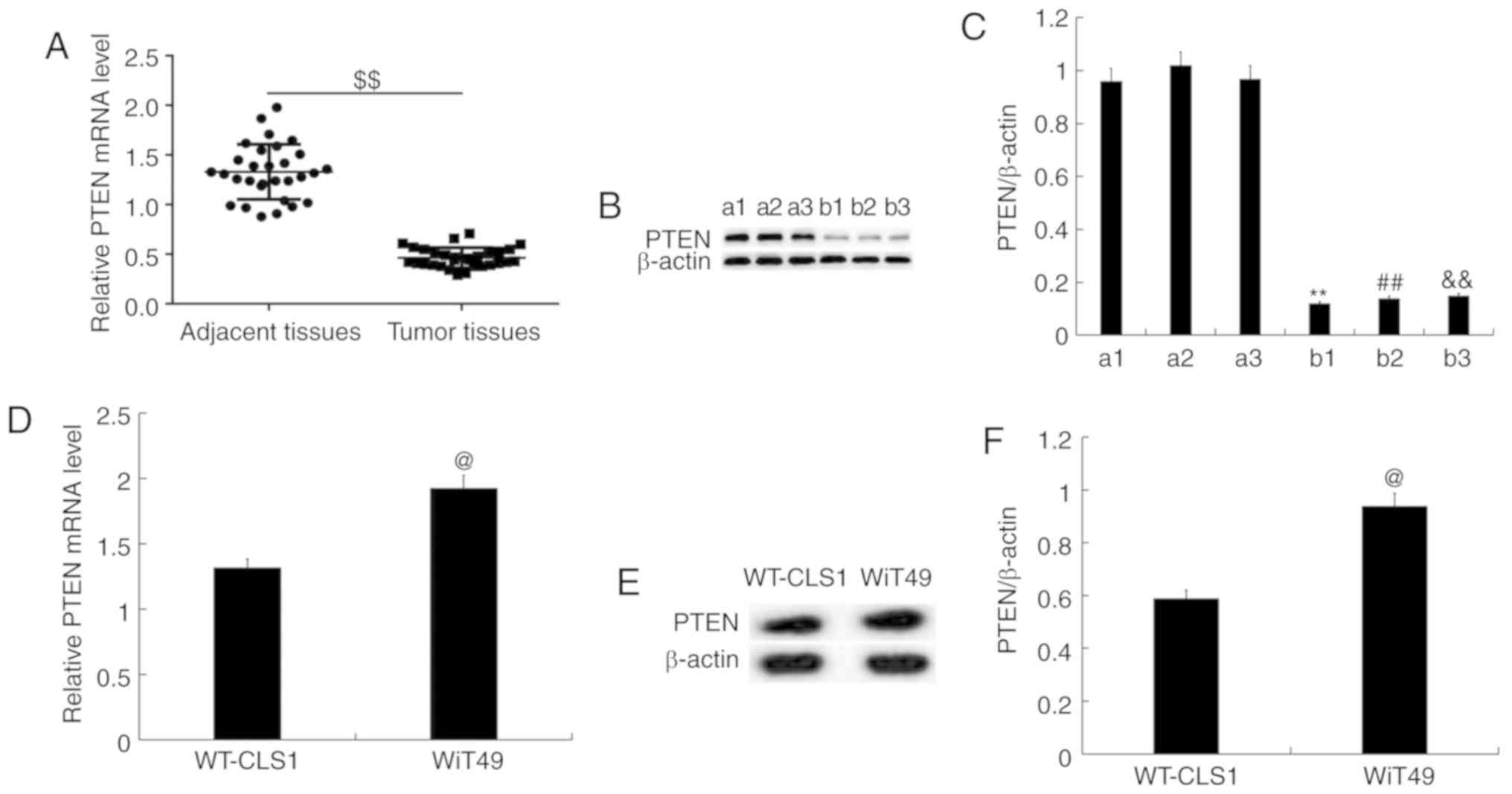
The expression level of PTEN in
nephroblastoma tissues
In order to examine the expression of PTEN in
nephroblastoma tissues, RT-qPCR and western blot analysis were
performed. The results demonstrated that compared with the adjacent
tissues, PTEN was less expressed in nephroblastoma tissues, at the
mRNA and protein expression level (three representative cases in
each group are shown; Fig. 3A-C) in
children. The mRNA (Fig. 3D) and
protein expression (Fig. 3E and F)
of PTEN in WT-CLS1 and WiT49 cells was determined using RT-qPCR and
western blotting, respectively. The results indicated that the mRNA
(Fig. 3D) and protein expression
(Fig. 3E and F) of PTEN was higher
in WiT49 cells than in WT-CLS1 cells. WiT49 cells were then
selected for further study.
Effect of miR-130b-3p on proliferation
and apoptosis of nephroblastoma cells
WiT49 cells were transfected with inhibitor control
or miR-130b-3p inhibitor for 48 h and RT-qPCR was performed to
detect the efficiency of the inhibition. The results revealed that
miR-130b-3p inhibitor significantly reduced miR-130b-3p expression
in WiT49 cells (Fig. 4A).
Furthermore, WiT49 cells were transfected with control-small
interfering (si) RNA or PTEN-siRNA for 48 h, and subsequently,
RT-qPCR and western blotting were performed to detect the
efficiency of siRNA. The results demonstrated that PTEN-siRNA
reduced both mRNA and protein levels of PTEN in WiT49 cells
(Fig. 4B-D). Finally, in order to
test whether miR-130b-3p had an effect on PTEN expression in WiT49
cells, WiT49 cells were transfected with: Inhibitor control,
miR-130b-3p inhibitor, miR-130b-3p inhibitor + control-siRNA, or
miR-130b-3p inhibitor + PTEN-siRNA for 48 h. RT-qPCR and western
blotting were used to detect the expression of PTEN. It was
revealed that miR-130b-3p inhibitor markedly increased PTEN
expression levels at both mRNA and protein level, which was
reversed by PTEN-siRNA (Fig.
4E-G).
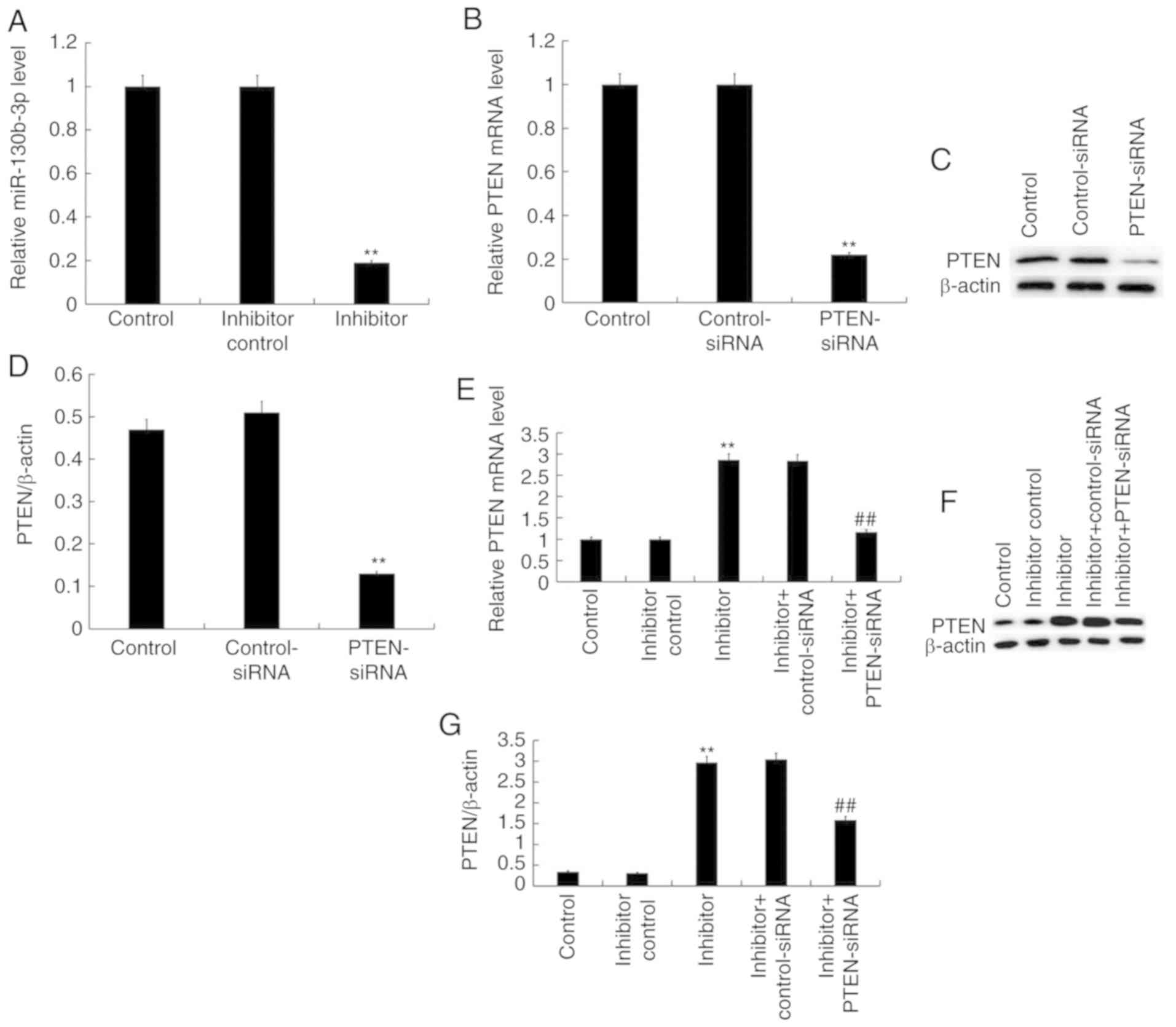 | Figure 4.miR-130b-3p regulates the expression
of PTEN in WiT49 cells. (A) The mRNA expression levels of
miR-130b-3p after WiT49 cells transfected with control, inhibitor
control and miR-130b-3p inhibitor for 48 h. (B) mRNA expression
levels of PTEN following transfection with control, control-siRNA
or PTEN-siRNA. (C) PTEN protein expression levels and (D)
normalized to β-actin in different groups. (E) PTEN mRNA expression
levels after transfection with control, inhibitor control,
miR-130b-3p inhibitor, inhibitor + control-siRNA or inhibitor +
PTEN-siRNA (F) PTEN expression levels and (G) normalized to β-actin
in different groups. Expression levels were measured by reverse
transcription-quantitative polymerase chain reaction/western
blotting. Data presented as the mean ± standard deviation.
**P<0.01 vs. control, ##P<0.01 vs. inhibitor. miR,
microRNA; PTEN, Phosphatase and tensin homolog; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; siRNA, small
interfering RNA. |
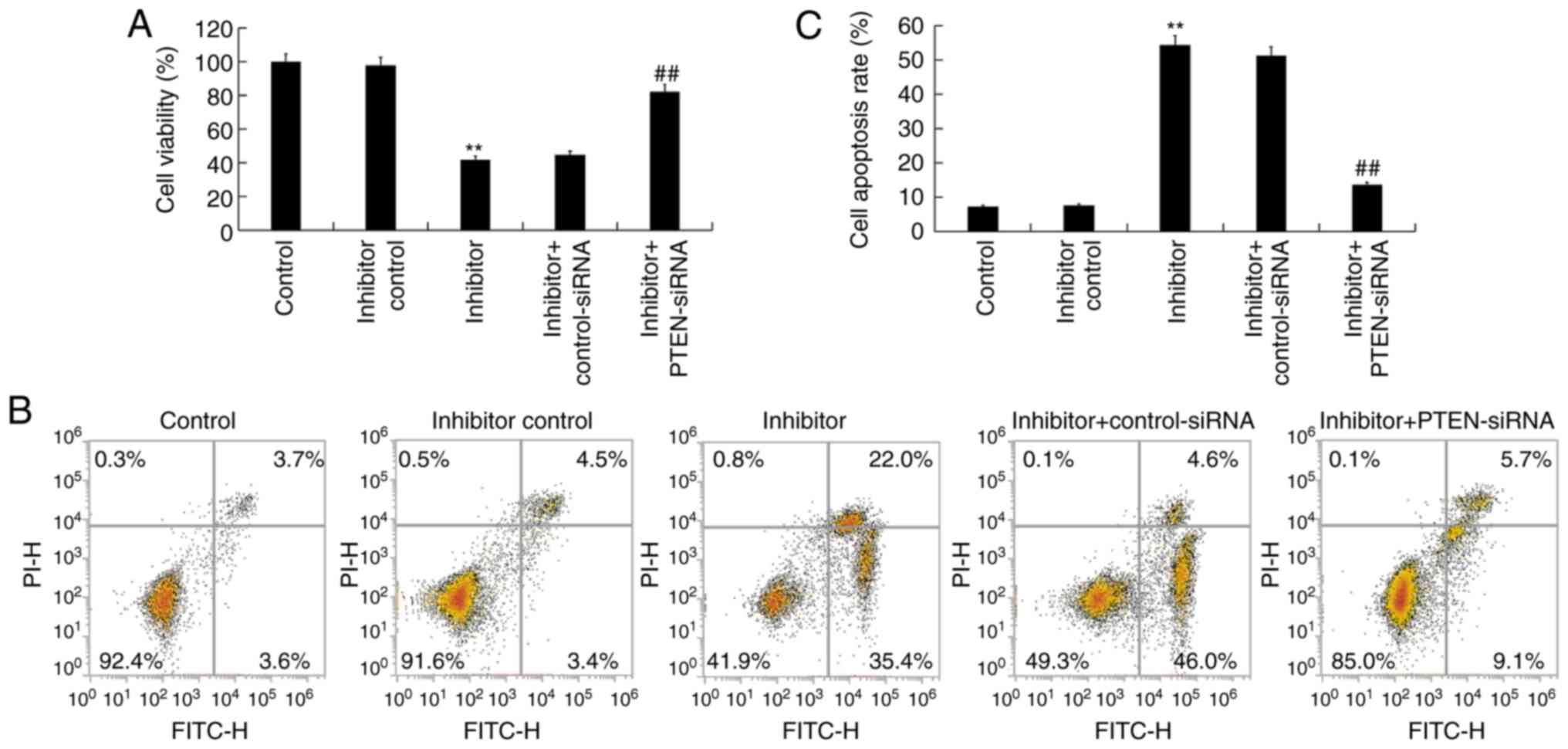
In order to shed light on the function of
miR-130b-3p in nephroblastoma, the effect of miR-130b-3p on the
cell proliferation of WiT49 cells was investigated. The results
demonstrated that miR-130b-3p inhibitor significantly decreased
WiT49 cell viability, which was reversed by PTEN-siRNA (Fig. 5A). To further determine whether
miR-130b-3p could regulate WiT49 cell apoptosis, flow cytometry was
performed to analyze apoptosis. The results revealed that
miR-130b-3p inhibitor significantly induced WiT49 cell apoptosis,
and this induction was reversed by PTEN-siRNA (Fig. 5B and C).
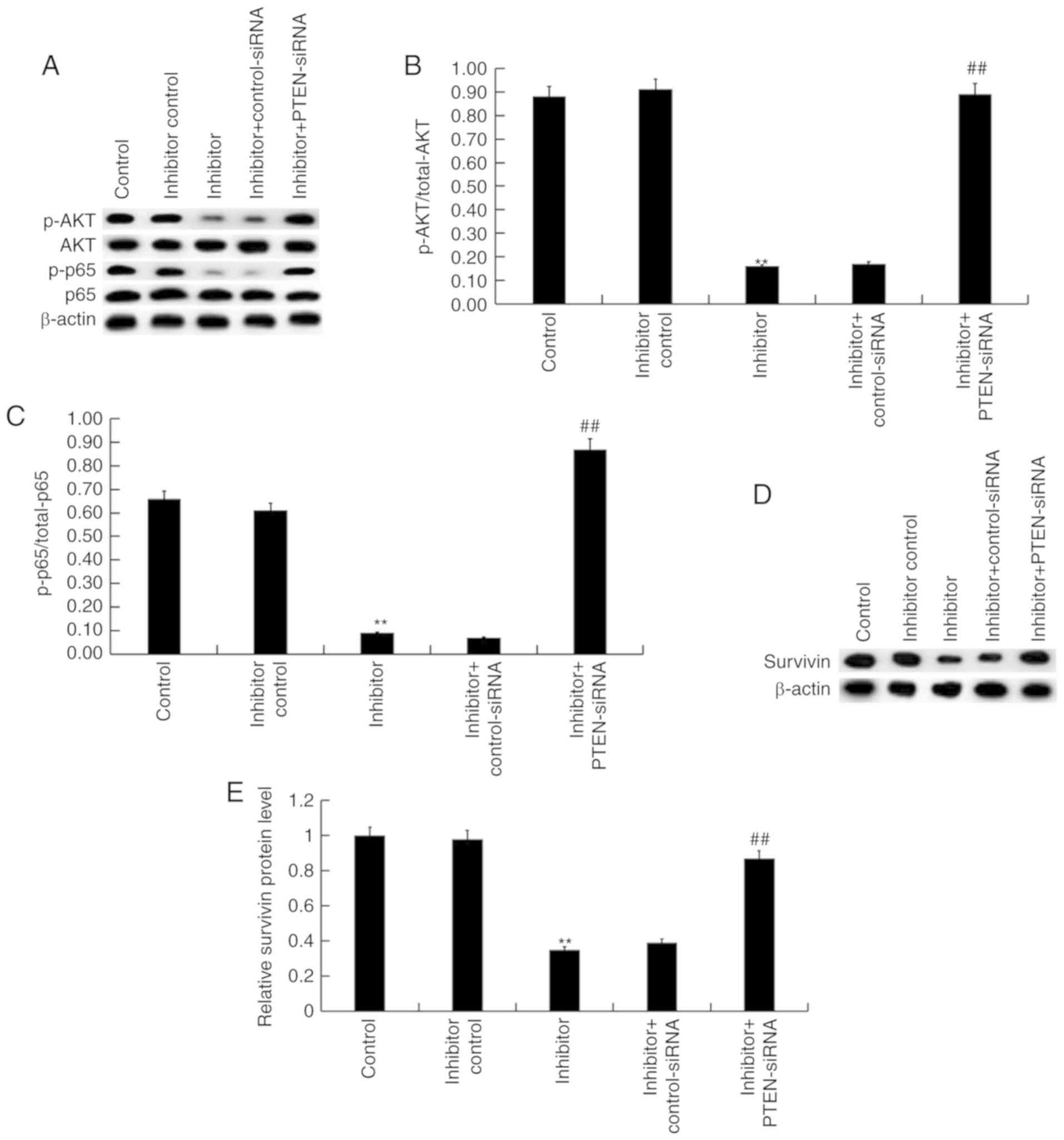
Effect of miR-130b-3p on
Akt/p-NF-κB/survivin signaling pathway in nephroblastoma cells
Finally, to explore the molecular mechanism of the
effect of miR-130b-3p inhibitor on nephroblastoma cells,
Akt/p-NF-κB/survivin signaling pathway was analyzed. As shown in
Fig. 6A, compared with the control
group, the protein levels of p-Akt, p-NF-κB (p-p65) were markedly
decreased by miR-130b-3p inhibitor, and these decreases were
eliminated by PTEN silencing. The ratio of p-Akt/total-Akt and
p-p65/p65 significantly decreased following treatment with the
miR-130b-3p inhibitor, and these decreases were ameliorated
following PTEN silencing (Fig. 6B and
C). Furthermore, survivin protein levels were significantly
decreased by miR-130b-3p inhibitor treatment. However, these
decreases were reversed following PTEN silencing (Fig. 6D and E).
Discussion
miRNAs have emerged as promising biomarkers for
tumors (20–22). It has been reported that aberrant
expression of miRNAs participated in physiological and pathological
processes of a variety of human cancers, including proliferation
(23), invasion (24), apoptosis (25) and chemotherapy resistance (26). In addition, miRNA dysregulation is
causally involved in the initiation and progression of cancer
(27–29). Furthermore, miRNAs can act as either
oncogenes or tumor suppressors through interaction with specific
targets (30,31).
A series of microarray chips have been used to
detect the miRNA expression of nephroblastoma and these studies
have indicated the abnormal expression levels of various miRNAs in
nephroblastoma, including the upregulated genes, miR-378 and
miR-18b, and the downregulated genes, miR-193a-5p and miR-199a-5p
(5,32). Zhu et al (33) demonstrated that miR-92a-3p was
downregulated in nephroblastoma, and it inhibited nephroblastoma
cell proliferation, colony formation, migration and invasion of
nephroblastoma by targeting Notch1. The present study revealed that
miR-130b-3p was upregulated in nephroblastoma tissues. A large
number of studies have indicated that miR-130b-3p was associated
with various cancers, including breast cancer (34), lung cancer (35), bladder carcinoma (36), and human epithelial ovarian cancer
(37).
A previous study demonstrated that miR-130b-3p
targets PTEN to mediate chemoresistance, proliferation and
apoptosis via Wnt/β-catenin pathway in lung cancer cells (35). It has been reported that miR-130b-3p
targets PTEN to promote proliferation and decrease apoptosis by the
PI3K/Akt signaling pathway in breast cancer cells (34). The results of the present study
confirmed that PTEN was a direct target gene of miR-130b-3p in
nephroblastoma cells, which were consistent with a previous study
(38). The expression of PTEN was
markedly decreased in nephroblastoma tissues and cells.
Furthermore, the present study demonstrated that the inhibitor of
miR-130b-3p, inhibited proliferation, promoted apoptosis and
repressed the Akt/p-NF-κB/survivin signaling pathway in
nephroblastoma cells. It is worth mentioning that all the effects
of miR-130b-3p inhibitor on nephroblastoma cells were eliminated by
PTEN silencing.
In conclusion, the present study indicated that
miR-130b-3p was upregulated in nephroblastoma, and its
downregulation could inhibit proliferation and induce apoptosis
through repressing Akt/p-NF-κB/survivin signaling pathway by
targeting PTEN in nephroblastoma cells. These findings may provide
important theoretical basis and therapeutic strategies for
nephroblastoma. However, this is a preliminary study of the role of
miR-130b-3p in nephroblastoma and the effect of miR-130b-3p
overexpression on nephroblastoma cells is still unclear. Therefore,
in order to fully understand the role of miR-130b-3p in
nephroblastoma, and to ascertain whether miR-130b-3p has a similar
effect on nephroblastoma in vivo requires further study.
Furthermore, the association of miR-130b-3p expression with the
clinical characteristics and prognosis of children with
nephroblastoma is still unclear. Thus, further in vivo and
clinical studies are required to prove the role of miR-130b-3p in
nephroblastoma.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data sets used and/or generated during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
YH designed the current study. YH and JY analyzed
the data and prepared the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the Zaozhuang Municipal Hospital (Zaozhuang, China)
and informed consent was obtained from each child and their
guardian prior to enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Charlton J, Pavasovic V and
Pritchard-Jones K: Biomarkers to detect Wilms tumors in pediatric
patients: Where are we now? Future Oncol. 11:2221–2234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cone EB, Dalton SS, Van Noord M, Tracy ET,
Rice HE and Routh JC: Biomarkers for Wilms tumor: A systematic
review. J Urol. 196:1530–1535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qi C, Hu Y, Yang F, An H, Zhang J, Jin H
and Guo F: Preliminary observations regarding the expression of
collagen triple helix repeat-containing 1 is an independent
prognostic factor for Wilms' tumor. J Pediatr Surg. 51:1501–1506.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brok J, Pritchard-Jones K, Geller JI and
Spreafico F: Review of phase I and II trials for Wilms' tumour-Can
we optimise the search for novel agents? Eur J Cancer. 79:205–213.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu X, Li Z, Chan MT and Wu WK: The roles
of microRNAs in Wilms' tumors. Tumour Biol. 37:1445–1450. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X, Liu F, Lin B, Luo H, Liu M, Wu J, Li
C, Li R, Zhang X, Zhou K and Ren D: miR-150 inhibits proliferation
and tumorigenicity via retarding G1/S phase transition in
nasopharyngeal carcinoma. Int J Oncol. 50:1097–1108. 2017.
View Article : Google Scholar
|
|
7
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mallory AC and Vaucheret H: MicroRNAs:
Something important between the genes. Curr Opin Plant Biol.
7:120–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rogers K and Chen X: Biogenesis, turnover,
and mode of action of plant microRNAs. Plant Cell. 25:2383–2399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao ZQ, Yin TK, Li YX, Zhang JH and Gu
JJ: miR-130b regulates the proliferation, invasion and apoptosis of
glioma cells via targeting of CYLD. Oncol Rep. 38:167–174. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun B, Li L, Ma W, Wang S and Huang C:
MiR-130b inhibits proliferation and induces apoptosis of gastric
cancer cells via CYLD. Tumour Biol. 37:7981–7987. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li BL, Lu C, Lu W, Yang TT, Qu J, Hong X
and Wan XP: miR-130b is an EMT-related microRNA that targets DICER1
for aggression in endometrial cancer. Med Oncol. 30:4842013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Di Cristofano A and Pandolfi PP: The
multiple roles of PTEN in tumor suppression. Cell. 100:387–390.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y,
Yuan L, Zhang C, Hong M, Wang S and Li X: MiR-106b induces cell
radioresistance via the PTEN/PI3K/Akt pathways and p21 in
colorectal cancer. J Transl Med. 13:2522015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou X, Yang X, Sun X, Xu X, Li X, Guo Y,
Wang J, Li X, Yao L, Wang H and Shen L: Effect of PTEN loss on
metabolic reprogramming in prostate cancer cells. Oncol Lett.
17:2856–2866. 2019.PubMed/NCBI
|
|
20
|
Shin VY and Chu KM: MiRNA as potential
biomarkers and therapeutic targets for gastric cancer. World J
Gastroenterol. 20:10432–10439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rocci A, Hofmeister CC and Pichiorri F:
The potential of miRNAs as biomarkers for multiple myeloma. Expert
Rev Mol Diagn. 14:947–959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Galardi A, Colletti M, Businaro P,
Quintarelli C, Locatelli F and Di Giannatale A: MicroRNAs in
neuroblastoma: Biomarkers with therapeutic potential. Curr Med
Chem. 25:584–600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan Y, Anbalagan D, Lee LH, Samy RP,
Shanmugam MK, Kumar AP, Sethi G, Lobie PE and Lim LH: ANXA1
inhibits miRNA-196a in a negative feedback loop through NF-κB and
c-Myc to reduce breast cancer proliferation. Oncotarget.
7:27007–27020. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Li Q, Wang Q, Jiang Z and Zhang L:
Silencing of miRNA-218 promotes migration and invasion of breast
cancer via Slit2-Robo1 pathway. Biomed Pharmacother. 66:535–540.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li P, Xie XB, Chen Q, Pang GL, Luo W, Tu
JC, Zheng F, Liu SM, Han L, Zhang JK, et al: MiRNA-15a mediates
cell cycle arrest and potentiates apoptosis in breast cancer cells
by targeting synuclein-γ. Asian Pac J Cancer Prev. 15:6949–6954.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: MicroRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Watson JA, Bryan K, Williams R, Popov S,
Vujanic G, Coulomb A, Boccon-Gibod L, Graf N, Pritchard-Jones K and
O'Sullivan M: miRNA profiles as a predictor of chemoresponsiveness
in Wilms' tumor blastema. PLoS One. 8:e534172013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu S, Zhang L, Zhao Z, Fu W, Fu K, Liu G
and Jia W: microRNA-92a-3p inhibits the cell proliferation,
migration and invasion of Wilms tumor by targeting NOTCH1. Oncol
Rep. 40:571–578. 2018.PubMed/NCBI
|
|
34
|
Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou
H and Jia L: MicroRNA-130b targets PTEN to mediate drug resistance
and proliferation of breast cancer cells via the PI3K/Akt signaling
pathway. Sci Rep. 7:419422017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Q, Zhang B, Sun L, Yan Q, Zhang Y,
Zhang Z, Su Y and Wang C: MicroRNA-130b targets PTEN to induce
resistance to cisplatin in lung cancer cells by activating
Wnt/β-catenin pathway. Cell Biochem Funct. 36:194–202. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lv M, Zhong Z, Chi H, Huang M, Jiang R and
Chen J: Genome-wide screen of miRNAs and targeting mRNAs reveals
the negatively regulatory effect of miR-130b-3p on PTEN by PI3K and
integrin β1 signaling pathways in bladder carcinoma. Int J Mol Sci.
18:782017. View Article : Google Scholar
|
|
37
|
Zhou D, Zhang L, Sun W, Guan W, Lin Q, Ren
W, Zhang J and Xu G: Cytidine monophosphate kinase is inhibited by
the TGF-β signalling pathway through the upregulation of
miR-130b-3p in human epithelial ovarian cancer. Cell Signal.
35:197–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu T, Cao R, Li S, Fu M, Ren L, Chen W,
Zhu H, Zhan Q and Shi R: MiR-130b plays an oncogenic role by
repressing PTEN expression in esophageal squamous cell carcinoma
cells. BMC Cancer. 15:292015. View Article : Google Scholar : PubMed/NCBI
|