Introduction
The increase in antimicrobial abuse, bone marrow and
organ transplantation, invasive treatments, and incidences of HIV
infections and cancer in the past decades has resulted in an
increase in the incidence of mycoses due to emerging filamentous
fungi (1). Most filamentous fungi
grow slowly, therefore it often takes several days for the
laboratory diagnosis of a fungal infection (2–4).
Furthermore, some filamentous fungi, such as Sporothrix schenckii,
Sporothrix globosa, Trichophyton interdigitale, Trichophyton
mentagrophytes, Arthroderma benhamiae and Trichophyton
rubrum cannot be clearly differentiated by microscopic
examination after traditional culturing and staining. Their
overlapping phenotypic characteristics can be confusing, and
identification requires growth of the organisms in culture for at
least one week, thus delaying the diagnosis (5–9).
It is crucial to develop rapid identification
methods and effective treatments for infections by filamentous
fungi (10). Molecular techniques
such as PCR, ribosomal DNA (rDNA) internal transcribed spacer (ITS)
sequence analysis, or 18S ribosomal RNA (rRNA) gene sequencing can
accurately identify many filamentous fungi at the species level
(11). However, these methods are
not only time-consuming and expensive but also require expertise
(12,13). Matrix-assisted laser
desorption/ionization (MALDI) time-of-flight mass spectrometry
(MALDI-TOF MS) offers a time-saving and highly accurate means for
rapid identification of microbial agents, especially bacteria
(14–17). In recent years, the application of
MALDI-TOF MS has also emerged as a means to identify isolates from
fungi, and it is a faster and more robust diagnostic technique
compared with the other established protocols for the
identification of yeasts isolated from carious clinical specimens
(18,19). After the successful MALDI-TOF MS
identification of bacteria and yeasts in clinical laboratories,
increased interest has emerged in applying this method to
filamentous fungi (20,21). There are however some disadvantages
to this method. MALDI-TOF identification has been hindered by the
absence of filamentous fungi from in-house MALDI-TOF MS libraries.
Consequently, several isolates have been mis-identified or
unidentified because of the absence of the important reference
spectra in the databases (22,23).
Additionally, the high cost of equipment prevents this technique
from being used in developing countries (24). In light of this, this present study
attempted to identify 123 strains of filamentous fungi by using
conventional methods and MALDI-TOF MS. When these methods gave
discordant results, the discrepancies were further confirmed by ITS
sequence analysis.
The Clinical and Laboratory Standards Institute has
developed a reference broth (M38A2) microdilution (BMD) approach
for testing the antifungal susceptibility of various filamentous
fungi (25). Unfortunately, this
method is cumbersome. By contrast, commercially available Etest
strips containing defined concentration gradients of drugs provide
a simple and easy approach for testing the susceptibility of
filamentous fungi. In the present study, the Etest method was
employed to investigate the antifungal activities of five
antifungal agents (fluconazole, amphotericin B, voriconazole,
itraconazole and caspofungin) against 79 strains of filamentous
fungi in vitro.
Materials and methods
Fungal isolates
A total of 123 filamentous fungi were collected in
this study. Of these 123 fungi, 19 standard strains were purchased
from The China Medical Culture Collection Center (Table I). The remaining 104 isolates were
from clinical specimens: 21 isolates were obtained from The
Clinical Laboratory of the Third Affiliated Hospital of Soochow
University, and the remaining 83 strains were kindly provided by
the Clinical Laboratory of The Jiangsu Province Hospital and
Nanjing General Hospital. These clinical isolates consisted of
sputum, urine, bronchial washing fluid and skin.
 | Table I.A total of 19 standard strains
obtained from China Medical Culture Collection Center. |
Table I.
A total of 19 standard strains
obtained from China Medical Culture Collection Center.
| Standard
strain | Preservation
number |
|---|
| Aspergillus
clavatus | CMCC(F)A4a |
| Aspergillus
chevalieri | CMCC(F)A28 |
| Aspergillus
flavus | CBS13161 |
| Aspergillus
fumigatus | ATCC 3626 |
| Aspergillus
nidulans | CMCC(F)A7a |
| Aspergillus
niger | CMCC(F)A3 |
| Aspergillus
tamarii | CMCC(F)A13 |
| Aspergillus
terreus | ATCC 3633 |
| Aspergillus
ustus | CMCC(F)A15a |
| Aspergullus
versicolor | CBS 245.65 |
| Beauveria
bassiana | CMCC(F)B13b |
| Microsporum
gypseum | CBS 118893 |
| Paecilomyces
lilacinus | CMCC 35539 |
| Penicillium
chrysogenum | CMCC(F)B31 |
| Rhizopus
microsporus | CMCC(F)B46 |
| Sporothrix
schenckii | ATCC 49.12 |
| Talaromyces
marneffei | CMCC(F)B33r |
| Trichophyton
rubrum | ATCC 4438 |
| Trichophyton
tonsurans | CBS 171.65 |
Phenotypic identification
All standard strains and clinical isolates were
subcultured on Sabouraud dextrose agar (SDA) at 28°C for 2–6 days.
The fungi were first identified by skilled mycologists on the basis
of macroscopic and light microscopic features using conventional
methods, including lactophenol cotton blue and KOH staining.
Briefly, the size, shape, pigment, texture and growth conditions of
each isolate were recorded. Subsequently, the spores and hyphae
were smeared on glass slides, stained with lactophenol cotton blue
(Baso Diagnostic, Inc.) or 10% KOH, and observed under an Olympus
IX53 microscope at a final magnification of ×1,000.
Identification by MALDI-TOF MS
Protein analysis was performed using a MALDITOF
Microflex LT mass spectrometer with a N2 laser set at 337 nm. The
spectra were recorded in positive ion mode at a frequency of 60 Hz
laser within a m/z range of 2,000 to 20,000. For each spectrum, a
total of 240 shots from different positions of the metal plate were
collected. The acceleration voltage, extraction voltage, lens
voltage and delayed extraction time were set as 20 kV, 18.00 kV, 6
kV and 150 ns, respectively. In addition, the flow rate was
maintained at 70 l/min. The fungal isolates were cultured and
prepared for MALDI-TOF MS (Bruker Corporation) in accordance with
the manufacturer's instructions. Briefly, the fungal isolates were
incubated in liquid Sabouraud medium (Hangzhou Binhe Microbial
Reagent Co., Ltd.) on a shaker at 28°C for 24–48 h until abundant
mycelia were observed. Subsequently, the mixture was transferred
into a microcentrifuge tube and centrifuged at 25°C for 2 min at
15,493 × g to remove the supernatant. Next, the pellet was vortexed
first with 300 µl of water and then with 900 µl of absolute ethanol
(Sigma-Aldrich; Merck KGaA). The suspension was then centrifuged at
25°C for 2 min at 15,493 × g to remove the supernatant. The pellet
was air dried for 5 min and then vortexed first with ~10–30 µl of
70% formic acid (Sigma-Aldrich; Merck KGaA) and then with the same
amount of acetonitrile. The suspension was then centrifuged for at
25°C 2 min at 15,493 × g. A total of 1.0 µl of the supernatant was
added to a 96-spot polished steel target plate and allowed to dry.
A saturated solution of 1.0 µl of MALDI-TOF MS matrix (a saturated
solution of α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile and
2.5% trifluoroacetic acid) was applied to each sample, and the
mixture was allowed to dry at room temperature (26–28).
Mass spectra were acquired by MALDI-TOF MS and analysed using
FlexControl 3.1 and the Bruker filamentous fungi library 1.0
(Bruker Daltonics). The spectrometer was calibrated using the
bacterial test standard (BTS; Bruker Daltonics). Each sample was
analysed in triplicate. Before each analysis, quality assurance was
performed by using Aspergillus fumigatus (ATCC®
3626™; American Type Culture Collection) and Aspergillus
niger (CMCC (F)A3; National Center for Medical Culture
Collections) reference strains. Identification scores ≥2.000
indicated species-level identification, scores of 1.700–1.999
indicated genus-level identification, while scores <1.700
indicated no identification (29–31).
Results showing ‘no peaks’ required reanalysis.
ITS sequence analysis
Isolates with identification discrepancy between
phenotyping and MALDI-TOF MS findings were further analysed by ITS
sequencing. A total of 15 filamentous fungi were identified by
rDNA-ITS sequence analysis. Fungal genomic DNA was extracted from
mycelium with a HiPure Fungal DNA kit (Magen) according to the
manufacturer's instructions. The total DNA was amplified by PCR
using forward ITS1, 5′-TCCGTAGGTGAACCTGCGG-3′ and reverse ITS4,
5′-TCCTCCGCTTATTGATATGC-3′ universal primers (32) synthesized by Genewiz, Inc. PCR was
performed with the following conditions: An initial denaturation at
95°C for 5 min followed by 35 cycles of 30 sec at 94°C, 30 sec at
52°C and 30 sec at 72°C, with a final extension step at 72°C for 5
min. All reactions were performed in 2 µl volumes composed of 10X
reaction buffer, 1 µl of template, 0.5 µl of each dNTP, 0.5 µl of
each primer, 0.2 µl of TransStart Tag DNA Polymerase (Beijing
Transgen Biotech Co., Ltd.) and water added to a total volume of 20
µl. The products were electrophoresed in 1% agarose gel containing
0.01% Genecolour™ nucleic acid stain (Gene Bio Tech Co., Ltd.). All
PCR-amplified products were sequenced with an ABI 3730 Sequencer
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The ITS
sequences of the isolates were compared with reference sequences
from GenBank by using the associated BLAST online tool (National
Center for Biotechnology Information). Strains were identified on
the basis of 99% similarity or higher.
Antifungal susceptibility testing
Etest gradient strips with fluconazole (range,
0.016–256 µg/ml), amphotericin B, voriconazole, itraconazole and
caspofungin (range, 0.002–32 µg/ml) were purchased from Autobio
Diagnostics Co., Ltd. The Etest assay was performed in accordance
with the manufacturer's instructions. Fungi were grown on SDA at
35°C for 3–7 day(s) to ensure adequate sporulation from mature
fungi. Conidial suspensions were created from harvested cultures,
and the turbidity was adjusted to 0.5 McF. These suspensions were
directly inoculated onto the surface of the antimicrobial test
fungal medium (BIO-KONT Co., Ltd.) using swabs, and the plates were
incubated at 35°C until confluent lawns of filamentous fungi were
formed. Subsequently, the surface of each of the plates was dried,
and the antifungal agent strips were placed on the plates. The
Etest minimum inhibitory concentration (MIC) was recorded as the
lowest concentration of an antifungal drug for which the elliptical
zone of growth inhibition intersected the Etest strip.
Microcolonies within the ellipse were ignored. The Etest MIC was
recorded after incubation for 24 or 48 h at 35°C. For most fungal
species, the final Etest MIC readings were recorded at 24 h for
echinocandins (caspofungin) and at 48 h for other drugs.
Seventy-nine strains were randomly selected and further tested
using the Etest MIC method, and quality control for this assay was
performed using Candida parapsilosis (ATCC®
22019™; American Type Culture Collection).
Statistical analysis
Distinct sequences were annotated by Blast search
against the NCBI non-redundant database with a cut-off E-value of
10−5. A low E-value represents high confidence in the
annotation.
Results
Identification of filamentous fungi by
phenotypic methods
For most of the evaluated filamentous fungi, the
conventional methods take an average of 4 days and cost $2 each for
completing the identification process. After 3–7 days of
incubation, all isolates showed different sizes, shapes, pigments
and textures on SDA plates. The phenotypic characteristics of
filamentous fungi varied among each other. The Aspergillus
species (spp.) grew rapidly and formed loose colonies on SDA
plates, with most species exhibiting characteristic colours and
shapes. For example, Aspergillus flavus formed fluffy
greyish-green colonies consisting predominantly of vegetative
hyphae, while A. niger usually formed characteristic black
colonies on SDA plates. The Penicillium spp. grew more
slowly than the Aspergillus spp. On SDA, most
Penicillium spp. produced radiated sulcata with a velvety
colony surface, and some displayed coloured exudates. For example,
Talaromyces marneffei produced a soluble red pigment, which
diffused into the agar, lending a red or pink appearance to the
reverse side of the colony. Dermatophytes showed radial growth, and
many typical isolates of common dermatophytes were identified
directly on the SDA plates. Microsporum canis secreted a
yellow pigment, and T. rubrum also usually secreted a yellow
pigment. Fig. 1 shows 12 typical
colony morphologies of filamentous fungi. Of the 123 filamentous
fungi analysed in this study, 113 (91.9%) were correctly identified
by phenotypic methods, while 5 T. interdigitale, 3 S.
globosa and 2 Trichophyton tonsurans were
morphologically confused with T. mentagrophytes, S.
schenckii and T. rubrum, respectively.
 | Figure 1.Twelve types of colony morphology of
filamentous fungi. (A) Aspergillus tamarii (B)
Aspergillus flavus, (C) Rhizopus microsporus, (D)
Talaromyces marneffei, (E) Aspergillus nidulans, (F)
Aspergillus terreus, (G) Aspergillus ustus, (H)
Sporothrix schenckii, (I) Aspergillus chevalieri, (J)
Trichophyton rubrum, (K) Purpureocillium lilacinus,
(L) Beauveria bassiana. Scale bar, 1 cm. |
Identification by MALDI-TOF MS
The MALDI-TOF MS method took an average of 15 min
and US$0.3 for identifying filamentous fungi (starting from the
time the culture became available for testing). Of the 123
filamentous fungi analysed by MALDI-TOF MS, 114 (92.70%) isolates
could be identified at the genus level (identification scores,
1.700–1.999), and 80 (65.0%) were correctly identified at the
species level (identification scores ≥2.000). However, this method
failed to identify six strains (4.9%; scores <1.700).
Penicillium chrysogenum and T. mentagrophytes were
misidentified as Penicillium camemberti and Trichophytum
equinum, respectively (Table
II). Fig. 2 shows characteristic
MALDI Biotyper spectra of five representative filamentous fungi.
These results showed that MALDI-TOF MS rapidly and accurately
identified most filamentous fungi and is, therefore, a reliable
tool for the diagnosis and treatment of fungal infections.
 | Table II.Results of identification of
filamentous fungi by MALDI-TOF MS (n=123). |
Table II.
Results of identification of
filamentous fungi by MALDI-TOF MS (n=123).
|
|
| MALDI-TOF MS
results, no. (%) |
|---|
|
|
|
|
|---|
|
|
| Unreliable ID |
|
|
|---|
|
|
|
|
|
|
|---|
| Strain | No. of
isolates | No ID
generated | Incorrect ID (ID
generated) | Genus level ID
only | Species level
ID |
|---|
| Aspergillus
fumigates | 31 | 0 | 0 | 0 | 31 (100) |
| Aspergillus
flavus | 16 | 0 | 0 | 0 | 16 (100) |
| Aspergillus
niger | 7 | 0 | 0 | 0 | 7
(100) |
| Aspergillus
nidulans | 1 | 0 | 0 | 0 | 1
(100) |
| Aspergillus
terreus | 1 | 0 | 0 | 0 | 1
(100) |
| Aspergillus
tamarii | 1 | 0 | 0 | 1 (100) | 0 |
| Aspergillus
clavatus | 1 | 0 | 0 | 0 | 1
(100) |
| Aspergillus
ustus | 1 | 0 | 0 | 0 | 1
(100) |
| Aspergillus
versicolor | 2 | 0 | 0 | 0 | 2
(100) |
| Trichophyton
rubrum | 23 | 0 | 0 | 16 (69.6) | 7
(30.4) |
| Epidermophyton
floccosum | 5 | 0 | 0 | 5 (100) | 0 |
| Microsporum
canis | 5 | 0 | 0 | 4 (80) | 1
(20) |
| Trichophyton
tonsurans | 6 | 0 | 0 | 3 (50) | 3
(50) |
| Microsporum
gypseum | 6 | 0 | 0 | 3 (50) | 3
(50) |
| Sporothrix
schenckii | 1 | 0 | 0 | 1 (100) | 0 |
| Sporothrix
globosa | 3 | 3 | 0 | 0 | 0 |
| Rhizopus
microspores | 1 | 0 | 0 | 0 | 1 (100) |
| Penicillium
chrysogenum | 1 | 0 | 1 (Penicillium
camemberti) | 0 | 0 |
| Purpureocillium
lilacinus | 1 | 0 | 0 | 1 (100) | 0 |
| Beauveria
bassiana | 1 | 1 | 0 | 0 | 0 |
| Aspergillus
chevalieria | 1 | 1 | 0 | 0 | 0 |
| Talaromyces
marneffeia | 1 | 1 | 0 | 0 | 0 |
| Trichophyton
interdigitale | 5 | 0 | 0 | 0 | 5 (100) |
| Trichophyton
entagrophytes | 2 | 0 | 2 (Trichophytum
equinum) | 0 | 0 |
| Total | 123 | 6 (4.9) | 3 (2.4) | 34 (27.6) | 80 (65.0) |
ITS sequence analysis
Fifteen isolates of filamentous fungi were further
identified by rDNA-ITS sequencing analysis. The fungal rDNA ITS
were amplified by PCR and then subjected to agarose electrophoresis
(Fig. 3). Of the 15 isolates, five
were confirmed as T. interdigitale, three as S.
globosa, two as T. mentagrophytes, two as T.
tonsurans, and the remaining as T. marneffei, Aspergillus
chevalieri and Beauveria bassiana (Table III). These results indicated that
rare or ambiguous fungi were accurately identified by rDNA-ITS
sequencing.
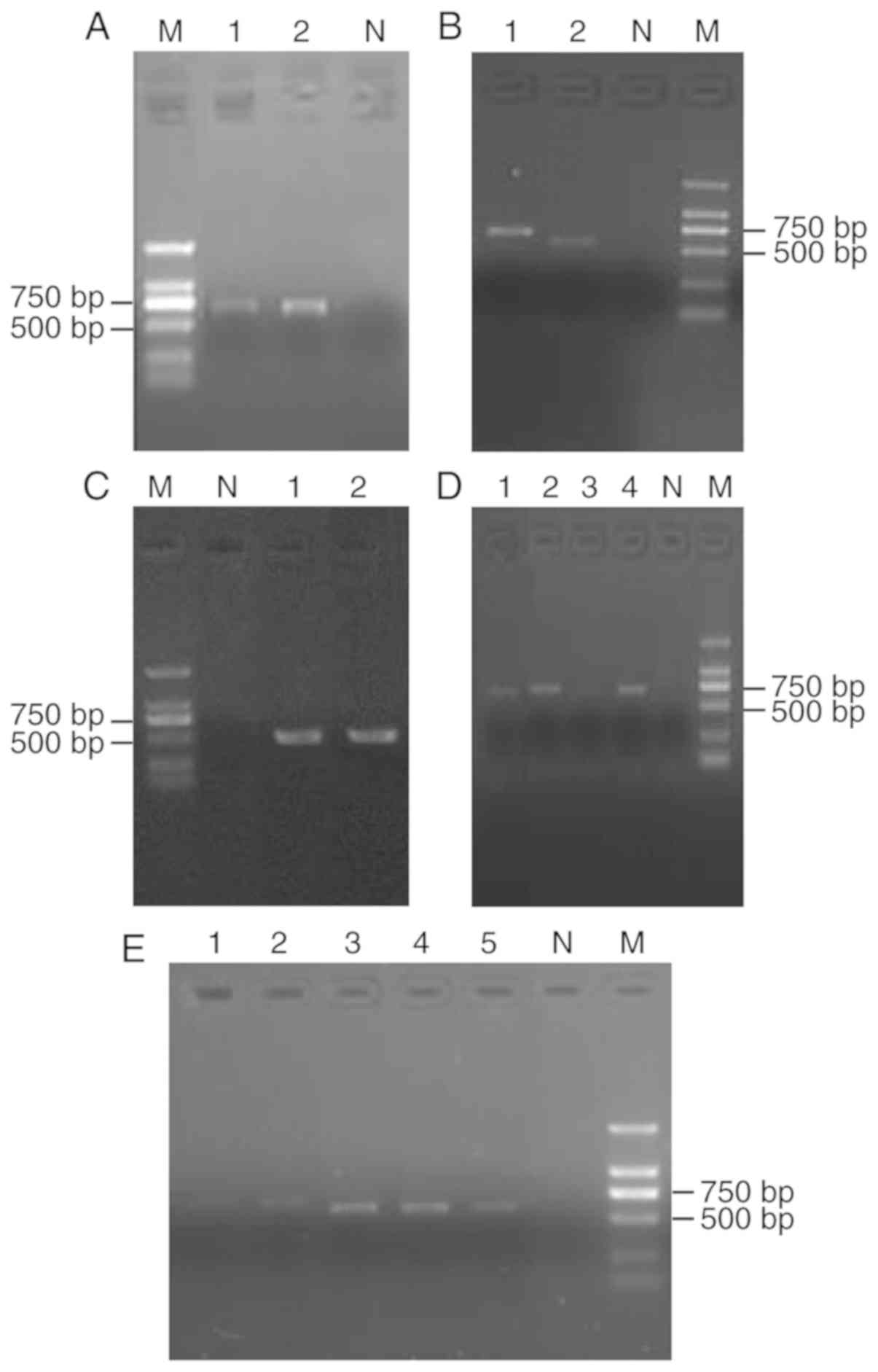 | Figure 3.PCR products of the fungal ribosomal
DNA internal transcribed spacer sequence. Lane M, Marker. (A) Lane
1, Trichophyton interdigitale; lane 2, Trichophyton
mentagrophytes; lane N, negative control. (B) Lane 1,
Trichophyton tonsurans; lane 2, Beauveria bassiana;
lane N, negative control. (C) Lane 1, Aspergillus
chevalieri; lane 2, Talaromyces marneffei; lane N,
negative control. (D) Lane 1, T. tonsurans; lane 2, T.
interdigitale, lane 3, T. interdigitale; lane 4, T.
mentagrophytes; lane N, negative control. (E) Lane 1, T.
interdigitale; lane 2, T. interdigitale; lane 3,
Sporothrix globosa; lane 4, S. globosa; lane 5, S.
globosa; lane N, negative control. bp, base pairs. |
 | Table III.Filamentous fungi identified by ITS
sequencing analysis (n=15). |
Table III.
Filamentous fungi identified by ITS
sequencing analysis (n=15).
| Strain | Result | Max score | Query cover | E-value | Identity | Accession |
|---|
|
134-194Vo | Trichophyton
interdigitale | 1262 | 100% | 0.0 | 99% | KP308373.1 |
| 20157639 | T.
interdigitale | 1260 | 100% | 0.0 | 99% | KP308373.1 |
| 23-20101126 | Sporothrix
globosa | 835 | 99% | 0.0 | 100% | JX997737.1 |
| 24-20101149 | S.
globosa | 837 | 99% | 0.0 | 100% | JX997737.1 |
| 25-20134489 | S.
globosa | 835 | 99% | 0.0 | 99% | LC317795.1 |
| 10-2157180 | Trichophyton
tonsurans | 1249 | 99% | 0.0 | 99% | AB220045.1 |
| 130–048
Vo | T.
interdigitale | 1254 | 100% | 0.0 | 99% | LC317434.1 |
| 131–104
Vo | T.
interdigitale | 1157 | 100% | 0.0 | 100% | JX122224.1 |
| 2157649 | Trichophyton
mentagrophytes | 1253 | 99% | 0.0 | 99% | KM355551.1 |
| 133–053
Vo | T.
interdigitale | 1266 | 99% | 0.0 | 100% | KP308373.1 |
| 2157536 | T.
mentagrophytes | 1256 | 99% | 0.0 | 99% | AB566303.1 |
| 13-2157226 | T.
tonsurans | 1188 | 99% | 0.0 | 99% | AB220045.1 |
| CMCC (f)B13b | Beauveria
bassiana | 1042 | 99% | 0.0 | 99% | JF429894.1 |
| CMCC (f)B33r | Talaromyces
marneffei | 1046 | 100% | 0.0 | 100% | KY115196.1 |
| CMCC (f)A28 | Aspergillus
chevalieri | 1022 | 100% | 0.0 | 99% | KX463363.1 |
Antifungal susceptibility
analysis
The results of the quality control tests were within
the control limits for the five antifungal drugs. Fluconazole
showed the highest MIC that caused 90% inhibition
(MIC90) values against the 79 evaluated filamentous
fungi: >256 µg/ml against A. fumigatus, A. flavus, A. niger,
T. rubrum, T. tonsurans, Aspergillus nidulans, Aspergillus terreus,
Aspergillus tamarii, Aspergillus clavatus, Aspergillus ustus,
Aspergillus versicolor and Microsporum gypseum; 24 µg/ml
against M. canis; and 12 µg/ml against Epidermophyton
floccosum. By contrast, voriconazole showed the lowest
MIC90 values against the 79 filamentous fungi: ≤0.38
µg/ml against A. fumigatus, A. flavus, A. niger, T. rubrum, T.
tonsurans, M. canis, E. floccosum, A. nidulans, A. terreus, A.
tamarii, A. versicolor and M. gypseum; 1.5 µg/ml against
A. clavatus; and 2 µg/ml against A. ustus. The
MIC90 values of caspofungin against the 79 filamentous
fungi were the second lowest, next to those of voriconazole: ≤1
µg/ml against A. fumigatus, A. flavus, A. niger, T. rubrum, T.
tonsurans, M. canis, E. floccosum, A. nidulans, A. terreus, and
A. tamarii; 6 µg/ml against M. gypseum; 8 µg/ml
against A. ustus and A. versicolor; and 12 µg/ml
against A. clavatus. For T. tonsurans, the
MIC90 values of itraconazole corresponded with those of
caspofungin. Against E. floccosum, the MIC90 and
MIC that caused 50% inhibition (MIC50) of amphotericin B
both corresponded with those of caspofungin. Against A.
fumigatus and T. rubrum, the MIC90 values of
amphotericin B corresponded with those of itraconazole.
Additionally, the MIC of amphotericin B against A. terreus
and that of itraconazole against both A. niger and A.
clavatus were >32 µg/ml. The in vitro
susceptibilities of the 79 filamentous fungi are summarized in
Table IV.
 | Table IV.Results of the Etest in vitro
susceptibility assay for filamentous fungi. |
Table IV.
Results of the Etest in vitro
susceptibility assay for filamentous fungi.
|
|
MIC50/MIC90, µg/ml
(range) |
|---|
|
|
|
|---|
| Strain | Amphotericin B | Itraconazole | Voriconazole | Fluconazole | Caspofungin |
|---|
| Aspergillus
fumigatus (n=19) | 2/4 (1–6) | 3/4 (1.5–6) | 0.094/0.125
(0.064–0.125) | >256/>256
(>256) | 0.5/1 (0.25–2) |
| Aspergillus
flavus (n=14) | 3/>32
(1->32) | 1.5/3 (0.5–3) | 0.125/0.19
(0.048–0.25) | >256/>256
(>256) | 0.38/0.75
(0.38–1) |
| Aspergillus
niger (n=5) | 4/4 (2–4) | 16/>32
(16->32) | 0.25/0.25
(0.25–1) | >256/>256
(>256) | 0.25/0.75
(0.19–0.75) |
| Trichophyton
rubrum (n=18) | 1.5/2 (1–4) | 0.75/2
(0.25–2) | 0.048/0.38
(0.004–0.64) | 16/>256
(3->256) | 1/1
(0.25–2.00) |
| Trichophyton
tonsurans (n=6) | 3/3 (2–3) | 0.38/0.38
(0.25–0.38) | 0.024/0.024
(0.016–0.024) | 16/>256
(16->256) | 0.25/0.38
(0.25–0.38) |
| Microsporum
canis (n=5) | 1/2 (1–2) | 0.38/0.38
(0.19–0.38) | 0.094/0.094
(0.064–0.094) | 24/24 (12–24) | 0.5/0.5
(0.048–0.5) |
| Epidermophyton
floccosum (n=5) | 0.125/0.19
(0.094–0.19) | 0.19/0.25
(0.125–0.25) | 0.004/0.006
(0.002–0.006) | 8/12 (6–12) | 0.125/0.19
(0.002–0.19) |
| Aspergillus
nidulans (n=1) | 3 | 0.75 | 0.048 | >256 | 1 |
| Aspergillus
terreus (n=1) | >32 | 1 | 0.25 | >256 | 0.25 |
| Aspergillus
tamarii (n=1) | 2 | 0.5 | 0.25 | >256 | 1 |
| Aspergillus
clavatus (n=1) | 6 | >32 | 1.5 | >256 | 12 |
| Aspergillus
ustus (n=1) | 8 | 12 | 2 | >256 | 8 |
| Aspergillus
versicolor (n=1) | 8 | 1.5 | 0.125 | >256 | 8 |
| Microsporum
gypseum (n=1) | 3 | 2 | 0.006 | >256 | 6 |
Discussion
Because of immune deficiency, some individuals, such
as those with malignant tumours, HIV infection/acquired immune
deficiency syndrome or infectious diseases treated with
extended-spectrum antimicrobials as well as transplant recipients,
are susceptible to fungal infections (33,34).
These infections can be classified as exogenous and endogenous
types on the basis of the pathogen source. Candida spp. are
still the main pathogens in endogenous opportunistic fungal
infections, whereas Aspergillus spp., Cryptococcus
spp., dermatophytes, zygomycetes and dimorphic fungi are the
predominant sources of exogenous fungal infections (35,36).
At present, identification of fungi, especially
filamentous fungi, is very challenging. Conventional methods such
as macroscopic observation (phenotyping on the basis of growing
conditions, colony shape, pigments and texture) and direct or
post-staining microscopic examination, are not only time consuming
but can also easily misidentify the fungi. Although β-1,3-D glucan
and galactomannan tests can rapidly provide evidence of fungal
infection, the specificities of these tests are limited because of
false negative or false positive results. MALDI-TOF MS has been
proven to be a remarkably flexible means for rapid identification
and classification of bacteria and yeasts in clinical microbiology
(37), and therefore presents a
strong challenger to established microscopic and molecular biology
methods. However, it has not been widely applied for identifying
filamentous fungi because of the complex phylogenetic relationships
between species and even more complicated morphology, which can
make it difficult to extract fungal proteins.
To the best of the authors' knowledge, there is no
standardised procedure for routine identification of filamentous
fungi in the clinical laboratory. The limited number of existing
studies in this regard have reported varying extraction procedures
for fungi (26). Current extraction
procedures include bead beating, ultrasonication, and chemical
lysis. Here, a simple, economic, and efficient method for
extracting filamentous fungi in a clinical laboratory setting has
been reported in order to achieve mass spectra of good quality. In
the present study, fungi were propagated in liquid cultures and
extraction was performed using the ethanol-formic acid method.
In the present study, MALDI-TOF MS achieved 65.0%
(80/123) identification of filamentous fungi at the species level,
with a particular high identification rate for Aspergillus
spp. (96.8%; 60/62), which is in agreement with the findings of
previous studies (17,38). However, the identification rate of
non-Aspergillus spp. was lower; of the 61
non-Aspergillus isolates, only 20 (32.8%) were correctly
identified at the species level. The low performance of MALDI-TOF
MS in identifying non-Aspergillus spp. may be because of the
difficult steps involved in protein extraction. Therefore, it is
necessary to identify novel techniques for protein extraction from
non-Aspergillus spp. MALDI-TOF MS could not identify some
filamentous fungi such as S. globosa and T.
marneffei, which is likely to be due to their limited reference
spectra in the Bruker spectrum database or the highly conserved
nature of ribosomal proteins among these species. MALDI-TOF MS also
failed to identify other species such as B. bassiana and
A. chevalieri because they do not exist in the Bruker
spectrum database. Furthermore, the MALDI-TOF Bruker Biotyper
generated wrong identifications for P. chrysogenum and T.
mentagrophytes. Therefore, it is vital to update the database
in order to extend the identification to all fungal species.
The Etest method overcomes the drawback of the BMD
method in that the BMD method can be a tedious operation. It also
offers other advantages, including direct and easy operation and
that it is likely to be available at clinical laboratories that
test relatively few filamentous fungi. Consequently, it would be
beneficial to validate the Etest method for antifungal
susceptibility testing of filamentous fungi. In testing the
susceptibility of filamentous fungi against amphotericin B,
triazoles and echinocandins, Etest findings show a strong
correlation with BMD results (39).
In the present study, it was found that incubating the filamentous
fungi in RPMI basal medium for 24 or 48 h was sufficient for visual
interpretation of the MIC without cumbersome microscopy.
In the present study, voriconazole (a triazole)
showed the most efficient in vitro activity against A.
ustus, with a MIC of 2 µg/ml. However, Lamoth and Alexander
(39) have previously reported a
voriconazole MIC of >16 µg/ml against A. ustus. In the
present study, the antifungal activity of caspofungin was inferior
to that of voriconazole, which contradicts the results reported by
Shi et al (40). Itraconazole
and amphotericin B exhibited slightly higher MIC values against
most filamentous fungi except A. terreus, A. niger and A.
clavatus. The MIC value of amphotericin B against A.
terreus and those of itraconazole against A. niger and
A. clavatus were >32 µg/ml, which suggested that the
Etest method effectively detected drug-resistant isolates. The MIC
of amphotericin B against A. terreus in this study (>32
µg/ml) was similar to that reported in a previous study (41). Furthermore, the data from the present
study were similar to the BMD results for the testing of A.
terreus against amphotericin B that were found in a study by
Vaezi et al (42). A large
number of tests have shown that A. terreus is a poor target
for amphotericin B and can, therefore, be reported as being
amphotericin-B-resistant without further antifungal testing
(43).
Fluconazole is by no means the most commonly used
antifungal drug for treating filamentous fungi. Although clinical
isolates of most filamentous fungi show a broad in vitro
resistance to fluconazole, fluconazole is still second-line drugs
of choice for prevention and treatment of certain mycoses,
including sporotrichosis (9,44,45).
Fluconazole showed the highest MIC50 and
MIC90 values against all filamentous fungi tested in the
present study, which might indicate that the majority of commonly
used drugs offer greater resistance than fluconazole. It is
interesting to note the activity of azoles against filamentous
fungi is variable. Although fluconazole shows weak activity against
most filamentous fungi, new-generation azoles such as voriconazole
might still exhibit favourable activity, highlighting the need for
guidance in drug selection. In general, the in vitro
activities of voriconazole and caspofungin in the present study
were more effective than those of fluconazole, itraconazole and
amphotericin B, consistent with previous findings (39,46). On
the basis of these observations, the Etest method appears to be
suitable for testing the in vitro activity of voriconazole,
caspofungin, amphotericin B and itraconazole against filamentous
fungi.
In conclusion, it was found that MALDI-TOF MS-based
identification of filamentous fungi is less expensive and easier
than phenotypic identification and ITS sequence analysis, and its
fast turnaround time allows the analysis to be performed more
rapidly. As indicated above, it is suitable as a first-line test
for identifying filamentous fungi in routine clinical laboratories.
However, conventional identification cannot be abandoned and will
continue to be an alternative when MALDI-TOF MS fails to provide a
definitive result. With the increasing use of MALDI-TOF MS in
diagnostic laboratories and the further expansion of online
databases of filamentous fungi libraries, a transition to more
accurate and rapid identification of filamentous fungi is likely to
occur. Etests for direct susceptibility testing for common
filamentous fungi has been reported as a rapid antifungal
susceptibility testing tool that can provide results in 24–48 h
(47). The present in vitro
antifungal susceptibility results showed that voriconazole
possessed the strongest antifungal activity among the tested drugs
and can be used against a broad range of filamentous fungi, while
caspofungin possessed better in vitro activity than
fluconazole, itraconazole and amphotericin B. Therefore,
voriconazole can still be used as the first-line drug for treating
serious infections caused by filamentous fungi, while caspofungin
could serve as a treatment option for fungal infections. The Etest
method is considered an appropriate alternative to guide directed
antifungal therapy for routine clinical laboratories. It is
important to assess the MIC values of more drugs by comparing
clinical outcomes and perfecting interpretive clinical breakpoints,
which can help determine whether the use of an antifungal drug is
appropriate.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81572052) and Natural
Science Foundation of Jiangsu Province, China (grant no.
BK20151178).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YP performed the experiments and prepared the
manuscript. QZ and CX analyzed the data. WS was responsible for
study conception and design. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kontoyiannis DP, Marr KA, Park BJ,
Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW,
Brown JM, et al: Prospective surveillance for invasive fungal
infections in hematopoietic stem cell transplant recipients,
2001–2006: Overview of the Transplant-Associated Infection
Surveillance Network (TRANSNET) Database. Clin Infect Dis.
50:1091–1100. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alcazar-Fuoli L, Mellado E,
Alastruey-Izquierdo A, Cuenca-Estrella M and Rodriguez-Tudela JL:
Aspergillus section Fumigati: Antifungal
susceptibility patterns and sequence-based identification.
Antimicrob Agents Chemother. 52:1244–1251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grumbt M, Monod M and Staib P: Genetic
advances in dermatophytes. FEMS Microbiol Lett. 320:79–86. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de Respinis S, Tonolla M, Pranghofer S,
Petrini L, Petrini O and Bosshard PP: Identification of
dermatophytes by matrix-assisted laser desorption/ionization
time-of-flight mass spectrometry. Med Mycol. 51:514–521. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abastabar M, Mirhendi H,
Rezaei-Matehkolaei A, Shidfar MR, Kordbacheh P and Makimura K:
Restriction analysis of β-tubulin gene for differentiation of the
common pathogenic dermatophytes. J Clin Lab Anal. 28:91–96. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cafarchia C, Iatta R, Latrofa MS, Gräser Y
and Otranto D: Molecular epidemiology, phylogeny and evolution of
dermatophytes. Infect Genet Evol. 20:336–351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Marimon R, Cano J, Gené J, Sutton DA,
Kawasaki M and Guarro J: Sporothrix brasiliensis, S.
globosa, and S. mexicana, three new Sporothrix
species of clinical interest. J Clin Microbiol. 45:3198–3206. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nenoff P, Herrmann J and Gräser Y:
Trichophyton mentagrophytes sive interdigitale? A dermatophyte in
the course of time. J Dtsch Dermatol Ges. 5:198–202. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gräser Y, Scott J and Summerbell R: The
new species concept in dermatophytes-a polyphasic approach.
Mycopathologia. 166:239–256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Drogari-Apiranthitou M, Mantopoulou FD,
Skiada A, Kanioura L, Grammatikou M, Vrioni G, Mitroussia-Ziouva A,
Tsakris A and Petrikkos G: In vitro antifungal susceptibility of
filamentous fungi causing rare infections: Synergy testing of
amphotericin B, posaconazole and anidulafungin in pairs. J
Antimicrob Chemother. 67:1937–1940. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu J, Yu Y, Cai Z, Bartlam M and Wang Y:
Comparison of ITS and 18S rDNA for estimating fungal diversity
using PCR-DGGE. World J Microbiol Biotechnol. 31:1387–1395. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen JH, Yam WC, Ngan AH, Fung AM, Woo WL,
Yan MK, Choi GK, Ho PL, Cheng VC and Yuen KY: Advantages of using
matrix-assisted laser desorption ionization-time of flight mass
spectrometry as a rapid diagnostic tool for identification of
yeasts and mycobacteria in the clinical microbiological laboratory.
J Clin Microbiol. 51:3981–3987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Del Chierico F, Masotti A, Onori M,
Fiscarelli E, Mancinelli L, Ricciotti G, Alghisi F, Dimiziani L,
Manetti C, Urbani A, et al: MALDI-TOF MS proteomic phenotyping of
filamentous and other fungi from clinical origin. J Proteomics.
75:3314–3330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Triest D, Stubbe D, De Cremer K, Piérard
D, Normand AC, Piarroux R, Detandt M and Hendrickx M: Use of
matrix-assisted laser desorption ionization-time of flight mass
spectrometry for identification of molds of the Fusarium genus. J
Clin Microbiol. 53:465–476. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ling H, Yuan Z, Shen J, Wang Z and Xu Y:
Accuracy of matrix-assisted laser desorption ionization-time of
flight mass spectrometry for identification of clinical pathogenic
fungi: A meta-analysis. J Clin Microbiol. 52:2573–2582. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chao QT, Lee TF, Teng SH, Peng LY, Chen
PH, Teng LJ and Hsueh PR: Comparison of the accuracy of two
conventional phenotypic methods and two MALDI-TOF MS systems with
that of DNA sequencing analysis for correctly identifying
clinically encountered yeasts. PLoS One. 9:e1093762014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bille E, Dauphin B, Leto J, Bougnoux ME,
Beretti JL, Lotz A, Suarez S, Meyer J, Join-Lambert O, Descamps P,
et al: MALDI-TOF MS Andromas strategy for the routine
identification of bacteria, mycobacteria, yeasts,
Aspergillus spp. and positive blood cultures. Clin Microbiol
Infect. 18:1117–1125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shokohi T, Aslani N, Ahangarkani F,
Meyabadi MF, Hagen F, Meis JF, Boekhout T, Kolecka A and Badali H:
Candida infanticola and Candida spencermartinsiae
yeasts: Possible emerging species in cancer patients. Microb
Pathog. 115:353–357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Aslani N, Janbabaei G, Abastabar M, Meis
JF, Babaeian M, Khodavaisy S, Boekhout T and Badali H:
Identification of uncommon oral yeasts from cancer patients by
MALDI-TOF mass spectrometry. BMC Infect Dis. 18:242018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ranque S, Normand AC, Cassagne C, Murat
JB, Bourgeois N, Dalle F, Gari-Toussaint M, Fourquet P, Hendrickx M
and Piarroux R: MALDI-TOF mass spectrometry identification of
filamentous fungi in the clinical laboratory. Mycoses. 57:135–140.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Normand AC, Cassagne C, Ranque S,
L'ollivier C, Fourquet P, Roesems S, Hendrickx M and Piarroux R:
Assessment of various parameters to improve MALDI-TOF MS reference
spectra libraries constructed for the routine identification of
filamentous fungi. BMC Microbiol. 13:762013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lau AF, Drake SK, Calhoun LB, Henderson CM
and Zelazny AM: Development of a clinically comprehensive database
and a simple procedure for identification of molds from solid media
by matrix-assisted laser desorption ionization-time of flight mass
spectrometry. J Clin Microbiol. 51:828–834. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bader O, Weig M, Taverne-Ghadwal L, Lugert
R, Gross U and Kuhns M: Improved clinical laboratory identification
of human pathogenic yeasts by matrix-assisted laser desorption
ionization time-of-flight mass spectrometry. Clin Microbiol Infect.
17:1359–1365. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yonetani S, Ohnishi H, Ohkusu K, Matsumoto
T and Watanabe T: Direct identification of microorganisms from
positive blood cultures by MALDI-TOF MS using an in-house saponin
method. Int J Infect Dis. 52:37–4. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Clinical and Laboratory Standards
Institute (CLSI), . Reference Method for Broth Dilution Antifungal
Susceptibility Testing of Filamentous Fungi; Approved standard.
(2nd). Clinical and Laboratory Standards Institute. (Wayne, PA).
M38–A2. 2008.
|
|
26
|
Cassagne C, Ranque S, Normand AC, Fourquet
P, Thiebault S, Planard C, Hendrickx M and Piarroux R: Mould
routine identification in the clinical laboratory by
matrix-assisted laser desorption ionization time-of-flight mass
spectrometry. PLoS One. 6:e284252011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McMullen AR, Wallace MA, Pincus DH, Wilkey
K and Burnham CA: Evaluation of the Vitek MS matrix-assisted laser
desorption ionization-time of flight mass spectrometry system for
identification of clinically relevant filamentous fungi. J Clin
Microbiol. 54:2068–2073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Normand AC, Cassagne C, Gautier M, Becker
P, Ranque S, Hendrickx M and Piarroux R: Decision criteria for
MALDI-TOF MS-based identification of filamentous fungi using
commercial and in-house reference databases. BMC Microbiol.
17:252017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Becker PT, de Bel A, Martiny D, Ranque S,
Piarroux R, Cassagne C, Detandt M and Hendrickx M: Identification
of filamentous fungi isolates by MALDI-TOF mass spectrometry:
Clinical evaluation of an extended reference spectra library. Med
Mycol. 52:826–834. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang Y, Zhang M, Zhu M, Wang M, Sun Y, Gu
H, Cao J, Li X, Zhang S, Wang J, et al: Comparison of two
matrix-assisted laser desorption ionization-time of flight mass
spectrometry systems for the identification of clinical filamentous
fungi. World J Microbiol Biotechnol. 33:1422017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Horká M, Kubesová A, Salplachta J,
Zapletalová E, Horký J and Slais K: Capillary and gel
electromigration techniques and MALDI-TOF MS - suitable tools for
identification of filamentous fungi. Anal Chim Acta. 716:155–162.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tarumoto N, Sakai J, Kodana M, Kawamura T,
Ohno H and Maesaki S: Identification of disseminated cryptococcosis
using MALDI-TOF MS and clinical evaluation. Med Mycol J.
57:E41–E46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pagano L, Akova M, Dimopoulos G, Herbrecht
R, Drgona L and Blijlevens N: Risk assessment and prognostic
factors for mould-related diseases in immunocompromised patients. J
Antimicrob Chemother. 66 (Suppl 1):i5–i14. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Herbrecht R, Bories P, Moulin JC, Ledoux
MP and Letscher-Bru V: Risk stratification for invasive
aspergillosis in immunocompromised patients. Ann NY Acad Sci.
1272:23–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pemán J, Cantón E, Quindós G, Eraso E,
Alcoba J, Guinea J, Merino P, Ruiz-Pérez-de-Pipaon MT,
Pérez-del-Molino L, Linares-Sicilia MJ, et al FUNGEMYCA Study
Group, : Epidemiology, species distribution and in vitro antifungal
susceptibility of fungaemia in a Spanish multicentre prospective
survey. J Antimicrob Chemother. 67:1181–1187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pana ZD, Farmaki E and Roilides E: Host
genetics and opportunistic fungal infections. Clin Microbiol
Infect. 20:1254–1264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schubert S and Kostrzewa M: MALDI-TOF MS
in the Microbiology Laboratory: Current Trends. Curr Issues Mol
Biol. 23:17–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
De Carolis E, Posteraro B, Lass-Flörl C,
Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM,
Sanguinetti M, et al: Species identification of Aspergillus,
Fusarium and Mucorales with direct surface analysis by
matrix-assisted laser desorption ionization time-of-flight mass
spectrometry. Clin Microbiol Infect. 18:475–484. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lamoth F and Alexander BD: Comparing Etest
and broth microdilution for antifungal susceptibility testing of
the most-relevant pathogenic molds. J Clin Microbiol. 53:3176–3181.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi JY, Xu YC, Shi Y, Lü HX, Liu Y, Zhao
WS, Chen DM, Xi LY, Zhou X, Wang H, et al: In vitro susceptibility
testing of Aspergillus spp. against voriconazole,
itraconazole, posaconazole, amphotericin B and caspofungin. Chin
Med J (Engl). 123:2706–2709. 2010.PubMed/NCBI
|
|
41
|
Heo MS, Shin JH, Choi MJ, Park YJ, Lee HS,
Koo SH, Lee WG, Kim SH, Shin MG, Suh SP, et al: Molecular
identification and amphotericin B susceptibility testing of
clinical isolates of Aspergillus from 11 hospitals in Korea. Ann
Lab Med. 35:602–610. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vaezi A, Fakhim H, Arastehfar A, Shokohi
T, Hedayati MT, Khodavaisy S, Rezaei-Matehkolaei A, Badiee P, Hagen
F, Lass-Flörl C, et al: In vitro antifungal activity of
amphotericin B and 11 comparators against Aspergillus
terreus species complex. Mycoses. 61:134–142. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Arendrup MC, Cuenca-Estrella M, Lass-Flörl
C and Hope WW; European Committee on Antimicrobial Susceptibility
Testing Subcommittee on Antifungal Susceptibility Testing
(EUCAST-AFST), : EUCAST technical note on Aspergillus and
amphotericin B, itraconazole, and posaconazole. Clin Microbiol
Infect. 18:E248–E250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Esquivel BD, Smith AR, Zavrel M and White
TC: Azole drug import into the pathogenic fungus Aspergillus
fumigatus. Antimicrob Agents Chemother. 59:3390–3398. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brilhante RS, Rodrigues AM, Sidrim JJ,
Rocha MF, Pereira SA, Gremião ID, Schubach TM and de Camargo ZP: In
vitro susceptibility of antifungal drugs against Sporothrix
brasiliensis recovered from cats with sporotrichosis in Brazil.
Med Mycol. 54:275–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gheith S, Saghrouni F, Bannour W, Ben
Youssef Y, Khelif A, Normand AC, Piarroux R, Ben Said M, Njah M and
Ranque S: In vitro susceptibility to amphotericin B, itraconazole,
voriconazole, posaconazole and caspofungin of Aspergillus
spp. isolated from patients with haematological malignancies in
Tunisia. Springerplus. 3:192014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pinto E, Lago M, Branco L, Vale-Silva LA
and Pinheiro MD: Evaluation of Etest performed in Mueller-Hinton
agar supplemented with glucose for antifungal susceptibility
testing of clinical isolates of filamentous fungi. Mycopathologia.
177:157–166. 2014. View Article : Google Scholar : PubMed/NCBI
|

















