Introduction
Cholangiocarcinoma (CCA) is a deadly malignancy of
the biliary tree. Due to the high frequency of diagnosis at a late
stage, metastasis and recurrence, surgical treatment for CCA is
associated with poor outcomes. The 5-year survival rate is 5–10% in
patients with CCA (1). Therefore,
understanding the molecular mechanisms involved in the
tumorigenesis of CCA is a critical step for improving early
diagnosis, reducing mortality, and developing effective targeted
therapies. Long non-coding RNAs (lncRNAs) are autonomously
transcribed non-coding RNAs of >200 nt in length, which have an
important role in the regulation of gene transcription through the
recruitment of chromatin-modifying enzymes (2). Accumulating studies have indicated that
lncRNAs may interact with microRNAs (miRNAs/miRs) as competing
endogenous RNAs (ceRNAs), and regulate the expression of target
genes, which may have a role in tumor occurrence and progression
(3). Therefore, lncRNAs are
considered to be potential diagnostic and prognostic biomarkers of
malignancy (4,5). Based on these facts, Salmena et
al (3) provided the ceRNA
hypothesis and constructed a large-scale ceRNA regulatory network,
which may explain tumor processes and present opportunities for
novel therapies.
In previous studies, regulatory ceRNA networks
composed of lncRNAs, miRNAs and mRNAs have been used to study
molecular mechanisms of tumor occurrence and progression. Numerous
studies have indicated that ceRNA regulatory lncRNA-miRNA-mRNA
networks are implicated in the occurrence and progression of
gastric, breast, pancreatic and liver cancer (6–9).
Recently, lncRNA actin filament associated protein 1
(AFAP1)-antisense (AS)1 was reported to promote the growth and
metastasis of CAA (10). AFAP1-AS1
expression was upregulated in CCA tumor samples and its knockdown
reduced cell stress filament integrity, suggesting that it may be a
diagnostic and prognostic biomarker for CCA. Another study
indicated that the expression of lncRNA colon cancer-associated
transcript 1 (CCAT1) was significantly upregulated in CAA samples,
and promoted cell migration and invasion by suppressing miR-152
(11). Based on the aforementioned
studies, it may be hypothesized that dysregulation of certain
lncRNAs may promote CAA by regulating key pathways. Therefore,
identification of a CAA-associated ceRNA network may be useful for
understanding the role of ceRNAs in the genesis of CAA and
therapeutic outcomes (12). In the
present study, a ceRNA network of CAA was constructed, including 16
lncRNAs, 55 miRNAs and 373 mRNAs. Based on the network, survival
analysis of all genes suggested that fucosyltransferase 4 (FUT4)
and huntingtin-interacting protein 1 related (HIP1R) were
associated with overall survival. These results provided further
insight into the mechanisms of the pathogenesis of CAA, and may
provide potential novel therapeutic markers for CAA treatment.
Materials and methods
Patients and The Cancer Genome Atlas
(TCGA) data retrieval
The microarray data for 45 CCA samples were obtained
from TCGA data portal (https://portal.gdc.cancer.gov/), with search results
up to October 14th, 2018 included. The RNA and miRNA sequencing
(seq) data were obtained from the IlluminaHiseq_RNASeq and the
IlluminaHiSeq_miRNASeq sequencing platforms. All data are open
access and free to download. The sequencing data included the
corresponding RNA-seq and miRNA-seq data. The human samples were
divided into 2 groups: The CCA samples (n=36) and adjacent
non-tumor samples (n=9). All of the protocols were in accordance
with the guidelines of TCGA and no further ethical approval was
required, since all of the data were collected from TCGA
(https://cancergenome.nih.gov/publications/publicationguidelines).
Identification of differentially
expressed (DE) lncRNAs, miRNAs and miRNAs
The raw data of the microarray datasets were
preprocessed via background correction and normalization. Prior to
the analysis, all unexpressed RNAs were filtered out by using R
language (version 3.2.5; http://cran.r-project.org/). According to the R
language results, those genes with mean read count ≤1 were deleted.
The lncRNAs and mRNAs were identified using the Ensembl database
(version 89; http://www.ensembl.org/index.html). Subsequently, the
DE mRNAs, lncRNAs and miRNAs of the two groups were obtained using
edgeR software (13). The false
discovery rate (FDR) was used for multiple comparisons of
statistically significant P-values. The fold change (FC) was used
for measuring the differential expression levels of genes, where
|log2FC|≥2 and FDR adjusted to P<0.01 were considered to
indicate a significant DE mRNA or lncRNA; the standard for miRNAs
was a fold change ≥2.5 and an FDR adjusted to P<0.01. Finally,
all of the DE RNAs were analyzed and a volcano map was generated
using the R platform.
Generation of the ceRNA regulatory
network of CCA
To investigate the association between ceRNAs in
patients with CAA, the lncRNA-miRNA-mRNA regulatory network was
constructed, which was established through the following procedures
based on the results of the DE analysis. First, the regulatory
interactions between lncRNAs and miRNAs were predicted using the
miRcode database (14).
Subsequently, miRNA-targeted mRNAs were extracted from the
miRTarBase (15), miRDB (16) and TargetScan (17) databases. Finally, using Cytoscape
3.5.1 (http://www.cytoscape.org/), the ceRNA
regulatory network was generated and visualized.
Functional enrichment analysis
In order to better understand the mechanisms of CCA
tumorigenesis, Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analyses of DE mRNAs were performed
using the Database for Annotation, Visualization and Integrated
Discovery (DAVID; http://www.david.abcc.ncifcrf.gov/). A GO term or KEGG
pathway with FDR <0.05 was considered statistically significant.
The enriched GO terms and pathways of the DE mRNAs with the most
significant P-values were ranked by their enrichment score (-log
P-value).
Survival analysis
The R survival package (version 2.41–3; http://CRAN.R-project.org/package=survival) was used
for the survival analysis. The Kaplan-Meier method was used to
estimate cumulative survival rates and the log-rank test was then
used to compare the differences in overall survival between the
different groups. P<0.05 was considered to indicate statistical
significance.
Results
DE lncRNAs, mRNAs and miRNAs in
CCA
The present study investigated the differential RNA
expression in 36 CCA tissues and 9 adjacent non-tumor tissues. The
integrated analysis identified 318 DE lncRNAs, 3,851 DE mRNAs and
87 DE miRNAs using the edgeR package. Of the 318 DE lncRNAs, 205
were upregulated (64.5%) and 113 (35.5%) were downregulated. A
total of 1,549 (40.2%) mRNAs were identified to be upregulated and
2,302 (59.8%) were downregulated out of the 3,851 DE mRNAs. For the
87 DE miRNAs, 41 (47.1%) were upregulated and the remaining 46
(52.9%) were downregulated. The top 20 upregulated and
downregulated lncRNAs, mRNAs and miRNAs are listed in Tables I–III, respectively. In addition, the
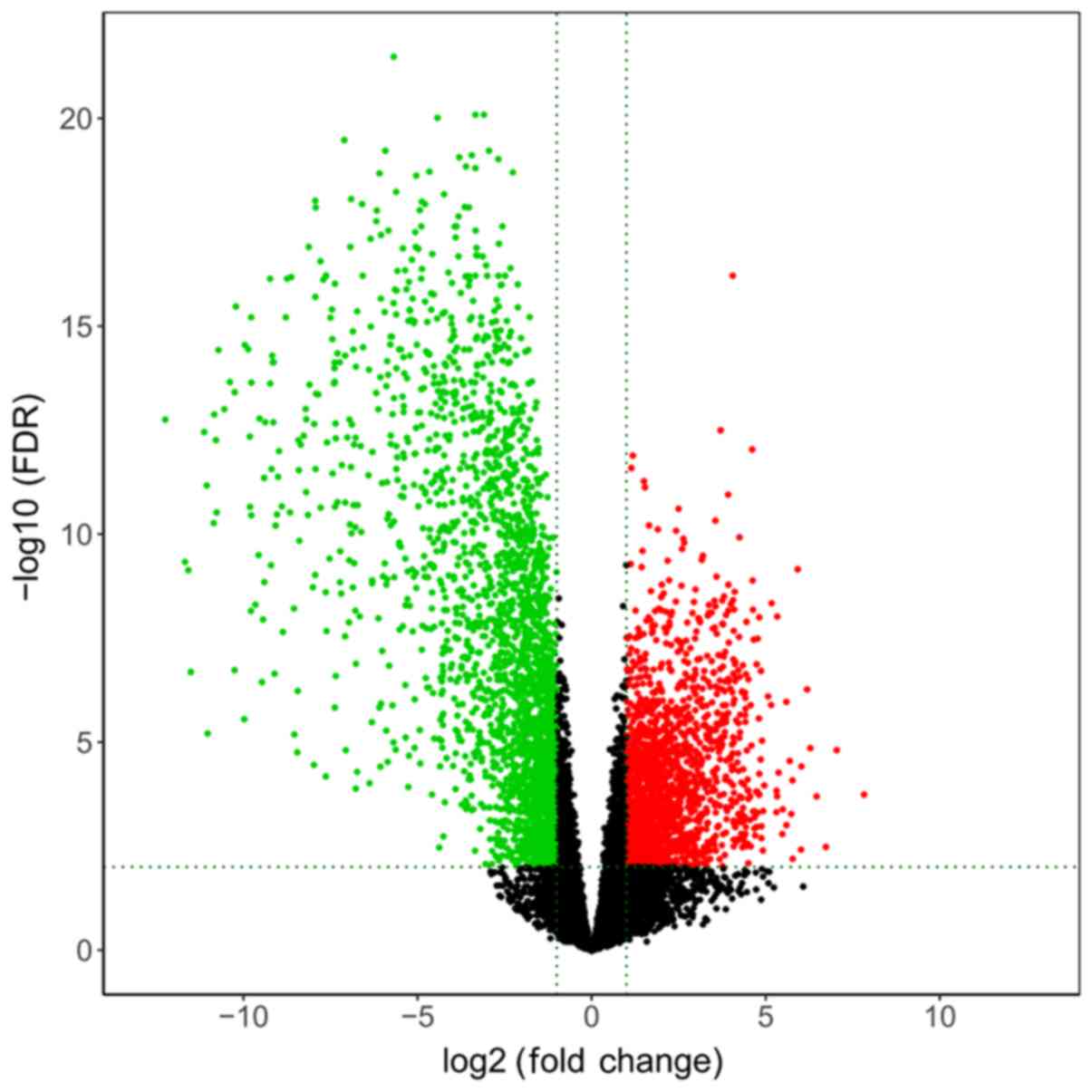
distributions of all DE mRNAs are presented in a volcano plot in
Fig. 1.
 | Table I.The top 20 up- and downregulated
lncRNAs (ranked by P-values). |
Table I.
The top 20 up- and downregulated
lncRNAs (ranked by P-values).
| A, Top 20
upregulated lncRNAs |
|---|
|
|---|
| Ensembl ID | logFC | P-value | FDR |
|---|
|
ENSG00000261659 |
3.198316385 |
1.55×10−11 |
3.30×10−10 |
|
ENSG00000253210 |
2.985738665 |
1.21×10−10 |
2.13×10−09 |
|
ENSG00000261183 |
4.001322806 |
2.92×10−10 |
4.81×10−09 |
|
ENSG00000228109 |
3.369141828 |
3.07×10−10 |
5.02×10−09 |
|
ENSG00000265688 |
3.446807939 |
4.91×10−10 |
7.64×10−09 |
|
ENSG00000261068 |
4.766573772 |
2.45×10−09 |
3.24×10−08 |
|
ENSG00000172965 |
2.575378258 |
3.51×10−09 |
4.50×10−08 |
|
ENSG00000273230 |
2.560329476 |
4.77×10−09 |
5.97×10−08 |
|
ENSG00000234741 |
2.018672373 |
1.11×10−08 |
1.30×10−07 |
|
ENSG00000261801 |
3.691796251 |
1.53×10−08 |
1.74×10−07 |
|
ENSG00000285255 |
4.217290775 |
1.65×10−08 |
1.86×10−07 |
|
ENSG00000226711 |
2.650291782 |
3.45×10−08 |
3.56×10−07 |
|
ENSG00000261437 |
4.32349391 |
6.22×10−08 |
6.02×10−07 |
|
ENSG00000277283 |
1.914981951 |
1.10×10−07 |
1.01×10−06 |
|
ENSG00000243479 |
5.595465468 |
1.18×10−07 |
1.08×10−06 |
|
ENSG00000244041 |
1.90448619 |
1.32×10−07 |
1.20×10−06 |
|
ENSG00000269680 |
3.185294307 |
1.55×10−07 |
1.39×10−06 |
|
ENSG00000273759 |
2.435273695 |
1.66×10−07 |
1.47×10−06 |
|
ENSG00000257556 |
2.979912742 |
1.96×10−07 |
1.71×10−06 |
|
ENSG00000255381 |
1.597506825 |
2.13×10−07 |
1.85×10−06 |
|
| B, Top 20
downregulated lncRNAs |
|
| Ensembl
ID | logFC | P-value | FDR |
|
|
ENSG00000225756 |
−4.439300193 |
5.12×10−18 |
6.55×10−16 |
|
ENSG00000251165 |
−5.214981183 |
6.12×10−17 |
5.09×10−15 |
|
ENSG00000261572 |
−3.875298917 |
1.23×10−15 |
6.95×10−14 |
|
ENSG00000263400 |
−4.480906234 |
1.29×10−15 |
7.23×10−14 |
|
ENSG00000215386 |
−3.687757377 |
1.76×10−15 |
9.49×10−14 |
|
ENSG00000264575 |
−2.497423285 |
2.22×10−15 |
1.17×10−13 |
|
ENSG00000267390 |
−3.142413979 |
2.31×10−15 |
1.20×10−13 |
|
ENSG00000234456 |
−3.123344646 |
5.80×10−15 |
2.68×10−13 |
|
ENSG00000261012 |
−7.173215127 |
6.10×10−14 |
2.19×10−12 |
|
ENSG00000235609 |
−2.872714774 |
1.54×10−13 |
4.97×10−12 |
|
ENSG00000261578 |
−4.233216461 |
1.78×10−13 |
5.66×10−12 |
|
ENSG00000228794 |
−1.905394222 |
8.41×10−13 |
2.31×10−11 |
|
ENSG00000259370 |
−4.369092089 |
3.65×10−12 |
8.74×10−11 |
|
ENSG00000215256 |
−2.110042475 |
3.88×10−12 |
9.20×10−11 |
|
ENSG00000223797 |
−2.072567868 |
5.02×10−12 |
1.17×10−10 |
|
ENSG00000275494 |
−2.27756684 |
1.10×10−11 |
2.41×10−10 |
|
ENSG00000269386 |
−2.361574508 |
2.36×10−11 |
4.81×10−10 |
|
ENSG00000273616 |
−2.942297335 |
3.18×10−11 |
6.25×10−10 |
|
ENSG00000267675 |
−5.990122102 |
5.41×10−11 |
1.03×10−09 |
|
ENSG00000260274 |
−1.989229751 |
5.54×10−11 |
1.05×10−09 |
 | Table III.The top 20 up- and downregulated
miRNAs (ranked by P-values). |
Table III.
The top 20 up- and downregulated
miRNAs (ranked by P-values).
| A, Top 20
upregulated miRNAs |
|---|
|
|---|
| miRNA | logFC | P-value | FDR |
|---|
| hsa-miR-183-5p |
4.836903706 |
6.38×10−17 |
1.40×10−14 |
| hsa-miR-182-5p |
4.483302702 |
6.43×10−17 |
1.40×10−14 |
| hsa-miR-21-5p |
2.485307292 |
6.28×10−14 |
6.52×10−12 |
| hsa-miR-96-5p |
5.172417117 |
1.81×10−13 |
1.31×10−11 |
| hsa-miR-27a-3p |
1.975273987 |
3.22×10−13 |
1.99×10−11 |
| hsa-miR-34c-3p |
3.911770715 |
2.54×10−10 |
4.76×10−09 |
| hsa-miR-222-3p |
2.267762864 |
3.08×10−10 |
5.34×10−09 |
| hsa-miR-23a-3p |
1.494080752 |
4.43×10−10 |
7.12×10−09 |
| hsa-miR-34c-5p |
4.181782951 |
1.10×10−08 |
1.45×10−07 |
| hsa-miR-92b-3p |
2.769232681 |
1.75×10−08 |
2.23×10−07 |
| hsa-miR-330-5p |
1.866060112 |
2.30×10−08 |
2.78×10−07 |
| hsa-miR-454-3p |
1.608154597 |
2.67×10−08 |
3.13×10−07 |
|
hsa-miR-181d-5p |
2.43995135 |
3.65×10−08 |
4.17×10−07 |
| hsa-let-7e-5p |
1.391443094 |
4.99×10−08 |
5.42×10−07 |
| hsa-miR-221-3p |
1.752600485 |
8.05×10−08 |
8.27×10−07 |
|
hsa-miR-181b-5p |
1.952571231 |
9.66×10−08 |
9.26×10−07 |
|
hsa-miR-200b-3p |
2.634730289 |
9.82×10−08 |
9.26×10−07 |
|
hsa-miR-24-2-5p |
1.597889448 |
1.14×10−07 |
1.05×10−06 |
|
hsa-miR-301a-3p |
1.537288068 |
2.14×10−07 |
1.94×10−06 |
| hsa-miR-99b-3p |
1.648708541 |
2.45×10−07 |
2.13×10−06 |
|
| B, Top 20
downregulated miRNAs |
|
| miRNA | logFC | P-value | FDR |
|
|
hsa-miR-148a-3p |
−2.8329 |
5.07×10−15 |
7.33×10−13 |
|
hsa-miR-4662a-5p |
−3.27785 |
7.51×10−14 |
6.52×10−12 |
| hsa-miR-101-3p |
−2.08208 |
4.85×10−13 |
2.63×10−11 |
| hsa-miR-505-3p |
−2.01987 |
2.01×10−12 |
9.68×10−11 |
|
hsa-miR-378a-3p |
−3.24209 |
3.83×10−12 |
1.66×10−10 |
|
hsa-miR-148a-5p |
−2.53305 |
7.56×10−12 |
2.98×10−10 |
|
hsa-miR-378a-5p |
−2.94829 |
1.74×10−11 |
6.30×10−10 |
|
hsa-miR-125b-2-3p |
−2.4123 |
2.24×10−11 |
7.48×10−10 |
| hsa-miR-483-3p |
−4.78428 |
2.72×10−11 |
8.43×10−10 |
| hsa-miR-122-3p |
−7.01304 |
3.26×10−11 |
9.43×10−10 |
| hsa-miR-194-3p |
−3.2353 |
3.93×10−11 |
1.07×10−09 |
| hsa-miR-139-3p |
−2.76339 |
5.01×10−11 |
1.28×10−09 |
| hsa-let-7c-5p |
−2.24221 |
6.46×10−11 |
1.56×10−09 |
| hsa-miR-99a-3p |
−2.34474 |
8.98×10−11 |
1.97×10−09 |
| hsa-miR-675-3p |
−5.17096 |
9.09×10−11 |
1.97×10−09 |
| hsa-miR-139-5p |
−3.13233 |
1.08×10−10 |
2.24×10−09 |
| hsa-miR-378c |
−2.82711 |
2.22×10−10 |
4.38×10−09 |
| hsa-miR-885-5p |
−6.05348 |
2.63×10−10 |
4.76×10−09 |
| hsa-miR-99a-5p |
−2.41861 |
3.84×10−10 |
6.42×10−09 |
| hsa-miR-483-5p |
−4.28222 |
4.93×10−10 |
7.65×10−09 |
Construction of a ceRNA regulatory
network in CCA
To better understand the characteristics of the
lncRNAs and to further clarify the interactions between the lncRNAs
and miRNAs, an lncRNA-miRNA-mRNA-associated regulatory network was
constructed. First, the 318 DE lncRNAs derived from the miRcode
database were used; these were applied to the Perl program, which
identified 56 pairs of interacting lncRNAs and miRNAs. From the 87
DE miRNAs retrieved, it was predicted that 55 of them were able to
interact with 16 DE lncRNAs. mRNAs targeted by these 55 DE miRNAs
were selected in all three databases (miRTarBase, miRDB and
TargetScan). The targeted mRNAs with DE miRNAs retrieved from the
miRcode database were cross-checked. The 373 DE targeted mRNAs were
then selected. Finally, the ceRNA regulatory network for CCA was
created by incorporating 16 DE lncRNAs, 55 DE miRNAs and 373 DE
mRNAs (Fig. 2).
Functional enrichment analysis
GO analysis revealed that there were 427 enriched GO
terms with statistical significance in the category biological
process (BP), 44 in the category cellular component (CC) and 105 in
the category molecular function (MF). The top enriched terms for
BP, CC and MF are small molecule catabolic process, mitochondrial
matrix and coenzyme binding, respectively (Fig. 3). KEGG analysis revealed 49 pathways
associated with mRNAs, including ‘cell metabolism’, ‘proliferation’
and ‘sustained angiogenesis’. The top 15 pathways are provided in
Fig. 4 and most of them are
associated with metabolism.
Survival analysis and the ceRNA
network
To identify the prognostic characteristics of the
ceRNA network, each gene of the 16 lncRNAs, 55 miRNAs and 373 mRNAs
were analyzed using Kaplan-Meier survival analysis separately. The
results revealed that only two DE mRNAs in the network (FUT4 and
HIP1R) were associated with overall survival. The Kaplan-Meier
survival analysis indicated that high levels of FUT4 (P<0.005)
and HIP1R (P<0.001) were positively associated with overall
survival (Fig. 5).
Discussion
CCA is the second most common type of primary liver
malignancy. Although surgical resection remains the only potential
treatment, it is frequently unfeasible due to the advanced tumor
stage at the initial diagnosis, with a 5-year survival rate of
<5–10% (18). Diagnostic and
therapeutic biomarkers are urgently required for patients with CCA.
In recent years, accumulating evidence has indicated that lncRNAs
have a key role in cancer. It has been suggested that lncRNAs may
serve as sponges for miRNA, reducing their regulatory effect on
mRNAs (19). This function
introduces an extra layer of complexity in the miRNA-target
interaction network, the dysregulation of which may contribute to
the development and progression of multiple diseases (19). Although recent studies have reported
that certain lncRNAs, including AFAP1-AS1, CCAT1, nuclear
paraspeckle assembly transcript 1 and metastasis associated lung
adenocarcinoma transcript 1, may be associated with CCA, it remains
necessary to perform a more comprehensive analysis of the ceRNA
regulatory networks in CCA (20).
The present study systematically analyzed and
constructed a ceRNA regulatory network containing 16 DE lncRNAs, 55
DE miRNAs and 373 DE mRNAs, in an attempt to better understand the
pathogenesis of CCA. In this network, the lncRNAs primarily bound
to Homo sapiens (hsa)-miR-16-5p, hsa-miR-424-5p,
hsa-miR-130a-3p, hsa-miR-130b-3p and hsa-miR-454-3p. In addition,
hsa-miR-16-5p and hsa-miR-424-5p were the largest nodes that
interacted with 82 non-coding and coding RNAs in the current
network, suggesting that these two miRNAs may be critical to the
pathogenesis of CCA. The enriched GO terms of the DE mRNAs were
strongly linked to metabolism and included ‘small molecule
catabolic process’, ‘organic acid catabolic process’ and
‘carboxylic acid catabolic process’. Pathway analysis revealed that
‘carbon metabolism’, ‘peroxisome proliferator-activated receptor
(PPAR) signaling pathway’, ‘bile secretion’ and ‘fat digestion’
were significantly enriched, and these have been reported to exert
a pivotal influence on CCA (21,22).
Of the 16 DE lncRNAs identified in the ceRNA
network, the complementary mRNAs mainly bound to H19, F-box and
leucine rich repeat protein 19-AS1, HLA complex group 18, PVT1 and
small nucleolar RNA host gene 1. H19 and PVT1 are the most widely
studied lncRNAs among the 16 DE lncRNAs. A growing number of
studies have highlighted the fact that the oncofetal lncRNA gene
H19 is a critical factor in embryonic development, fibrosis and
tumorigenesis (23,24). Accumulating evidence has revealed
that the tumor suppressor protein and cell cycle regulator p53
negatively regulates H19 in tumor cells. Yang et al
(25) reported that H19 was
associated with p53 inactivation, which may contribute to
suppression of apoptosis and increased cell proliferation in
gastric cancer. Furthermore, it was reported that H19-derived
miR-675 has an important role in inhibiting p53 and p53-dependent
protein expression in bladder cancer cells in vivo (26). H19 also has crucial roles in tumor
metastasis through the regulation of epithelial to mesenchymal
transition (EMT), by functioning as an miRNA sponge in colorectal
cancer (27,28). A study has indicated that oxidative
stress caused by infection is linked to inflammation and the
occurrence of CAA (29). In
addition, H19 is considered to be involved in the progression of
CCA by causing partial inactivation of interleukin-6 and downstream
inflammatory responses triggered by oxidative stress (30). Similar to H19, overexpression of PVT1
has also been indicated to be associated with poor prognosis in a
variety of human malignancies (31).
In non-small cell lung cancer, PVT1 contributes to lung
adenocarcinoma cell proliferation, and knockdown markedly reduces
cell proliferation and induces apoptosis in vitro and in
vivo (32). This phenomenon may
be partly mediated through enhancer of zeste 2 polycomb repressive
complex 2 subunit-associated suppression of the large tumor
suppressor kinase 2/MDM2/p53 pathway. PVT1 has been further
observed to be involved in the regulation of EMT, which is a vital
step in the progression, invasion and metastasis of esophageal
cancer (33). Furthermore, PVT1 was
reported to promote liver fibrosis by downregulating patched 1
expression via binding to miR-152 and contributing to EMT (34). Huang et al (35) reported that increased expression of
PVT1 is associated with tumor progression, and suggested that PVT1
may be an independent prognostic factor for poor prognosis in
pancreatic cancer patients. Furthermore, a meta-analysis reported
that increased PVT1 expression was significantly associated with
factors of poor prognosis, including positive lymph node
metastasis, positive distant metastasis, advanced
tumor-nodes-metastasis stage and poor degree of differentiation,
suggesting that PVT1 may be a potential biomarker in various cancer
types (36). However, studies on the
association between H19 or PVT1 and CCA have rarely been
performed.
PPARs belong to the ligand-inducible nuclear hormone
receptor superfamily (37). By
forming heterodimers with retinoid X receptor, PPARs modulate the
expression of lipid metabolism-, adipogenesis-, inflammation- and
anti-cancer-associated genes in various human cancer types
(37). In the present study, pathway
analysis revealed that the PPAR signaling pathway was significantly
enriched and exerted a pivotal influence in patients with CCA. Of
all PPAR isoforms, PPARγ is considered to be most associated with
tumors through the activation of different pathways. PPAR ligands
have been reported to promote differentiation and apoptosis in
numerous malignancies, including CCA, breast cancer and ovarian
cancer (38–40). Han et al (41) reported that ligands of PPARγ inhibit
the proliferation of CCA cells through p53-dependent mechanisms.
Furthermore, the PPARγ ligand 15-deoxy-δ-12,14-PGJ2 may induce
apoptosis in CCA cell lines, although apoptosis-associated protein
expression varies between different cell lines (41). In addition, accumulating evidence
suggests that the mechanisms of the EMT promote the occurrence of
CCA (42). PPARγ is able to
upregulate the expression of Sprouty 4 through Wnt7A/Fzs9
signaling, and suppresses EMT (42).
On the contrary, ectopic PPARγ expression in CCA cell lines may
promote cell proliferation via the Smad pathway, which is a vital
step in EMT (43,44). Suzuki et al (45) reported on the administration of the
PPARγ agonist pioglitazone in a 73-year-old male patient diagnosed
with diabetes, CCA and CCA-induced cholangiohepatitis. In this
case, not only the diabetes was controlled, but also the
cholangiohepatitis, which was thought to be linked to CCA
progression. Furthermore, Asukai et at (46) reported that pioglitazone had a
synergistic effect with gemcitabine and alleviated gemcitabine
resistance in CCA gemcitabine-resistant cells by reducing
miR-130a-3p expression in vitro. However, further studies of
how PPARs are involved in CCA pathogenesis will be required.
The prognostic value of the ceRNA network was also
analyzed. The results revealed that high expression of FUT4
(P<0.005) and HIP1R (P<0.001) was positively correlated with
overall survival. However, according to previous studies, increased
FUT4 expression was observed to be associated with poor prognosis
in breast cancer, lung adenocarcinoma and colorectal cancer, which
was contrary to the present results (47–49).
Another study suggested that lower HIP1R protein expression is
associated with a poor overall survival rate and progression-free
survival in patients with diffuse large B-cell lymphoma (50). However, the opposite appears to be
the case for prostate cancer: HIP1R was reported to increase the
migratory and invasive properties of prostate cancer cells, and
this may be suppressed by the miRNA-23b/27b cluster (51). To the best of our knowledge, the
roles of FUT4 or HIP1R in CCA have not been assessed by any
previous study.
The present study had several limitations. In the
present study, only the data from TCGA database were considered to
create the network. Furthermore, the network was not validated by
any other database or in vitro analysis. Therefore, further
confirmation of key RNAs and investigation of the possible
molecular biological mechanisms of CCA in human CCA tissues will be
performed in the future. Furthermore, the ceRNA network established
in the present study requires broad validation by future
studies.
In conclusion, in the present study, an integrated
analysis of DE lncRNAs, miRNAs and mRNAs in CCA as performed and a
ceRNA network was constructed. This ceRNA network may provide novel
insight for further mechanistic investigation and may lead to
improvements in the survival and prognosis of patients with CAA.
Further experimental verification is required.
Acknowledgements
The authors would like to thank Professor Qulian
Guo, Department of Anesthesia, Xiangya Hospital, Central South
University, Changsha, Hunan, for his encouragement and
guidance.
Funding
This study was supported by the National Science
Foundation for Young Scientists of China (grant no. 81700275).
Availability of data and materials
All data were collected from The Cancer Genome Atlas
(TCGA, http://portal.gdc.cancer.gov/)
Authors' contributions
WG concieved and designed the study. FX designed the
experiments and wrote the first draft of the manuscript. YZ
participated in data analysis, intepreted the data and revised the
manuscript. GQ collected and analyzed the data. YH and LL prepared
all figures and tables; WG and LL revised the manuscript and
approved the final version. All authors reviewed the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cai JQ, Cai SW, Cong WM and Chen MS:
Diagnosis and treatment of cholangiocarcinoma: A consensus from
surgical specialists of China. J Huazhong Univ Sci Technolog Med
Sci. 34:469–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi P: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou X, Liu J and Wang W: Construction and
investigation of breast-cancer-specific ceRNA network based on the
mRNA and miRNA expression data. Let Syst Biol. 8:96–103. 2014.
|
|
7
|
Arun K, Arunkumar G, Bennet D,
Chandramohan SM, Murugan AK and Munirajan AK: Comprehensive
analysis of aberrantly expressed lncRNAs and construction of ceRNA
network in gastric cancer. Oncotarget. 9:18386–18399. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
9
|
Peng H, Lu M and Selaru FM: The
genome-wide gene expression profiling to predict competitive
endogenous RNA network in hepatocellular cancer. Genomics Data.
4:93–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi X, Zhang H, Wang M, Xu X, Zhao Y, He
R, Zhang M, Zhou M, Li X, Peng F, et al: LncRNA AFAP1-AS1 promotes
growth and metastasis of cholangiocarcinoma cells. Oncotarget.
8:58394–58404. 2017.PubMed/NCBI
|
|
11
|
Zhang S, Xiao J, Chai Y, Du YY, Liu Z,
Huang K, Zhou X and Zhou W: LncRNA-CCAT1 promotes migration,
invasion, and EMT in intrahepatic cholangiocarcinoma through
suppressing miR-152. Dig Dis Sci. 62:3050–3058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chan WL, Huang HD and Chang JG: lncRNAMap:
A map of putative regulatory functions in the long non-coding
transcriptome. Comput Biol Chem. 50:41–49. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ashwini J, Marks DS and Erik L: miRcode: A
map of putative microRNA target sites in the long non-coding
transcriptome. Bioinformatics. 28:2062–2063. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: miRTarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
18
|
Li YY, Li H, Lv P, Liu G, Li XR, Tian BN
and Chen DJ: Prognostic value of cirrhosis for intrahepatic
cholangiocarcinoma after surgical treatment. J Gastrointest Surg.
15:608–613. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Batista PJ and Chang HY: Long Noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng B, Jeong S, Zhu Y, Chen L and Xia Q:
miRNA and lncRNA as biomarkers in cholangiocarcinoma(CCA).
Oncotarget. 8:100819–100830. 2017.PubMed/NCBI
|
|
21
|
Huang QX, Cui JY, Ma H, Jia XM, Huang FL
and Jiang LX: Screening of potential biomarkers for
cholangiocarcinoma by integrated analysis of microarray data sets.
Cancer Gene Ther. 23:48–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang W, Li Y, Song X, Xu J and Xie J:
Genome-wide analysis of long noncoding RNA and mRNA co-expression
profile in intrahepatic cholangiocarcinoma tissue by RNA
sequencing. Oncotarget. 8:26591–26599. 2017.PubMed/NCBI
|
|
23
|
Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li
TL, Cai JQ, Zhou HH and Zhu YS: H19 lncRNA mediates
17β-estradiol-induced cell proliferation in MCF-7 breast cancer
cells. Oncol Rep. 33:3045–3052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li L, Chen W, Wang Y, Tang L and Han M:
Long non-coding RNA H19 regulates viability and metastasis, and is
upregulated in retinoblastoma. Oncol Lett. 15:8424–8432.
2018.PubMed/NCBI
|
|
25
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu C, Chen Z, Fang J, Xu A, Wei Z and
Wang Z: H19-derived miR-675 contributes to bladder cancer cell
proliferation by regulating p53 activation. Tumor Biol. 5:263–270.
2016. View Article : Google Scholar
|
|
27
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Y, Wang Z, Jiang X and Cui Y:
Overexpression of long noncoding RNA H19 indicates a poor prognosis
for cholangiocarcinoma and promotes cell migration and invasion by
affecting epithelial-mesenchymal transition. Biomed Pharmacother.
92:17–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boonyanugomol W, Chomvarin C, Sripa B,
Bhudhisawasdi V, Khuntikeo N, Hahnvajanawong C and Chamsuwan A:
Helicobacter pylori in Thai patients with cholangiocarcinoma
and its association with biliary inflammation and proliferation.
HPB (Oxford). 14:177–184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou DD, Liu XF, Lu CW, Pant OP and Liu
XD: Long non-coding RNA PVT1: Emerging biomarker in digestive
system cancer. Cell Prolif. 50:e123982017. View Article : Google Scholar
|
|
32
|
Wan L, Sun M, Liu GJ, Wei CC, Zhang EB,
Kong R, Xu TP, Huang MD and Wang ZX: Long non-coding RNA PVT1
promotes non-small cell lung cancer cell proliferation through
epigenetically regulating LATS2 expression. Mol Cancer Ther.
15:1082–1094. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shuang Y, Ning Q, Zhang G, Hong S, Zhen W
and Li Y: Construction of differential mRNA-lncRNA crosstalk
networks based on ceRNA hypothesis uncover key roles of lncRNAs
implicated in esophageal squamous cell carcinoma. Oncotarget.
7:85728–85740. 2016.PubMed/NCBI
|
|
34
|
Zheng J, Yu F, Dong P, Wu L, Zhang Y, Hu Y
and Zheng L: Long non-coding RNA PVT1 activates hepatic stellate
cells through competitively binding microRNA-152. Oncotarget.
7:62886–62897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Huang C, Yu W, Wang Q, Cui H, Wang Y,
Zhang L, Han F and Huang T: Increased expression of the lncRNA PVT1
is associated with poor prognosis in pancreatic cancer patients.
Minerva Med. 106:143–149. 2015.PubMed/NCBI
|
|
36
|
Lu D, Luo P, Wang Q, Ye Y and Wang B:
lncRNA PVT1 in cancer: A review and meta-analysis. Clin Chim Acta.
474:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Semple RK, Chatterjee VK and O'Rahilly S:
PPAR gamma and human metabolic disease. J Clin Invest. 116:581–589.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Colincassin C, Yao X, Cerella C, et al:
PPAR, breast cancer. Molecular Carcinogenesis. 54:393–404.
2013.PubMed/NCBI
|
|
39
|
Morgado M and Carson DD: PPARγ Modulation
of cytokine-stimulated MUC16 (CA125) expression in breast and
ovarian cancer-derived cells. J Cell Biochem. 118:163–171. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu D, Davis BB, Wang ZH, Zhao SP, Wasti B,
Liu ZL, Li N, Morisseau C, Chiamvimonvat N and Hammock BD: A potent
soluble epoxide hydrolase inhibitor, t-AUCB, acts through PPARγ to
modulate the function of endothelial progenitor cells from patients
with acute myocardial infarction. Int J Cardiol. 167:1298–1304.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han C, Demetris AJ, Michalopoulos GK, Zhan
Q, Shelhamer JH and Wu T: PPARgamma ligands inhibit
cholangiocarcinoma cell growth through p53-dependent GADD45 and p21
pathway. Hepatology. 38:167–177. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tennis MA, Van Scoyk MM, Freeman SV,
Vandervest KM, Nemenoff RA and Winn RA: Sprouty-4 inhibits
transformed cell growth, migration and invasion, and
epithelial-mesenchymal transition, and Is Regulated by Wnt7A
through PPARgamma in non-small cell lung cancer. Mol Cancer Res.
8:833–843. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Han C, Demetris AJ, Liu Y, Shelhamer JH
and Wu T: Transforming growth factor-β (TGF-β) activates cytosolic
phospholipase A2α (cPLA2α)-mediated prostaglandin E2 (PGE)2/EP1 and
peroxisome proliferator-activated receptor-γ (PPAR-γ)/Smad
signaling pathways in human liver cancer cells. JBC. Aug
4–2004.(Epub ahead of print). doi: 10.1074/jbc.M404852200.
View Article : Google Scholar
|
|
44
|
Reka AK, Kurapati H, Narala VR, Bommer G,
Chen J, Standiford TJ and Keshamouni VG: Peroxisome
proliferator-activated receptor-gamma activation inhibits tumor
metastasis by antagonizing Smad3-mediated epithelial-mesenchymal
transition. Mol Cancer Ther. 9:3221–3232. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Suzuki S, Mori J, Yamazaki M, Sato A,
Hosoda W and Hashizume K: Beneficial effects of pioglitazone on
cholangiohepatitis induced by bile duct carcinoma. Internal Med.
46:1723–1727. 2007. View Article : Google Scholar
|
|
46
|
Asukai K, Kawamoto K, Eguchi H, Konno M,
Asai A, Iwagami Y, Yamada D, Asaoka T, Noda T, Wada H, et al:
Micro-RNA-130a-3p Regulates Gemcitabine resistance via PPARG in
cholangiocarcinoma. Ann Surg Oncol. 24:2344–2352. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nagaraj NS and Singh OV: Integrating
genomics and proteomics-oriented biomarkers to comprehend lung
cancer. Expert Opin Med Diagn. 3:167–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang A, Lu C, Ning Z, Gao W, Xie Y, Zhang
N, Liang J, Abbasi FS, Yan Q and Liu J: Tumor-associated
macrophages promote Ezrin phosphorylation-mediated
epithelial-mesenchymal transition in lung adenocarcinoma through
FUT4/LeY up-regulation. Oncotarget. 8:28247–28259. 2017.PubMed/NCBI
|
|
49
|
Li Y, Sun Z, Liu B, Shan Y, Zhao L and Jia
L: Tumor-suppressive miR-26a and miR-26b inhibit cell
aggressiveness by regulating FUT4 in colorectal cancer. Cell Death
Dis. 8:e28922017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wong KK, Gascoyne DM, Brown PJ, Soilleux
EJ, Snell C, Chen H, Lyne L, Lawrie CH, Gascoyne RD, Pedersen LM,
et al: Reciprocal expression of the endocytic protein HIP1R and its
repressor FOXP1 predicts outcome in R-CHOP-treated diffuse large
B-cell lymphoma patients. Leukemia. 28:362–372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rice MA, Ishteiwy RA, Magani F, Udayakumar
T, Reiner T, Yates TJ, Miller P, Perez-Stable C, Rai P, Verdun R,
et al: The microRNA-23b/-27b cluster suppresses prostate cancer
metastasis via Huntingtin-interacting protein 1-related. Oncogene.
35:4752–4761. 2016. View Article : Google Scholar : PubMed/NCBI
|