Introduction
Liver cancer is one of the most frequently diagnosed
types of cancer that causes unacceptably high mortality rates
worldwide (1). Particularly in less
developed countries, such as China, the high incidence rate of
liver cancer is a heavy burden on public health (2). Although increased efforts have been
made regarding diagnosis and treatment of liver cancer (3,4), the
survival outcome of patients is still poor due to the high
prevalence of tumor metastasis at the time of diagnosis, and
surgical resection no longer being a treatment option for patients
with metastasis (5). Unclear
pathogenesis of liver cancer is one of the major causes of
treatment failure in patients with liver cancer (6,7).
Therefore, understanding the molecular mechanism underlying the
development of liver cancer may benefit the treatment strategy for
patients with liver cancer.
Rho associated coiled-coil containing protein kinase
2 (ROCK2) is a key regulator of cell polarity and actin
cytoskeleton, and may play a pivotal role in cancer (8,9).
Inhibition of ROCK2 has therapeutic effects on several types of
cancer, including hepatocellular carcinoma (HCC) (9). Several studies have demonstrated that
ROCK2 may serve as a potential therapeutic target for cancer
treatment (9,10). It has been well established that
microRNAs can regulate ROCK2 expression in human diseases,
including cancer (11,12). MicroRNA-466 was recently
characterized as a tumor suppressor in prostate cancer (13), however, its involvement in other
types of cancer, including HCC remains unknown. The present study
demonstrated that microRNA-466 may inhibit cancer cell migration
and invasion in HCC by indirectly mediating the downregulation of
ROCK2.
Materials and methods
Patient samples
The present study analyzed tumor tissue and adjacent
healthy tissue biopsies, as well as plamsa samples obtained from 62
patients with HCC (male, n=33; female, n=29; age range, 32–68
years; mean age, 48.4±4.6 years). In addition, plasma samples were
also obtained from 38 healthy volunteers (male, n=20; female, n=
18; age range, 31–67 years; mean age, 48.1±4.3 years). Patient
information from each group is summarized in Table I. All samples were obtained from
patients and healthy volunteers admited at The Fourth Hospital of
Hebei Medical University between May 2015 and May 2018. Inclusion
criteria were as follows: i) Patients diagnosed with HCC confirmed
by pathologcal examination; and ii) newly diagnosed HCC. Exclusion
criteria were as follows: i) Patients diagnosed with other
diseases; and ii) patients who received treatment ≥3 months prior
to the current study. The current study was approved by the Ethics
Committee of The Fourth Hospital of Hebei Medical University
(Shijiangzhuang, China) and all participants provided written
informed consent.
 | Table I.Basic information for each group of
participants. |
Table I.
Basic information for each group of
participants.
| Variable | Patients with
hepatocellular carcinoma (n=62) | Healthy controls
(n=38) |
|---|
| Sex
(male/female) | 33/29 | 20/18 |
| Age range
(years) | 32–68 | 31–67 |
| Mean age (years) | 48.4±4.6 | 48.1±4.3 |
| Clinical stage |
|
|
| I | 12 | N/A |
| II | 14 | N/A |
| III | 12 | N/A |
| IV | 24 | N/A |
Cell culture and transfection
Human HCC cell lines SNU-398 (ATCC®
CRL-2233™) and SNU-182 (ATCC® CRL-2235™) were purchased
from the American Type Culture Collection (ATCC). Cells were
cultured in RPMI-1640 medium (ATCC) supplemented with
heat-inactivated 10% fetal bovine serum (FBS; ATCC) and maintained
at 37°C in 5% CO2-humidified incubator.
Cells were transfected with 15 nM ROCK2 expression
pcDNA3 vectors or empty pcDNA3 vectors, purchased from GeneCopoeia,
using Lipofectamine® 3000 reagent (Thermo Fisher
Scientific, Inc.). Cells were transfected with 50 nM miRNA-466
mimics (5′-AUACACAUACACGCAACACACAU-3′) or negative control miRNA
(5′-UUCUCCGAACGUGUCACGUdTdT-3′), purchased from Applied Biosystems
(Thermo Fisher Scientific, Inc.), using Lipofectamine®
3000 reagent. Cells transfected with empty vector or control miRNA
were used as the negative control, while untransfected cells were
used as the control. Subsequent experiments were performed 24 h
post-transfections.
Target site analysis
TargetScan bioinformatics analysis (www.targetscan.org) was used to predict potential
targets of has-miRNA-466 on ROCK2 with default parameters (human
species).
Reverse transcription-quantitative PCR
(RT-q) PCR
Total RNA and miRNA was extracted from tissues as
well as SNU-398 and SNU-182 cells using the MPure™ Total RNA
Extraction kit (cat. no. 117022160; MP Biomedicals, LLC) and
miRNeasy Mini kit (cat. no. 217004; Qiagen, Inc.), repectively. To
detect the mRNA expression level of ROCK2, qPCR was subseqeuntly
performed using SYBR Green Master mix (Bio-Rad Laboratories, Inc.).
The following primer pairs were used for qPCR: ROCK2 forward,
5′-TCCCAACCAACTGTGAGGCATGT-3′ and reverse,
5′-TGTGGCACCTACGGCACTCT-3′; GAPDH forward, 5′-GCATCTTCTTTTGCGTCG-3′
and reverse, 5′-TGTAAACCATGTAGTTGAGGT-3′. To detect the expression
level of miRNA-466, qPCR was performed using a TaqMan Real-Time PCR
assay (Thermo Fisher Scientific, Inc.). The following primer pairs
were used for qPCR: miRNA-466 forward, 5′-GTCGTATCCAGTGCAGGGTCC-3′
and reverse, 5′-TTGTAGTCACTAGGGCAC-3′; U6 small nuclear RNA (U6)
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. qPCR reactions were performed on a 7500
Fast Real Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the following thermocycling conditions:
Initial denaturation at 95°C for 1 min; followed by 40 cycles of
95°C for 10 sec and 58.5°C for 35 sec. ROCK2 and miRNA-466
expression levels were quantified using the 2−ΔΔCq
method (14) and normalized to GAPDH
and U6, respectively.
Transwell migration and invasion
assays
Following transfection with ROCK2 and/or
microRNA-466, serum-free cell suspensions (3×104
cells/ml) were prepared and 0.1 ml cell suspension/well was added
to the upper chamber (Transwell membranes were pre-coated with
Matrigel prior to the invasion assay). Corning® HTS
Transwell-96 well plate (pore size, 8.0 µm; Corning, Inc.) was
used. The lower chamber was filled with RPMI-1640 culture medium
containing 20% FBS. Following 24-h incubation at 37°C, the cells
were stained with 0.5% crystal violet (Sigma-Aldrich; Merck KGaA)
for 15 min at room temperature. Stained cells were counted under a
light microscope (magnification, ×40; Olympus Corporation).
Western blot analysis
Total protein was extracted from SNU-398 and SNU-182
cells using a CelLytic™ MEM Protein Extraction kit (Sigma-Aldrich;
Merck KGaA). A BCA kit (Sangon Biotech Co., Ltd.) was used to
measure protein concentrations. Electrophoresis was performed using
SDS-PAGE (10% gel) with 30 µg protein per lane. The proteins were
transferred to a PVDF membrane followed by blocking in 5% non-fat
milk for 2 h at room temperature. The membranes were incubated with
primary antibodies against ROCK2 (1:1,500; cat. no. ABS436; EMD
Millipore) and GAPDH (1:1,200; cat. no. ab9485; Abcam) for 12 h at
4°C prior to incubation with horseradish peroxidase-labeled
secondary antibody (1:1,200; cat. no. MBS435036; MyBioSource, Inc.)
for 2 h at room temperature. Protein bands were visualized using
Pierce™ ECL Western Blotting Substrate (Pierce; Thermo Fisher
Scientific, Inc.). Protein expression was quantified using ImageJ
software (v1.46; National Institutes of Health).
Statistical analysis
Data were presented as the mean ± standard deviation
from three independent experiments. All statistical analyses were
perfromed using GraphPad Prism software (version 6.0; GraphPad
Software, Inc.). Pearson correlation coefficient was used to
examine the correlation between the expression of ROCK2 and
miRNA-466. Student's t-test was used to analyze differences between
two groups. Paired t-test was used to analyze differences between
tumor tissue and adjacent healthy tissue samples. One-way analysis
of variance followed by Tukey's post hoc test was used to analyze
differences among multiple groups. Receiver operating
characteristic analysis was used to evaluate the diagnostic value
of serum miRNA-466. P<0.05 was considered to indicate a
statistically significant difference.
Results
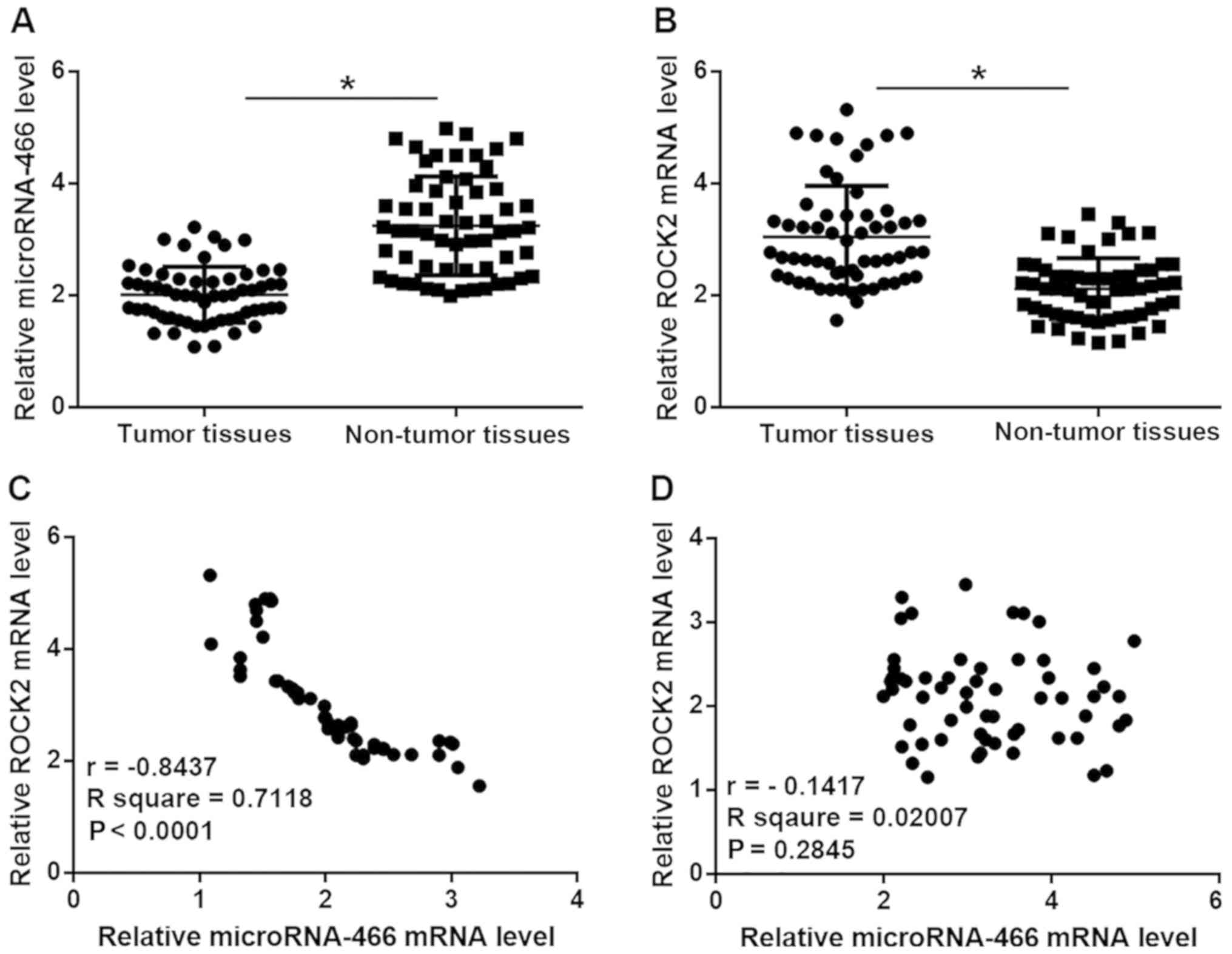
Expression levels of microRNA-466 and
ROCK2 are inversely correlated in tumor tissues but not in adjacent
healthy tissues in patients with HCC
The relative expression level of microRNA-466 was
significantly reduced, while the relative mRNA expression level of
ROCK2 was significantly increased in tumor tissue compared with
adjacent healthy tissue samples in patients with HCC (P<0.05;
Fig. 1A and B). In addition, Pearson
correlation coefficient analyses demonstrated that the expression
levels of microRNA-466 and ROCK2 were inversely correlated in tumor
tissue samples (P<0.0001; Fig.
1C), however, there was no correlation observed between the
expression levels of microRNA-466 and ROCK2 in adjacent healthy
tissue samples (Fig. 1D).
Plasma levels of microRNA-466 and
ROCK2 are inversely correlated in patients with HCC but not in
healthy controls
The relative expression level of microRNA-466 was
significantly reduced, while the relative mRNA expression level of
ROCK2 was significantly increased in plasma samples from patients
with HCC compared with healthy controls (P<0.05; Fig. 2A and B). Pearson correlation
coefficient analyses demonstrated that plasma levels of
microRNA-466 and ROCK2 were inversely correlated in patients with
HCC patients (P<0.0001; Fig. 2C),
however, there was no correlation observed between the plasma
expression levels of microRNA-466 and ROCK2 in the healthy control
group (Fig. 2D).
Diagnostic value of the reduced plasma
expression level of microRNA-466 in the detection of early stage
HCC
Receiver operating characteristic (ROC) curve
analysis was performed to evaluate the diagnostic value of serum
microRNA-466 to distinguish between patients with early stage HCC
(n=26) and healthy controls (n=38). The area under the curve was
0.9074 (95% confidence interval: 0.8381–0.9767) with a standard
error of 0.03534 (P<0.0001; Fig.
3).
MicroRNA-466 overexpression suppresses
ROCK2 expression in HCC cell lines
The relative expression level of microRNA-466 was
significantly increased in both HCC cell lines following
transfection with microRNA-466 mimic compared with control and
negative control groups (P<0.05; Fig.
4A). In addition, transfection with microRNA-466 mimic
significantly decreased the relative protein expression level of
ROCK2 in SNU-398 and SNU-182 HCC cell lines compared with control
and negative control groups (P<0.05; Fig. 4A). The relative mRNA expression level
of ROCK2 was significantly increased in both HCC cell lines
following transfection with ROCK2 expression vector compared with
control and negative control groups (P<0.05; Fig. 4B). Furthermore, overexpression of
ROCK2 did not significantly alter the expression level of
microRNA-466 (Fig. 4B). Taken
together, these results suggest that miRNA-466 may be an upstream
inhibitor of ROCK2 in HCC. However, bioinformatics analysis did not
predict a target site of microRNA-466 in ROCK2 (data not
shown).
MicroRNA-466 overexpression inhibits
HCC cell migration and invasion via ROCK2
MicroRNA-466 overexpression significantly suppressed
HCC cell migration and invasion, while ROCK2 overexpression
significantly enhanced HCC cell migration and invasion in both HCC
cell lines SNU-398 and SNU-182 compared with control (P<0.05;
Fig. 5A and B). Furthermore, ROCK2
overexpression partially reversed the inhibitory effect of
microRNA-466 overexpression on HCC cell migration and invasion
(Fig. 5A and B).
Discussion
MicroRNA-466 was recently characterized as a tumor
suppressor with a known biological function in prostate cancer
(13). The present study
demonstrated that microRNA-466 may function as a tumor suppressor
in HCC, the most common type of primary liver cancer, which
accounts for 90% of all cases of primary liver cancer (1–4). The
current study demonstrated that microRNA-466 may be involved in the
regulation of HCC cancer cell migration and invasion by
downregulating ROCK2 expression.
The development and progression of liver cancer is
associated with changes in a large set of human genes, including
miRNAs (15,16). Differential expression of specific
microRNAs can predict the survival and treatment outcome of
patients with liver cancer (15,16). The
present study revealed that microRNA-466 was significantly
downregulated in tumor tissue compared with adjacent healthy
control tissue samples obtained from patients with HCC. In
addition, plasma microRNA-466 was significantly downregulated in
patients with HCC compared with healthy controls. These results
suggest that downregulation of microRNA-466 may be involved in the
pathogenesis of HCC.
The treatment outcome of patients with early stage
HCC is generally satisfactory, while the outcome and survival of
patients with late-stage HCC is poor due to the existence of tumor
metastasis (17). Therefore,
improved diagnosis and treatment strategy following early diagnosis
is important for the treatment of HCC. In the current study, ROC
curve analysis demonstrated that reduced plasma expression level of
microRNA-466 may be used to effectively distinguish patients with
HCC from healthy controls, indicating the potential application of
plasma microRNA-466 to detect early stage HCC. However, multiple
biomarkers should be used to improve overall diagnostic specificity
due to the unknown expression pattern of microRNA-466 in other
diseases.
ROCK2 serves an oncogenic role and is overexpressed
in several types of human cancer, such as oral cancer and renal
cancer (18,19). Therefore, inhibition of ROCK2 may
serve as a potential therapeutic target for cancer treatment. The
present study demonstrated that microRNA-466 suppressed ROCK2
expression in HCC cells, which suggests that miRNA-466 may be an
upstream inhibitor of ROCK2 in HCC. However, the lack of reverse
correlation between expression levels of microRNA-466 and ROCK2 in
adjacent healthy tissue samples from patients with HCC patients, as
well as in the plasma samples from healthy controls, suggests the
inhibitory effect of microRNA-466 on ROCK2 expression may be
achieved indirectly. Therefore, other pathological factors, which
mediate the interaction between microRNA-466 and ROCK2, may exist
in patients with HCC. For example, microRNA-466 may directly target
specific genes involved in HCC, and those genes may interact with
ROCK2. However, studies are required to further understand the
interaction between microRNA-466 and ROCK2 in HCC.
The present study revealed that microRNA-466 may be
involved in the regulation of HCC cell migration and invasion via
ROCK2; however, microRNA-466 is not involved in HCC cell
proliferation (data not shown). By contrast, a recent study
demonstrated that microRNA-466 is involved in tumor growth in
prostate cancer (13), which
suggests that microRNA-466 may have different functions in
different types of cancer. In addition, bioinformatics analysis did
not identify a target binding site for microRNA-466 on ROCK2,
indicating an indirect interaction between microRNA-466 and ROCK2.
However, further studies are required to understand the underlying
molecular mechanism of microRNA-466 in regulating cell migration
and invasion in HCC.
In conclusion, the relative expression level of
microRNA-466 was downregulated, while the relative expression level
of ROCK2 was upregulated in HCC. The present findings suggest that
microRNA-466 may be involved in the regulation of cancer cell
migration and invasion in HCC by downregulating ROCK2
expression.
Acknowledgements
Not applicable.
Funding
The current study was supported by a grant from the
Key Project of Medical Research in Hebei Province (grant no.
20150335).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NA and FY designed experiments. NA, BL and LL
performed experiments. ZL, HJ and GY assisted in the experiments
and analyzed data. FY drafted the manuscript and all authors
approved the final version to be published.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of The Fourth Hospital of Hebei Medical University
(Shijiangzhuang, China) and all participants provided written
informed consent.
Patient consent for publication
All patients provided informed consent for
publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kudo M: Surveillance, diagnosis,
treatment, and outcome of liver cancer in Japan. Liver Cancer.
4:39–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao YY, Chen H, Zhou YY, Wang LT, Hou Y,
Xia XH and Ding Y: Intraorgan targeting of gold conjugates for
precise liver cancer treatment. ACS Appl Mater Interfaces.
9:31458–31468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology 127 (5 Suppl 1). S5–S16. 2004.
|
|
6
|
Palmer WC and Patel T: Are common factors
involved in the pathogenesis of primary liver cancers? A
meta-analysis of risk factors for intrahepatic cholangiocarcinoma.
J Hepatol. 57:69–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vigil D, Kim TY, Plachco A, Garton AJ,
Castaldo L, Pachter JA, Dong H, Chen X, Tokar B, Campbell SL and
Der CJ: ROCK1 and ROCK2 are required for non-small cell lung cancer
anchorage-independent growth and invasion. Cancer Res.
72:5338–5347. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li M, Ke J, Wang Q, Qian H, Yang L, Zhang
X, Xiao J, Ding H, Shan X, Liu Q, et al: Upregulation of ROCK2 in
gastric cancer cell promotes tumor cell proliferation, metastasis
and invasion. Clin Exp Med. 17:519–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2J. Gut. 61:278–289.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rath N and Olson MF: Rho-associated
kinases in tumorigenesis: Re-considering ROCK inhibition for cancer
therapy. EMBO Rep. 13:900–908. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kroiss A, Vincent S, Decaussin-Petrucci M,
Meugnier E, Viallet J, Ruffion A, Chalmel F, Samarut J and Allioli
N: Androgen-regulated microRNA-135a decreases prostate cancer cell
migration and invasion through downregulating ROCK1 and ROCK2.
Oncogene. 34:2846–2855. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng F, Jiang J, Yu Y, Tian R, Guo X, Li
X, Shen M, Xu M, Zhu F, Shi C, et al: Direct targeting of
SUZ12/ROCK2 by miR-200b/c inhibits cholangiocarcinoma
tumourigenesis and metastasis. Br J Cancer. 109:3092–3104. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Colden M, Dar AA, Saini S, Dahiya PV,
Shahryari V, Yamamura S, Tanaka Y, Stein G, Dahiya R and Majid S:
MicroRNA-466 inhibits tumor growth and bone metastasis in prostate
cancer by direct regulation of osteogenic transcription factor
RUNX2. Cell Death Dis. 8:e25722017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lepage C, Capocaccia R, Hackl M, Lemmens
V, Molina E, Pierannunzio D, Sant M, Trama A and Faivre J;
EUROCARE-5 Working Group, : Survival in patients with primary liver
cancer, gallbladder and extrahepatic biliary tract cancer and
pancreatic cancer in Europe 1999–2007: Results of EUROCARE-5. Eur J
Cancer. 51:2169–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dourado MR, de Oliveira CE,
Sawazaki-Calone I, Sundquist E, Coletta RD and Salo T:
Clinicopathologic significance of ROCK2 expression in oral squamous
cell carcinomas. J Oral Pathol Med. 47:121–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu Z, Hong Z, Ma M, Liu X, Chen L, Zheng
C, Xi X and Shao J: Rock2 promotes RCC proliferation by decreasing
SCARA5 expression through β-catenin/TCF4 signaling. Biochem Biophys
Res Commun. 480:586–593. 2016. View Article : Google Scholar : PubMed/NCBI
|