Introduction
Antibiotics remain one of the most important
discoveries in modern medicine and have proven to be powerful drugs
in controlling infectious diseases. Although antibiotics have been
successfully used for several decades, the widespread and
unrestricted use of antibiotics have led to the development of
antimicrobial resistance (1,2). Bacterial resistance to antimicrobial
agents, including multidrug resistance, is a major problem in the
treatment of infectious diseases. Bacteria have the ability to
rapidly acquire resistance genes through inter- and intra-species
genetic transference, thus exacerbating the worldwide problem of
resistant strains (3). The World
Health Organization considers antibiotic resistance to be one of
the greatest threats to the treatment of bacterial infectious
diseases (4,5). Approximately 50% of hospital acquired
infections worldwide are caused by multidrug-resistant pathogens
(6). The major mechanisms of
antibiotic resistance include the inactivation of the antibiotic,
target modification, alteration of drug access to the target
through drug efflux and decreased uptake (2). Methicillin-resistant Staphylococcus
aureus (MRSA) strains are some of the most tenacious
antibiotic-resistant pathogens and contribute extensively to
hospital-acquired infections (7).
Bacterial multidrug efflux pumps are the major drivers of microbial
resistance against several classes of antibiotics (2,3). As the
most well studied chromosomally-encoded efflux pump in pathogenic
Gram-positive bacteria [such as Staphylococcus (S.) aureus],
NorA multidrug efflux pump (NorA) is overexpressed in certain
resistant mutants that demonstrate reduced susceptibility to
quinolones, including norfloxacin (NFX) and ciprofloxacin (3,8,9).
The discovery of novel antibiotics to combat
resistant strains is therefore a major goal for the pharmaceutical
industry (4,10). However, since the 1960s, the rate of
development of novel antibiotic classes approved by the Food and
Drug Administration has markedly reduced (2). A potential strategy to overcome the
emergence of resistant strains is the discovery and development of
novel drugs capable of partly or completely inhibiting bacterial
resistance mechanisms, which have been termed resistance-modifying
agents (RMAs) (2,11). Plants provide a valuable source of
novel chemical entities, which can contribute to the discovery of
novel therapeutic agents. In fact, products derived from higher
plants represent ~25% of drugs in current clinical use (2,12).
Numerous plant-derived compounds act as bacterial RMAs against
S. aureus (2,13). For example, the indole alkaloid,
reserpine, is a known NorA efflux pump inhibitor and exhibited a
synergistic effect with ciprofloxacin (14).
Plants of the Mahonia genus, which is in the
Berberidaceae family, have been widely used for a long time in
Traditional Chinese Medicine as a treatment for tuberculosis,
periodontitis, dysentery, icterus, pharyngolaryngitis, eczema and
wounds (15). The majority of
Mahonia species exert their effects by suppressing pain,
inhibiting coughing and alleviating inflammation (15). The isolated compounds and crude
extracts from Mahonia species exhibit a wide spectrum of
in vitro and in vivo pharmacological effects,
including antimicrobial, anti-inflammatory, hepatoprotective,
antioxidant, antimutagenic and analgesic effects (15). Jatrorrhizine is one of the main
protoberberine alkaloids widely distributed among Mahonia
and Berberis species (15).
In the current study, the in vitro and in vivo
synergistic effects of jatrorrhizine and NFX against MRSA, and the
underlying mechanisms were investigated.
Materials and methods
Materials
Optical density was recorded on a Multiskan™ FC
Microplate Photometer (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and fluorescence was measured on an Infinite® M1000
PRO Multimode reader (Tecan Group, Ltd., Mannedorf, Switzerland).
Water was purified using a Milli-Q water purification system (EMD
Millipore, Billerica, MA, USA).
Institute of Cancer Research (ICR) mice were
obtained from Lingchang Biotechnology Co., Ltd. (Shanghai, China).
All procedures were performed according to the Qiqihar Medical
University guidelines, and approved by the Ethics Committee for
Animal Care and the Use of Laboratory Animals at Qiqihar Medical
University (Qiqihar, China). Specific pathogen-free ICR mice were
bred in the animal facility of Qiqihar Medical University.
Antimicrobial assay
The MRSA strain, SA1199B, derived from a
methicillin-susceptible S. aureus bloodstream isolate from a
rabbit endocarditis model, previously described by Kaatz et
al (16), which overexpresses
the gene encoding NorA, was used in the present study (17). Minimum inhibitory concentrations
(MICs) were determined following the guidelines of Clinical
Laboratory Standards Institute (18). Mueller-Hinton broth (MHB; Oxoid;
Thermo Fisher Scientific, Inc.), containing 20 mg/l Ca2+
and 10 mg/l Mg2+, was used in the assay. All bacterial
suspensions were adjusted to 5×105 Cfu/ml for the
bioassay, cells were seeded at a density of 5×104
cells/well in 96-well plates. Ofloxacine (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) served as the positive control and 2%
DMSO in MHB as the negative control. Controls and treatments were
assessed in duplicate. Following the incubation at 37°C for 18–24
h, 20 µl MTT solution (5 mg/ml; Biosharp, Solon, OH, USA) was added
into each well and incubated at 37°C for 2–4 h. Concentrations that
completely inhibited the visible bacterial growth (from light
yellow to deep purple) were recorded as MIC values against the
SA1199B strain. The optical density in each well was measured at
570 nm. The 50% effective concentrations (EC50s) were
calculated based on the percentage of bacteria growth inhibition
following the treatment of the bacteria with different
concentrations of compounds.
Synergy assays
Using the strain SA1199B, a checkerboard
microdilution assay was employed to examine for the presence of a
synergistic interaction of antimicrobes (19). Jatrorrhizine or reserpine (both
Dalian Meilun Biotech Co., Ltd., Dalian, China) in a concentration
range of 2–128 mg/l (2, 4, 8, 16, 32, 64 and 128 mg/l) was tested
in combination with NFX (Dalian Meilun Biotech Co., Ltd.) against
the tested S. aureus strain; the concentration range of NFX
was 0.5–256 mg/l (0.5, 1, 2, 4, 8, 16, 32, 64, 128 and 256 mg/l).
Cells were seeded at a density of 5×104 cells/well in
96-well plates. All treatments were tested in duplicate wells.
Following the incubation at 37°C for 18–24 h, 20 µl MTT solution (5
mg/ml; Biosharp) was added into each well and incubated at 37°C for
2–4 h. MIC values were used to evaluate the effects of the
combination by calculating the fractional inhibitory concentration
index (FICI) according to the following formula:
FICI=MIC(antibioticcombinedwithcompound)MIC(antibioticalone)+
MIC(compoundcombinedwithantibiotic)MIC(compoundalone)
‘Synergy’ was confirmed when the FICI was ≤0.5. When
the FICI ranged from 0.5–4.0 the effect was regarded as
‘indifferent’, whereas an ‘antagonistic’ effect was confirmed when
the FICI was >4.0.
Growth kinetics
The growth kinetics of SA1199B cells were assessed
to elucidate the effect of NFX in the presence of jatrorrhizine in
MHB, and evaluated using a time-growth curve method, as described
previously (14,20). A bacterial suspension in its
logarithmic phase (1×106 Cfu/ml) was used as the
inoculum. The growth kinetics were analysed following the treatment
of NFX at 16 mg/ml (1/4 MIC) alone and in combination with
jatrorrhizine at 16 mg/ml (1/4 MIC). The cfu/ml was determined by a
serial dilution method (21) on
Mueller-Hinton agar (MHA; Yeasen Biotech, Shanghai, China) plates
at 37°C for 2, 4, 6, 8, 10, 12 and 24 h.
Ethidium bromide (EtBr) efflux
assay
Jatrorrhizine was subjected for a bacterial drug
efflux assay as described previously (22). The efflux activity of strain SA1199B
was determined by measuring the accumulation of the fluorescent dye
EtBr (25 µM; Aladdin Shanghai Biochemical Technology Co., Ltd.,
Shanghai, China) in the absence or presence of known efflux
inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP; 100 µM;
J&K Chemical, Ltd., Beijing, China) and in the absence or
presence of jatrorrhizine (16 mg/l). Cells were seeded at a density
of 5×104 cells/well in 96-well plates. With an
excitation wavelength of 530 nm and emission wavelength of 600 nm,
the fluorescence of the suspension was monitored every 5 min for 1
h (slit width, 5 nm). The assay was performed in triplicate.
Total RNA extraction and
semi-quantitative reverse transcription-polymerase chain reaction
(RT-PCR)
Total RNA was isolated from the bacteria using the
TRIzol reagent (Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. The reverse transcription step was
conducted using the RevertAid™ First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.) to synthesise cDNA following the
manufacturer's protocol. The semi-quantitative PCR analysis was
performed using the 2X Taq PCR MasterMix (PC1120; Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) and with an ABI
7300 real-time fluorescent quantitative PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The PCR thermocycling
parameters were as follows: 95°C for 1 min followed by 40 cycles of
95°C for 15 sec, 58°C for 25 sec and 72°C for 20 sec, and a final
extension step of 72°C for 5 min. The relative gene expression was
calculated by the 2−ΔΔCq method (23). Each sample was assessed in duplicate
and all the samples were analysed in parallel for the expression of
the housekeeping gene 16s rRNA, which was used as an endogenous
control for normalisation of the expression level of target genes.
The fold induction was determined from the mean replicate values.
At the end of the RT-PCR assay, equal amounts of the final
amplified product of each sample was loaded in 2% agarose gel and
subjected to electrophoresis and visualized using ethidium bromide
(1610433; Bio-Rad Laboratories, Inc., Hercules, CA, USA) staining.
The densitometry analysis was performed with ImageJ version 1.48
software (National Institutes of Health, Bethesda, MD, USA). The
primer sequences for the analysis of NorA and 16s rRNA genes are
described in Table I.
 | Table I.Primers used for norA and 16s
rRNA. |
Table I.
Primers used for norA and 16s
rRNA.
| Gene | Primer | Sequence
(5′-3′) | Size (bp) |
|---|
| norA | Sense |
GTTGCTGCTTTCGCCTTATCTC | 200 |
| norA | Antisense |
GGCATAACCATACCAGCACTCA |
|
| 16s rRNA | Sense |
GCTCGTGTCGTGAGATGTTGG | 195 |
| 16s rRNA | Antisense |
TTTCGCTGCCCTTTGTATTGT |
|
Molecular modeling
The three-dimensional structure of jatrorrhizine was
built using ChemDraw Ultra 14.0 and ChemBio3D® Ultra
14.0 (CambridgeSoft Corp., Waltham, MA, USA). The structure was
constructed using the Prepare Ligands modules in Discovery Studio
software (version 2016; Biovia Corp., San Diego, CA, USA). The
amino acid sequence of NorA was investigated in the National Center
for Biotechnology Information database (GI: 153055; Accession:
AAA26658.1). The glycerol-3-phosphate transporter from
Escherichia coli (Protein Data Bank ID: 1PW4) was selected
from the Protein Data Bank database and used as template for NorA
in the docking studies (14). The
NorA homology model for molecular docking was created on the
I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/), which
is one of the computational tools for automated protein 3D
structure prediction (24–26), then refined by Fragment-Guided
Molecular Dynamics simulation (27).
The optimized model was further evaluated using server-based
structural verification from the UCLA-DOE Institute for Genomics
and Proteomics server. The probable binding site of the modeled
structure was identified using the Define and Edit Binding Site
Module, and then applied to the Flexible Docking Module for
flexible docking studies with Chemistry at HARvard Macromolecular
Mechanics force fields in Discovery Studio software.
Murine thigh infection model
The intramuscular infection model was based on
methods described previously (28)
using 4–6 week-old male ICR mice (18–20 g). A total of 70 mice were
provided with free access to food and water, and housed at room
temperature (25°C) with a 12-h light/dark cycle and humidity of
65–70%. They were rendered neutropenic by intraperitoneally (i.p.)
injecting them with 150 and 100 mg/kg cyclophosphamide at 4 days
and 1 day prior to inoculation, respectively. The colonies of MRSA
SA1199B grown on MHA plates were suspended in MHB and then
incubated at 37°C overnight. The animals were injected
intramuscularly with 1×105 CFU MRSA SA1199B and
antimicrobial treatment was started 1 h following bacterial
challenge. The mice were divided into five groups treated
(n=10/group) with a drug-free solution [0.5% w/v sodium
carboxymethyl-cellulose, 0.5 ml/20 g; oral administration (p.o.)],
jatrorrhizine (50 mg/kg, i.p.), NFX [100 mg/kg, p.o.], a
combination of jatrorrhizine (10, 25 or 50 mg/kg, i.p.) and NFX
(100 mg/kg, p.o.) or ofloxacine (50 mg/kg, p.o.; Dalian Meilun
Biotech Co., Ltd.). The mice that received the drug-free solution
were deemed the control group. A total of 24 h after infection, the
mice were euthanized, the muscle was removed and then homogenized
in 1 ml PBS to recover the bacteria. In order to count the viable
MRSA colonies, samples were serially diluted and plated onto MHA
plates, and incubated at 37°C for 24 h. The bacterial colonies (~1
mm) on agar plates counted visually were used for determing viable
bacteria in samples. Bacterial quantities are displayed as
ln(Cfu/ml) in all treatment groups.
Statistical analysis
The data are presented as mean ± standard deviation
of two or three separate experiments. Significant differences were
established by one-way analysis of variance and post hoc Tukey's
test using GraphPad Prism (version 5; GraphPad Software, Inc., La
Jolla, CA, USA). P<0.05 indicated that the difference between
groups was statistically significant.
Results
Jatrorrhizine or NFX alone exhibits
low antimicrobial activity
Jatrorrhizine was subjected to an antibacterial
assay against MRSA strain SA1199B. The MICs of jatrorrhizine and
NFX were measured as 64 mg/l; the two agents exhibited low
antibacterial activity against MRSA SA1199B (Table II). As the positive control, the
antibiotic ofloxacine substantially inhibited the growth of MRSA
SA1199B with an MIC of 1 mg/l. 2% DMSO (negative control) showed no
inhibition on bacterial growth. The EC50s of
jatrorrhizine and NFX were determined to be 27.1 and 21.6 mg/l,
respectively. The EC50 values were high, supporting the
antimicrobial activities of jatrorrhizine and NFX.
 | Table II.MIC and FICI of agents against
Staphylococcus aureus SA1199B. |
Table II.
MIC and FICI of agents against
Staphylococcus aureus SA1199B.
|
| MIC (mg/l) |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Agent | Alone | Combined with 16
mg/ljatrorrhizine | EC50
(mg/l) | FICI | FICI in combination
with jatrorrhizine |
|---|
| Jatrorrhizine | 64 | – | 27.1 | 0.250 | – |
| Norfloxacin | 64 | 8 | 21.6 | 0.125 | 0.375 |
| Reserpine | 128 | 16 | – | 0.125 | 0.250 |
| Ofloxacine | 1 | – | – | – | – |
Jatrorrhizine exhibits a synergistic
effect with NFX against MRSA
Jatrorrhizine alone exhibited little antibacterial
activity with an MIC of 64 mg/l (Table
II and Fig. 1). The strain
SA1199B was resistant to the antibiotic, NFX (MIC, 64 mg/l;
Table II). When the bacteria were
incubated with a combination of jatrorrhizine and NFX, the MIC of
NFX was markedly reduced compared with NFX alone. The synergistic
effect between jatrorrhizine and NFX against the strain SA1199B was
confirmed with an FICI of 0.375. However, reserpine demonstrated a
better synergistic effect with an FICI of 0.250.
MRSA proliferation is inhibited by
jatrorrhizine with NFX
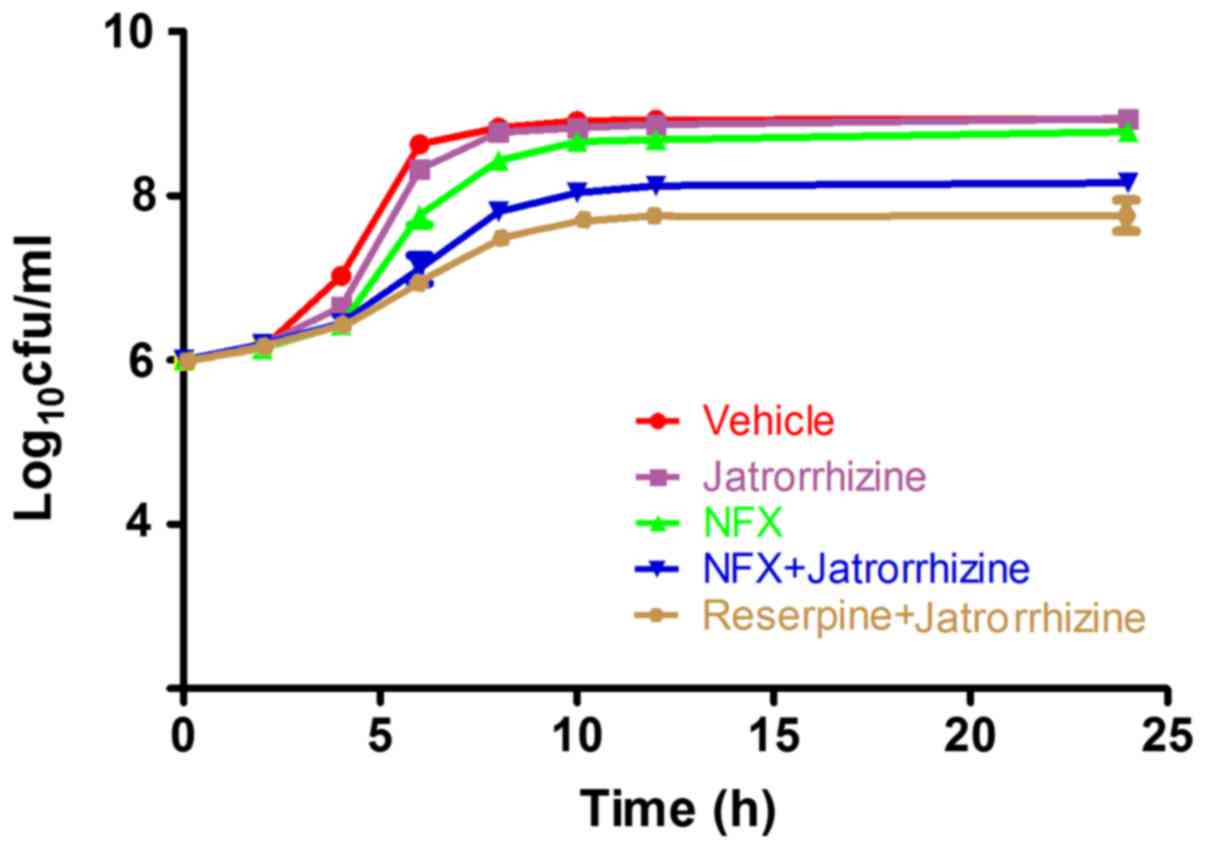
To further confirm the synergistic effect between
jatrorrhizine and NFX, time-growth experiments of the SA1199B
strain without treatment or treated with NFX, jatrorrhizine,
reserpine or a combination of two (Fig.
2). Jatrorrhizine did not significantly inhibit the SA1199B
strain at 16 mg/l (1/4 MIC), as shown in Fig. 2. The SA1199B strain did not markedly
increase with the first 4 h of 1/4 MIC NFX treatment; however,
proliferation markedly increased from 6–10 h. Compared with NFX
alone, the SA1199B strain treated with NFX (1/4 MIC) combined with
jatrorrhizine (1/4 MIC) grew at a slower rate. Jatrorrhizine
markedly enhanced the antibacterial activity of NFX against MRSA,
suggesting the synergistic effect between NFX and jatrorrhizine,
even the effect was weaker compared with reserpine in combination
with jatrorrhizine.
Jatrorrhizine inhibited bacterial EtBr
efflux
Measuring the accumulation of EtBr demonstrated that
jatrorrhizine inhibited the efflux pump of the SA1199B strain
(Fig. 3). Compared with the
vehicle-treated bacteria, in which fluorescence was quickly reduced
to <50%, the reduction in fluorescence was significantly slower
in the jatrorrhizine-treated bacteria (P<0.05) as the
fluorescence level plateaued at ~70%. The positive control CCCP
significantly (P<0.01) inhibited the efflux of EtBr cells with
the fluorescence retention of ~80%. Therefore, jatrorrhizine
inhibited bacterial drug efflux in the SA1199B strain, although its
inhibition was significantly weaker compared with the known efflux
inhibitor, CCCP (P<0.05). Drug efflux is one of the important
bacterial resistance mechanisms. Thus, jatrorrhizine may be a
resistance-modifying agent against bacteria resistance.
Inhibitory effect of jatrorrhizine on
induction of NorA at the transcriptional level
The expression of the NorA mRNA was investigated
(Fig. 4A). The expression of the
NorA mRNA in vehicle-treated bacteria was set as a 1-fold
induction. Jatrorrhizine (16 mg/l, 1/4 MIC) significantly inhibited
the expression of NorA compared with the vehicle (P<0.05;
Fig. 4B). Compared with the vehicle,
the expression of NorA was significantly increased in the SA1199B
strain following the incubation with NFX (16 mg/l, 1/4 MIC;
P<0.001) and a combination of NFX and jatrorrhizine (P<0.01).
However, the level of NorA mRNA expression in bacteria treated with
a combination of NFX and jatrorrhizine was significantly higher
compared with that of NFX-treated bacteria (P<0.01). Therefore,
jatrorrhizine significantly decreased the antibiotic-induced
expression of NorA at the mRNA level, thus assisting in the
treatment of MRSA.
NorA-jatrotthizine complex formed by
interactions
According to docking study, the NorA-jatrotthizine
complex was formed by NorA-jatrotthizine interactions cluding
hydrophobicity, as shown in Fig. 5A.
The functional groups in NorA involved in the jatrorrhizine binding
site were formed by Met263, Asn315, Phe259, Ala312, Tyr316, Leu269,
Thr270, Phe271, Arg380, Lys377, Glu268, Asn319 and Lys384 (Fig. 5B). It was observed that Ala312 and
Thr27 are associated with key interactions and hence serve an
important role in ligand binding. A hydrophobic cleft formed by
Met263, Asn315, Phe259, Tyr316, Leu269, Phe271, Lys377 and Lys384
provided extra stability to the complex. Also, the electrostatic
interaction between the benzene ring and polar groups of the amino
acid residues on Glu268 and Arg380 contributed extra stability to
the NorA-jatrotthizine complex. The docking results suggest that
jatrotthizine binds to NorA with hydrogen bonds, and hydrophobic
and electrostatic interactions, which may be associated with its
inhibitory efflux pump activity.
In vivo synergistic efficacy of
jatrorrhizine and NFX in a neutropenic murine thigh infection
model
To validate the in vivo synergistic
bactericidal activity of jatrorrhizine and NFX against the MRSA
strain, SA1199B, a mouse model of thigh infection was used. The
results revealed that ofloxacine (50 mg/kg), a positive control,
significantly decreased the bacterial burden after 24 h compared
with that of the PBS-treated animals (Fig. 6). No significant difference in the
bacteria count was identified between the groups treated with
jatrorrhizine (50 mg/kg), NFX (100 mg/kg), a combination of
jatrorrhizine NFX (10 and 100 mg/kg, respectively) or PBS. However,
the bacterial count in the groups treated with a combination of
jatrorrhizine (25 or 50 mg/kg) and NFX (100 mg/kg) significantly
decreased in what appears to be a dose-dependent manner (P<0.01
and P<0.001, respectively, suggesting that there is an in
vivo synergistic effect.
Discussion
Natural products, particularly those from plants,
are an important source for drug discovery (29). The efficiency of strategy that
combines antimicrobials with an agent capable of the inhibition of
resistance mechanisms has been developed with the successful
clinical use of β-lactams in combination with β-lactamase
inhibitors, such as clavulanic acid in combination with amoxicillin
(2). Efflux mechanisms are
widespread in bacteria and serve a role in susceptibility and
resistance to antibiotics; developing efflux-pump inhibitors to be
used in combination with existing antimicrobials is appealing and
may facilitate the recycling of existing antibiotics (3).
The MRSA strain SA1199B overexpresses the gene
encoding the NorA multidrug resistant efflux pump, which reduces
susceptibility of the bacteria to quinolones, such as NFX (2). The development of RMAs provides a
potential strategy to alleviate the spread of bacterial antibiotic
resistance (30). Several
protoberberine alkaloids including berberine, jatrorrhizine and
palmatine were previously measured against MRSA strains; only
jatrorrhizine exhibited a synergistic effect with NFX. In the
present study, the antibacterial activity of jatrorrhizine against
MRSA SA1199B was revealed to be an MIC of 64 mg/l, suggesting it is
a weak antibacterial agent. However, the synergy assay revealed
that jatrorrhizine and NFX against MRSA SA1199B had an FICI of
0.375. With an FICI <0.5, jatrorrhizine reduced the MIC of NFX,
which resulted in an 8-fold decrease in the bacterial count of MRSA
SA1199B. In order to confirm the synergistic effect, the growth
kinetics were measured in 24 h, which demonstrated that
jatrorrhizine synergistically inhibited the growth of MRSA with
NFX. As one of the most important antibacterial resistance
mechanisms, bacterial drug efflux prevents antimicrobials accessing
the targets in bacteria. In the current study, a bacterial drug
efflux assay demonstrated that jatrorrhizine significantly
inhibited the EtBr efflux of MRSA SA1199B.
NFX has been demonstrated to inhibit bacterial
activity by blocking nucleic acids synthesis (31). Incubating resistant bacteria with
substrates of the NorA efflux pump, such as NFX, can induce
norA overexpression at the mRNA and protein levels (32). According to the semi quantitative
RT-PCR analysis, the overexpression of NorA induced by NFX was
suppressed by jatrorrhizine at the mRNA level. This result suggests
that jatrorrhizine exhibited a synergistic effect with NFX against
MRSA through the suppression of NorA mRNA expression.
Molecular docking is a widely used technology, which
has served an important role in drug design (33). It is one of the fundamental tools to
elucidate the association between the structure of a ligand and its
biological activities. To further investigate the mechanism of the
synergistic antibacterial activity of jatrorrhizine and NFX, the
three-dimensional structure of NorA was generated and a molecular
docking study of the interactions between NorA and jatrorrhizine
was performed. It was revealed that jatrorrhizine extended into the
hydrophobic cleft formed by Met263, Asn315, Phe259, Tyr316, Leu269,
Phe271, Lys377 and Lys384 in the binding site. H-bonds and
electrostatic interactions between jatrorrhizine and amino acid
residues were formed in the binding site, contributing to a stable
NorA-jatrorrhizine complex. NorA in bacteria confers resistance to
NFX by effluxing NFX from cell. The interactions between NorA and
jatrorrhizine altered the function of the NorA efflux pump,
inhibiting NFX efflux. As a result, jatrorrhizine may be a
potential NorA efflux pump inhibitor.
NorA overexpression reduces the susceptibility of
MRSA to quinolones and the use of RMAs to inhibit NorA mRNA
expression can increase the susceptibility quinolones (2). Jatrorrhizine significantly increased
the antimicrobial effect of NFX against MRSA SA1199B infection in
mice, suggesting that there is an in vivo synergistic effect
between jatrorrhizine and NFX.
In conclusion, jatrorrhizine is a novel RMA against
MRSA. To the best of our knowledge, the current study is the first
to reveal that jatrorrhizine exhibits in vitro and in
vivo synergistic effects with NFX against MRSA. The effects are
mediated by the suppression of NorA mRNA expression and bacterial
drug efflux or the inhibition of bacterial drug efflux through the
binding of NFX and jatrorrhizine to the NorA efflux pump. These
data support the hypothesis that jatrorrhizine is a potential agent
for therapeutic use in infections caused by MRSA.
Acknowledgements
Not applicable.
Funding
The current study was supported by the fund of
scientific research project of Heilongjiang Provincial Education
Department (grant no. 2016-KYYWF-0839).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FD provided the conception and design of the study.
HY analyzed experimental data. YW, XW, JG, HW and HZ performed the
experiments. HY and FD were the major contributors in writing the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Medical Technology College of Qiqihar Medical
University (Qiqihar, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Russell AD: Biocide use and antibiotic
resistance: The relevance of laboratory findings to clinical and
environmental situations. Lancet Infect Dis. 3:794–803. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abreu AC, McBain AJ and Simoes M: Plants
as sources of new antimicrobials and resistance-modifying agents.
Nat Prod Rep. 29:1007–1021. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hooper DC: Efflux pumps and nosocomial
antibiotic resistance: A primer for hospital epidemiologists. Clin
Infect Dis. 40:1811–1817. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oluwatuyi M, Kaatz GW and Gibbons S:
Antibacterial and resistance modifying activity of Rosmarinus
officinalis. Phytochemistry. 65:3249–3254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wright GD: Bacterial resistance to
antibiotics: Enzymatic degradation and modification. Adv Drug Deliv
Rev. 57:1451–1470. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hoffman SB: Mechanisms of antibiotic
resistance. Compendium on Continuing Education for the Practising
Veterinarian-North American Edition. 23:464–472. 2001.
|
|
7
|
Gould IM, Reilly J, Bunyan D and Walker A:
Costs of healthcare-associated methicillin-resistant
Staphylococcus aureus and its control. Clin Microbiol
Infect. 16:1721–1728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Neyfakh AA, Borsch CM and Kaatz GW:
Fluoroquinolone resistance protein NorA of Staphylococcus
aureus is a multidrug efflux transporter. Antimicrob Agents
Chemother. 37:128–129. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Truong-Bolduc QC, Zhang X and Hooper DC:
Characterization of NorR protein, a multifunctional regulator of
norA expression in Staphylococcus aureus. J Bacteriol.
185:3127–3138. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alekshun MN and Levy SB: Molecular
mechanisms of antibacterial multidrug resistance. Cell.
128:1037–1050. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mallea M, Mahamoud A, Chevalier J,
Alibert-Franco S, Brouant P, Barbe J and Pagès JM:
Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump
in multidrug-resistant clinical isolates. Biochem J. 376:801–805.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Phillipson JD: Phytochemistry and
pharmacognosy. Phytochemistry. 68:2960–2972. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gibbons S: Anti-staphylococcal plant
natural products. Nat Prod Rep. 21:263–277. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalia NP, Mahajan P, Mehra R, Nargotra A,
Sharma JP, Koul S and Khan IA: Capsaicin, a novel inhibitor of the
NorA efflux pump, reduces the intracellular invasion of
Staphylococcus aureus. J Antimicrob Chemother. 67:2401–2408.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
He JM and Mu Q: The medicinal uses of the
genus Mahonia in traditional Chinese medicine: An
ethnopharmacological, phytochemical and pharmacological review. J
Ethnopharmacol. 175:668–683. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaatz GW, Seo SM and Ruble CA: Mechanisms
of fluoroquinolone resistance in Staphylococcus aureus. J
Infectious Dis. 163:1080–1086. 1991. View Article : Google Scholar
|
|
17
|
Ross JI, Farrell AM, Eady EA, Cove JH and
Cunliffe WJ: Characterisation and molecular cloning of the novel
macrolide-streptogramin B resistance determinant from
Staphylococcus epidermidis. J Antimicrob Chemother.
24:851–862. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Clinical Laboratory Standards Institute
(CLSI), . Performance Standards for Antimicrobial Susceptibility
Testing; Twentieth Informational Supplement. M100-S2030. CLSI;
Wayne, PA: pp. 54–62. 2010
|
|
19
|
Hemaiswarya S, Kruthiventi AK and Doble M:
Synergism between natural products and antibiotics against
infectious diseases. Phytomedicine. 15:639–652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Abascal K and Yarnell E: Herbs and drug
resistane: Part 2-Clinical implications of research on microbial
resistance to antibiotics. Complementary Therapies. 8:237–241.
2002. View Article : Google Scholar
|
|
21
|
Pearson RD, Steigbigel RT and Davis HT:
Method of reliable determination of minimal lethal antibiotic
concentrations. Antimicrobial Agents Chemotherapy. 18:699–708.
1980. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lechner D, Gibbons S and Bucar F: Plant
phenolic compounds as ethidium bromide efflux inhibitors in
Mycobacterium smegmatis. J Antimicrob Chemother. 62:345–348.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y: I-TASSER server for protein 3D
structure prediction. BMC Bioinformatics. 9:402008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roy A, Kucukural A and Zhang Y: I-TASSER:
A unified platform for automated protein structure and function
prediction. Nat Protoc. 5:725–738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang J, Yan R, Roy A, Dong X, Poisson J
and Yang Z: The I-TASSER Suite: Protein structure and function
prediction. Nat Methods. 12:7–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang J, Liang Y and Zhang Y: Atomic-level
protein structure refinement using fragment guided molecular
dynamics conformation sampling. Structure. 19:1784–1795. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aguinagalde L, Diez-Martinez R, Yuste J,
Royo I, Gil C, Lasa Í, Martín-Fontecha M, Marín-Ramos NI, Ardanuy
C, Liñares J, et al: Auranofin efficacy against MDR Streptococcus
pneumoniae and Staphylococcus aureus infections. J
Antimicrob Chemother. 70:2608–2617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Newman DJ and Cragg GM: Natural products
as sources of new drugs from 1981 to 2014. J Nat Prod. 79:629–661.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sibanda T and Okoh AI: The challenges of
overcoming antibiotic resistance: Plant extracts as potential
sources of antimicrobial and resistance modifying agents. African J
Biotechnol. 6:2886–2896. 2007.
|
|
31
|
Sriram D, Bal TR and Yogeeswari P: Design,
synthesis and biological evaluation of novel non-nucleoside HIV-1
reverse transcriptase inhibitors with broad-spectrum
chemotherapeutic properties. Bioorg Med Chem. 12:5865–5873. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zou D, Xie K, Wang H, Chen Y and Xie M:
Inhibitory effects of biochanin A on the efflux pump of
methicillin-resistant Staphylococcus aureus (MRSA). Wei
Sheng Wu Xue Bao. 54:1204–1211. 2014.(In Chinese). PubMed/NCBI
|
|
33
|
Fu Y, Wu X and Chen Z: A new approach for
flexible molecular docking based on swarm intelligence.
Mathematical Problems Engineering. 2015:1–10. 2015. View Article : Google Scholar
|