Introduction
Prostate cancer (PCa) is the second most frequently
diagnosed cancer and the fifth cause of cancer-related death in men
worldwide (1,2). Patients with PCa in the early stages
can be treated with prostatectomy, chemotherapy, or radiotherapy,
and the prognosis from these therapies is favorable (3,4);
however, most of the therapy methods have side effects and there is
a limited ability to treat patients in the late stages (5). Although androgen deprivation therapy is
the standard treatment for patients with PCa in the advanced
stages, owing to the high sensitivity of PCa cells to hormone
deprivation, the tumor will develop to an end-stage metastatic
disease, referred to as castration-resistant PCa, to overcome the
hormone inhibition following sustained treatment (4,6).
Therefore, patients with recurrent or metastatic PCa have lower
5-year survival rates (~30%) compared with patients with local or
regional PCa (~100%) (7–9). Therefore, exploring the molecular
mechanism underlying PCa development may aid in improving diagnosis
and prognosis, and further improving the survival rate of patients
with PCa.
MicroRNAs (miRNAs) are a class of single-stranded,
small, non-coding RNAs that can regulate ~60% of the protein-coding
gene at the post-transcriptional level (10,11).
miRNAs can target oncogenes or tumor suppressor genes to regulate
different biological processes, such as cell apoptosis, metastasis
and proliferation (12). Thus,
evaluation of miRNA function in cancers is a hotspot in molecular
oncology (10). For example,
miR-148a in colitis-associated tumorigenesis can act as a tumor
suppressant via inhibiting NF-κB/STAT3 signaling (13). miR-130a can function as an oncogenic
miRNA to promote cell survival and tumor growth through targeting
PTEN (14). Previous, studies have
also suggested that miR-1303 is related to the tumorigenesis and
development of several cancers including neuroblastoma, gastric
cancer and colorectal cancer (15–17);
however, there are no reports of the roles of miR-1303 in PCa.
The Wnt/β-catenin pathway is a highly conserved,
intercellular signaling cascade and a key regulator of various
cellular processes (18). Aberrant
activation of the Wnt/β-catenin pathway is closely associated with
the development of different cancers including PCa (19). A previous study has indicated that
the Wnt/β-catenin pathway has an important role in PCa cell
proliferation, invasion and differentiation (20). Li et al have demonstrated that
PHD finger protein 21B (PHF21B) overexpression activates the
Wnt/β-catenin pathway to promote PCa stem cell-like phenotype
(21). Flores et al have
suggested that targeting the Wnt/β-catenin pathway may enhance the
efficacy of taxane chemotherapy in patients with PCa in the
advanced stages of disease progression (22). Hence, it is clinically important to
understand the roles of the Wnt/β-catenin pathway in PCa.
In this present study, miR-1303 expression was
determined in PCa tissues and cell lines, and the effects of
miR-1303 on the proliferation, migration and invasion of PCa cells
were assessed. Subsequently, dickkopf Wnt signaling pathway
inhibitor 3 (DKK3) was identified as a direct target of miR-1303.
Finally, the Wnt/β-catenin pathway was found to be involved in the
potentiating effects of miR-1303 in the proliferation, migration
and invasion of PCa cells.
Materials and methods
Bioinformatics analysis
MicroRNA-mRNA binding sites were predicted using
computer-aided algorithms obtained from TargetScan (version 7.2;
http://www.targetscan.org/vert_72/)
(23). Given the critical roles of
the Wnt/β-catenin pathway in the development of PCa (19,20),
DKK3, as a key inhibitor of the Wnt/β-catenin pathway (24), was subsequently chosen as the target
for miR-1303.
Clinical sample collection
Primary PCa tissues and adjacent normal prostate
tissues were obtained from 30 patients with PCa. These patients
underwent surgery in Tongji Hospital (Shanghai, China) between
January 2012 and October 2013. Before surgery, no patients were
treated with radiotherapy or chemotherapy. Tissues were immediately
frozen at −80°C. The patients were followed up for 50 months after
surgery. The Human Research Ethics Committee of Tongji Hospital
approved this experiment, and informed consent was obtained from
each patient.
Cell lines and cell culture
PCa cell lines (DU145, PC-3, 22Rv1 and LNCAP) and a
human normal prostate epithelial cell line (RWPE-1) were purchased
from the Cellular Resource Center of Shanghai Institutes for
Biological Sciences, Chinese Academy of Sciences. Cells were
incubated in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 µg/ml streptomycin and 100 U/ml penicillin in a humidified
incubator containing 5% CO2 at 37°C.
Cell transfection
DU145 cells were seeded (4×105 cells/ml)
in a 6-well plate and incubated in DMEM medium with 10% FBS at 37°C
for 24 h prior to transfection. miR-1303 inhibitor, miR-1303
mimics, siRNA targeting DKK3 (siDKK3) and their corresponding
negative controls (NC) were obtained from Shanghai GenePharma Co.,
Ltd. A total of 100 nM siDKK3 (5′-CUCCACCCUCGUCAGACAUAUAUAA-3′), 30
nM miRNA-1303 mimics (5′-UUUAGAGACGGGGUCUUGCUCU-3′), 30 nM mimic
control (5′-CCUGACCUCAGGGUUGAAUGUU-3′), 100 nM miRNA-1303 inhibitor
(5′-AGAGCAAGACCCCGUCUCUAAA-3′) or 100 nM inhibitor control
(5′-AUUCACCUAAGGAUGACGUCCA-3′) were transfected into DU145 cells in
6-well plates using 2.5 µl Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. After 6 h transfection, the transfected
cells were incubated in DMEM medium with 10% FBS at 37°C with 5%
CO2 for another 48 h before harvest for subsequent
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from PCa
tissues (50 mg) and cells (5×106 cells/ml) according to
the manufacturer's protocol. SMARTScribe™ Reverse Transcriptase,
dNTP Mix and random primers (hexadeoxyribonucleotide mix: pd(N)6),
all which purchased from Takara Biotechnology Co., Ltd., were used
to generate cDNA from RNA.
For mature miR-1303 detection, miRNA was extracted
by miRNeasy Mini kit (Qiagen GmbH). The TaqMan microRNA Reverse
Transcription Kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) were used to reverse transcribe RNA into cDNA according to
the manufacturer's protocol, using the specific miRNA primers
(5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGAGCAAG-3′;
GenScript Biotechnology).
Quantitative PCR was performed using a
SYBR® Premix Ex Taq™ RT-PCR Kit (Takara Biotechnology
Co., Ltd.) and an ABI 7500 Fast Real-Time PCR system. Relative
expression mRNAs and miRNAs were evaluated using the
2−ΔΔCq method (25) and
normalized to β-actin and U6, respectively. The thermocycling
conditions for the qPCR were listed as follows: Initial
denaturation at 50°C for 2 min, 95°C for 10 min; followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. Primers sequences
were as follows: miR-1303 forward, 5′-ACGGGGTCTTGCTCTAAAAA-3′ and
reverse, 5′-CAGTGCGTGTCGTGGAGT-3′; DKK3 forward,
5′-CTGGGAGCTAGAGCCTGATG-3′; and reverse,
5′-TCATACTCATCGGGGACCTC-3′; β-actin forward,
5′-AGCCTCGCCTTTGCCGA-3′ and reverse, 5′-CTGGTGCCTGGGGCG-3′; U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Cell Counting kit 8 (CCK-8) assay
After 48 h transfection, cells were collected,
seeded (1×104 cells/well) in 96-well plates and
incubated in DMEM containing 10% FBS at 37°C for 24, 48 and 72 h.
According to the manufacturer's protocol of the CCK-8 detection kit
(Engreen Biosystem Co, Ltd.), CCK-8 solution (10 µl) was added to
the cells at the different time points and incubated for a further
4 h at 37°C. A microplate reader was used to measure the
proliferation of the cells at an absorbance of 450 nm.
Colony formation assay
After transfection, cells were trypsinized,
collected and added into 6-well plates (200 cells/well). The cells
were cultured in DMEM containing 10% FBS at 37°C under 5%
CO2 for 2 weeks, replacing the culture medium every 3
days. The cells were fixed with methanol at room temperature for 15
min, stained with 1% crystal violet for 30 min at room temperature
and images (containing >50 cells/colony) were captured using a
light microscope (magnification, ×100; Olympus Corporation). The
colony number was counted manually.
Wound healing assay
The transfected cells were incubated to 100%
confluence in 6-well plates. Subsequently, a straight line was
scratched using a sterile 100 µl pipette tip through the cell
monolayer. After removing the debris, the cells were cultured in
serum-free DMEM at 37°C under 5% CO2 for 24 h and then
images were captured with an Olympus BX51 light microscope
(magnification, ×100; Olympus Corporation). The degree of wound
closure was calculated using Image Pro-Plus version 6.0 (Media
Cybernetics, Inc.) with the following formula: Average width of
scratches = area of scratches/height of scratches; the degree of
wound closure = (Average width of scratches at 24 h/Average width
of scratches at 0 h) ×100%.
Invasion assay
Cellular invasion was determined in 24-well plates
using a Transwell chamber (8 µm pore; BD Biosciences) pre-coated
with Matrigel. Briefly, cells were transfected for 24 h as
aforementioned, suspended in serum-free DMEM and added into the
upper Transwell chamber. The medium including a chemoattractant
(10% FBS) was placed into the lower chamber. After incubation for
24 h at 37°C, the cells that invaded the chamber lower side were
fixed with 4% paraformaldehyde at room temperature for 30 min,
stained with 0.1% crystal violet at room temperature for 20 min,
and washed with PBS. Stained cells were counted and images were
captured with an Olympus BX51 light microscope (Olympus
Corporation) at ×200 magnification.
Luciferase reporter assay
The miR-1303 wild-type (WT) and mutant (MUT) target
sequences in the 3′-untranslated region (UTR) of the DKK3 gene were
obtained from Sangon Biotech Co., Ltd. WT and MUT DKK3 luciferase
reporter plasmids were generated through cloning the above two DNA
fragments into the psiCHECK-2 vector (Promega Corporation). The
cells were cultured at a density of 1.0×105 cells/well
in 24-well plates and co-transfected with DKK3-MUT or DKK3-WT
plasmids; and miR-1303 mimics or NC mimics using Lipofectamine 3000
(Invitrogen) at 37°C for 48 h, the Dual Luciferase Reporter Assay
kit (Promega Corporation) was utilized to detect the luciferase
activities, which were normalized to the corresponding
Renilla luciferase activity.
Western blotting
RIPA lysis buffer (Beyotime Institute of
Biotechnology) was used to extract cell lysates. Protein
concentration was quantified using Bicinchoninic Acid Assay, and
then 20 µg proteins were separated equally using 10% SDS-PAGE and
electro-transferred onto PVDF membranes. The membranes were blocked
using 5% non-fat milk at room temperature for 1 h and cultured
overnight with primary antibodies against DKK3 (cat. no.
10365-1-AP; dilution 1:1,000; ProteinTech Group, Inc.), β-catenin
(cat. no. 8480; dilution 1:1,000; Cell Signaling Technology, Inc.),
c-Myc (cat. no. 13987; dilution 1:1,000; Cell Signaling Technology,
Inc.), GAPDH (cat. no. 8884; dilution 1:1,000; Cell Signaling
Technology, Inc.) or cyclin D1 (cat. no. 55506; dilution 1:1,000;
Cell Signaling Technology, Inc.) at 4°C. After washing with TBS
buffer containing 0.05% Tween-20 (TBS-T), the membranes were
incubated with a horseradish peroxidase-conjugated goat anti-rabbit
secondary antibody (cat. no. SA00001-2, dilution 1:2,000;
ProteinTech Group, Inc.) at 25°C for 1 h. Subsequently, the
membranes were rinsed with blocking solution at room temperature
for 15 min. Protein bands visualized using an ECL system
(Immun-Star™ HRP chemiluminescent detection kit, Bio-Rad
Laboratories, Inc.). Densitometric analysis was determined by
Quantity One software (version 4.62; Bio-Rad Laboratories Inc.) and
normalized to the internal control, GAPDH.
Immunofluorescence microscopy
After 48 h of transfection, cells cultured
(1×106 cells/ml) on coverslips in a six-well plate for
24 h were fixed with 4% paraformaldehyde at 25°C for 15 min,
permeabilized with 0.05% Triton X-100 for 10 min, and blocked with
5% goat serum (Invitrogen; Thermo Fisher Scientific, Inc.) in PBS
for 1 h at room temperature. Cells were then incubated with primary
antibodies against β-catenin (1:100; Cell Signaling Technology,
Inc.) overnight at 4°C. Subsequently, the cells were cultured with
Alexa Fluor®488-conjugated donkey anti-rabbit IgG
antibody (cat. no. 4412, dilution 1:500 dilution; Cell Signaling
Technology, Inc.) in darkness for 1 h at room temperature. DAPI
(1:4,000; Beyotime Institute of Biotechnology) was applied to
counterstain the nuclei for 5 min at room temperature. The samples
were mounted on slides using an antifade reagent (Beijing Solarbio
Science & Technology Co., Ltd.) and examined under a
laser-scanning confocal microscope (Leica Microsystems GmbH) at
×200 magnification. The % positive β-catenin cells were analyzed
using the Image Pro-Plus software (version 6.0, Media Cybernetics,
Inc.,) and calculated using the following formula: % positive
β-catenin cells = (cells with nucleus positive for β-catenin/total
cells) ×100%.
Immunohistochemistry (IHC)
Tumor tissues were fixed with 4% paraformaldehyde
for 24 h at 25°C, dehydrated with an ethanol series and paraffin
embedded. The tissues were cut into 4 µm section. These sections
were deparaffinized with xylene and rehydrated using a graded
ethanol series. Antigen retrieval was performed at 100°C in citrate
buffer (pH 6.0). Each section was immersed in 3%
H2O2 at 25°C for 20 min to block endogenous
peroxidase activity, sealed with 5% normal goat serum (Beijing
Solarbio Biotechnology Co., Ltd.) at 23°C for 1 h and then
incubated with primary antibodies (ProteinTech Group, Inc.) against
Ki67 (cat. no. 27309-1-AP, dilution 1:4,000) and DKK3 (cat. no.
10365-1-AP, dilution 1:500) at 4°C overnight. Next, the sections
were cultured with horseradish peroxidase-conjugated secondary
antibodies (cat. no. ab150077; dilution 1:200; Abcam) for 30 min at
25°C, diaminobenzidine working solution was added for 6 min, and
counterstained with hematoxylin. Finally, the samples were
visualized and analyzed using a light microscope (Olympus
Corporation; magnification, ×200).
Tumor formation in nude mice
A total of 20 male BALB/C nude mice (age, 6 weeks;
weight, 18–22 g) were purchased from The Experimental Animal Center
of Tongji Hospital. The mice were randomly divided into two groups
(10/group), housed in a specific pathogen-free environment
(temperature, 21–23°C; humidity, 50–60%) under a 12-h light/dark
cycle and were allowed free access to water and food. DU145 cells
were harvested at the logarithmic growth period at a density of
1×107 cells/ml, and were subcutaneously injected into
the right side of the back ribs of the mice. When the tumor
diameter reached 6–8 mm, miR-1303 inhibitor/In
vivo-jetPEI® (Polyplus-transfection SA) or NC
inhibitor/In vivo-jetPEI® complexes were injected into
the xenograft tumor every 4 days for 30 days (a total of 8 times),
where In vivo-jetPEI® was the delivery agent
(concentration miRNA used, 10 µg; In vivo-jetPEI reagent used, 1.2
µl in 100 µl of 5% glucose per injection). The sizes of the tumors
were determined using a Vernier caliper every 6 days for 30 days.
The volume was calculated using the following formula: V = 0.5 L ×
W2, where L represents the long diameter and W
represents the short diameter. All mice were sacrificed on day 30
(the tumor volume <1,500 mm3; the tumor diameter
<1.5 cm). All procedures were approved by the Animal
Experimental Committee of Tongji Hospital.
Statistical analysis
The data are presented as the mean ± standard
deviation and were analyzed by SPSS software (version 13.0; SPSS
Inc.). All experiments were repeated at least three independent
times. Student's t-test was used to evaluate the differences
between two groups. One-way ANOVA followed by Tukey's test was
performed for multiple comparisons. The χ2 test was
applied to determine the association between miR-1303 expression
and clinical feature in patients with PCa. The relationship between
miR-1303 level and patient overall survival time was assessed by
the Kaplan-Meier method and log-rank test. P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-1303 is highly expressed in PCa
tissues and cell lines and predicts poor overall survival
To explore the role of miR-1303 in PCa, miR-1303
expression levels were determined in PCa tissues and cell lines
using RT-qPCR (Fig. 1). miR-1303 was
highly expressed in PCa tissues compared to the corresponding
adjacent normal tissues (P<0.001; Fig. 1A). Similarly, compared with human
normal prostate epithelial cell line RWPE-1, miR-1303 expression
levels were significantly higher in PCa cell lines PC-3, 22Rv1,
LNCAP and DU145 (P<0.05; Fig.
1B). As DU145 cells showed the greatest mean miR-1303
expression relative to RWPE-1 cells, they were chosen for all
subsequent experiments. These results suggested that miR-1303 was
highly expressed in PCa tissues and cell lines.
χ2 analyses were used to determine the
association between miR-1303 expression and clinical features in
PCa patients (Table I). The median
expression level of miR-1303 designated as a cut-off value was
utilized to divide the patients into miR-1303 high expression
(n=14) and miR-1303 low expression (n=16) groups. Statistical
analysis indicated that higher miR-1303 expression levels were
associated with higher Gleason scores (P=0.017) and more developed
tumor stages (P=0.024); no significant associations were made
between miR-1303 expression levels and age, prostate diameter,
lymph-node metastasis or the presentation of multiple lesions
(P>0.05; Table I). The
relationship between the miR-1303 level and patient overall
survival time was assessed by the Kaplan-Meier method and log-rank
test (Fig. 1C). Patients with high
levels of miR-1303 had a worse overall survival rate in comparison
with patients with low levels of miR-1303.
 | Table I.Association of miR-1303 and patient
clinicopathological features. |
Table I.
Association of miR-1303 and patient
clinicopathological features.
|
|
| miR-1303
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Number (n=30) | Low (n=16) | High (n=14) | χ2
P-value |
|---|
| Age (years) |
|
|
| 0.389 |
|
≥65 | 19 | 9 | 10 |
|
|
<65 | 11 | 7 | 4 |
|
| Prostate diameter
(cm) |
|
|
| 0.282 |
|
≥2.5 | 14 | 6 | 8 |
|
|
<2.5 | 16 | 10 | 6 |
|
| Gleason score |
|
|
| 0.017 |
| ≥7 | 19 | 7 | 12 |
|
|
<7 | 11 | 9 | 2 |
|
| Lymph-node
metastasis |
|
|
| 0.51 |
|
Yes | 11 | 5 | 6 |
|
| No | 19 | 11 | 8 |
|
| Tumor stage |
|
|
| 0.024 |
| T2 | 13 | 10 | 3 |
|
|
T3-T4 | 17 | 6 | 11 |
|
| Multiple
lesions |
|
|
| 0.919 |
|
Positive | 19 | 10 | 9 |
|
|
Negative | 11 | 6 | 5 |
|
miR-1303 promotes the proliferation,
migration and invasion of PCa cells
To investigate the role of miR-1303 in PCa cells,
DU145 cells were transfected with miR-1303 mimics, miR-1303
inhibitor or the corresponding negative controls, and CCK-8, colony
formation, wound healing and Matrigel assays were used to analyze
the effects on the cells (Fig. 2).
RT-qPCR analysis indicated that miR-1303 mimics significantly
increased miR-1303 expression in PCa cells (Fig. 2A), whereas miR-1303 inhibitor
decreased the expression levels of miR-1303 (both P<0.001),
which indicated the effective transfection of miR-1303 mimics or
inhibitor. The proliferation of PCa cells transfected with miR-1303
mimics or inhibitor was detected by CCK-8 and colony formation
assays. In the CCK-8 assay, the proliferation was enhanced in DU145
cells transfected with miR-1303 mimics, whereas the transfection of
miR-1303 inhibitor reduced the cell proliferation (both P<0.001;
Fig. 2B). Furthermore, colony
formation assay results demonstrated that the colony number of
DU145 cells was increased by miR-1303 mimics transfection, whereas
the number was decreased by miR-1303 inhibitor transfection (both
P<0.001; Fig. 2C). The migratory
ability of PCa cells was determined by wound healing assay
(Fig. 2D). miR-1303 mimics
accelerated wound closure in cell monolayers, whereas miR-1303
inhibitor suppressed wound closure compared with the respective
control groups (both P<0.05). The invasive ability of PCa cells
was measured by Matrigel assay (Fig.
2E). The invasive cell numbers were increased by miR-1303
mimics, but the number was reduced by miR-1303 inhibitor
transfection compared with respective control groups (P<0.001).
These results suggested that miR-1303 may promote the
proliferation, migration and invasion of PCa cells.
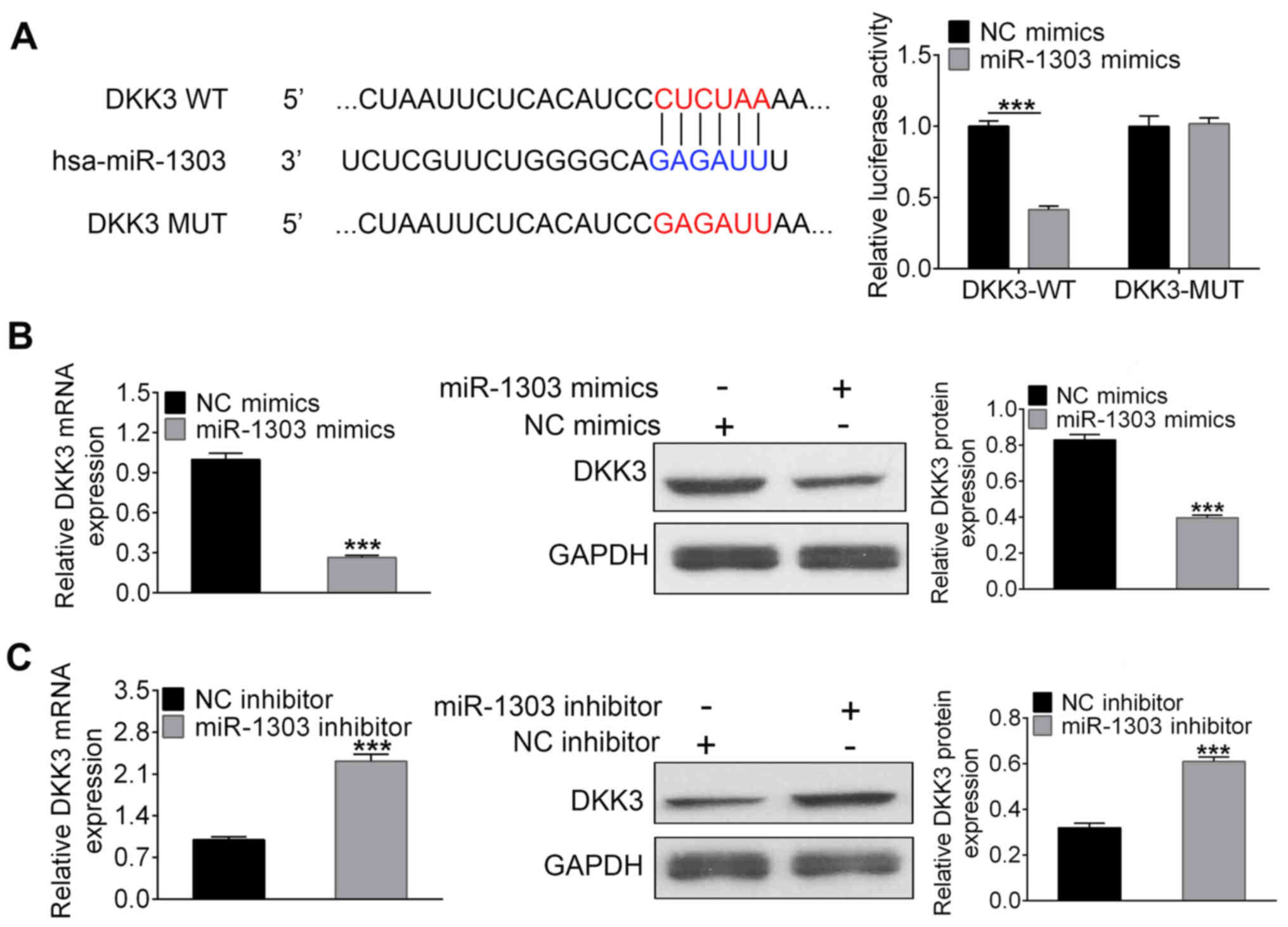
DKK3 is a direct target of
miR-1303
To investigate the molecular mechanism of miR-1303
in PCa tumorigenesis, TargetScan was utilized to identify putative
target genes of miR-1303. The results revealed that the 3′-UTR of
DKK3 contained a predicted target site for miR-1303. To verify this
relationship, luciferase reporter, RT-qPCR and western blotting
assays were performed (Fig. 3).
DKK3-WT or DKK3-MUT was co-transfected with miR-1303 mimics or NC
mimics into DU145 cells; miR-1303 mimics greatly decreased the
luciferase activity of DKK3-WT, but the luciferase activity in
DKK3-MUT had no obvious alterations (Fig. 3A). Thus, these results demonstrated
that DKK3 was a direct target of miR-1303. Furthermore, RT-qPCR and
western blotting showed that the mRNA and protein expression of
DKK3 were greatly decreased by miR-1303 mimics and increased by
miR-1303 inhibitors (all P<0.001; Fig. 3B and C, respectively). These data
demonstrated that miR-1303 negatively regulates DKK3
expression.
Knockdown of miR-1303 suppresses the
proliferation, migration and invasion of PCa cells by regulating
DKK3
Further analysis was performed to determine whether
the effects of miR-1303 on PCa cells were related to DDK3
expression (Fig. 4). The expression
levels of DKK3 mRNA in DU145 cells were successfully knocked down
by siDKK3 transfection compared with the control siRNA group
(P<0.001; Fig. 4A). Thus, the
transfection of siDKK3 was effective. In addition, western blotting
demonstrated that miR-1303 inhibitor considerably increased the
level of DKK3 as compared to NC inhibitor group, but siDKK3
co-transfection effectively inhibited the miR-1303 induced
upregulation of DKK3 (P<0.001; Fig.
4B). CCK-8 assay results indicated that the proliferation of
DU145 cells was decreased by miR-1303 inhibitor and this effect was
abolished after co-transfection with siDKK3 (P<0.001; Fig. 4C). In addition, the colony formation
assay demonstrated that the decrease of colony numbers of DU145
cells caused by miR-1303 inhibitor was reversed after siDKK3
co-transfection (P<0.001; Fig.
4D). The Matrigel assay and wound healing assay showed that
miR-1303 inhibitor suppressed the invasion and migration of DU145
cells, respectively, whereas siDKK3 and miR-1303 inhibitor
co-transfection significantly improved the invasive and migratory
abilities compared to miR-1303 alone transfections (P<0.01;
Fig. 4E and F). Collectively, these
data suggested that the knockdown of miR-1303 inhibited the
proliferation, migration and invasion of PCa cells by regulating
DKK3 expression.
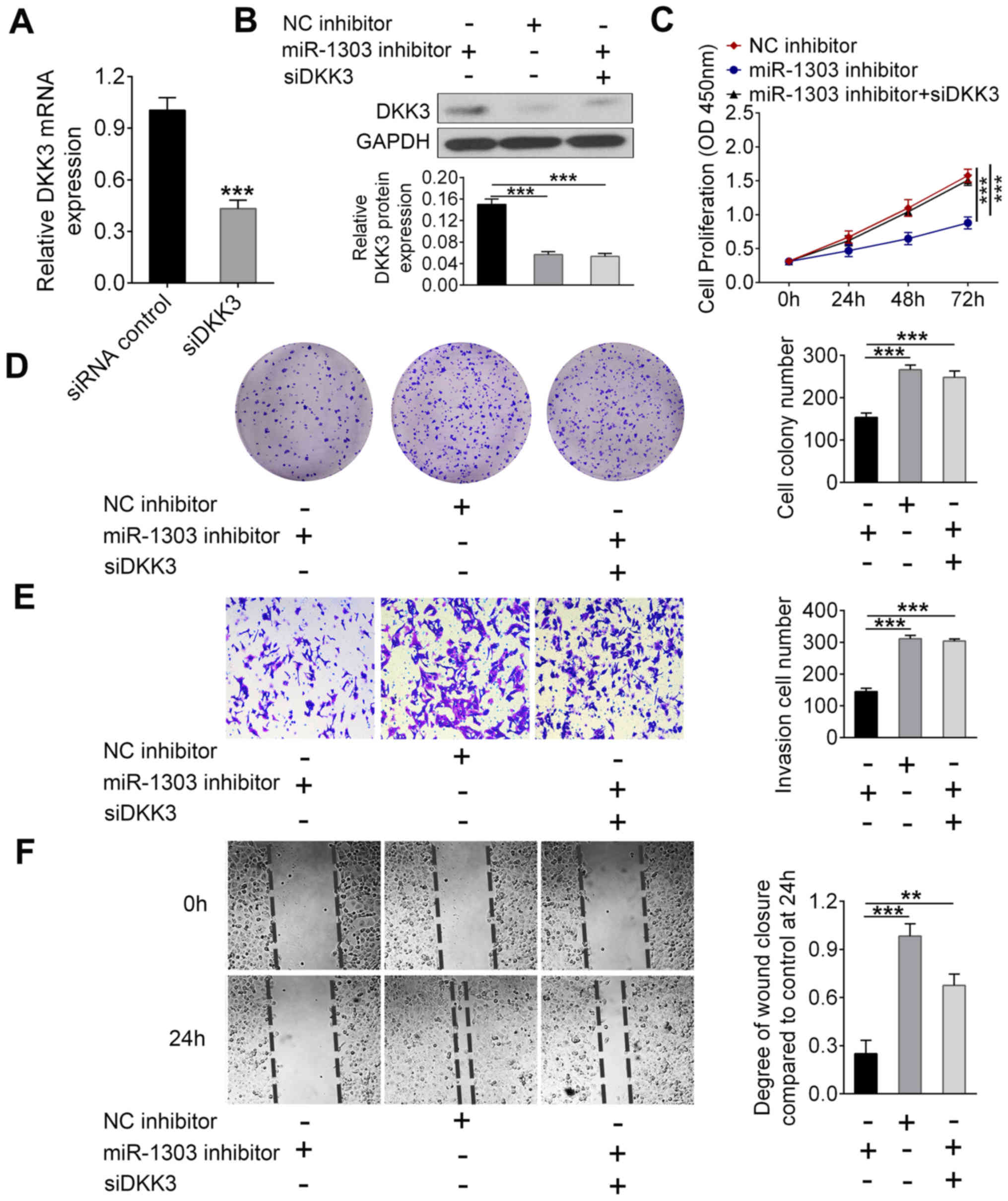 | Figure 4.miR-1303 promotes proliferation,
migration and invasion of prostate cancer cells through regulating
DKK3 expression. DU145 cells were transfected with a combination of
miR-1303 inhibitor, siDKK3 or controls. (A) Reverse
transcription-quantitative PCR was used to examine the level of
DKK3 mRNA expression. (B) Western blotting was used to detect the
protein expression level of DKK3. (C) Cell counting kit-8 and (D)
colony formation assays were utilized to detect the proliferation
of DU145 cells. (E) Matrigel assay and (F) wound healing assay
evaluated the cellular invasion and migration, respectively.
Magnifications, ×100 for (D) and (F); ×200 for (E). ***P<0.001,
**P<0.01 vs. miR-1303 inhibitor. DKK3, dickkopf Wnt signaling
pathway inhibitor 3; miR-1303, microRNA-1303; NC, negative control;
OD, optical density; siDKK3, siRNA for DKK3. |
Knockdown of miR-1303 inhibits the
activity of the Wnt/β-catenin signaling pathway by targeting DKK3
in PCa cells
Since it has been reported that DKK3 can function as
an inhibitor of Wnt/β-catenin signaling pathway (26), the effect of miR-1303 inhibitor on
the Wnt/β-catenin pathway was investigated by western blotting
(Fig. 5A). Compared with the NC
inhibitor, the expression of β-catenin and its down-stream
molecules c-Myc and cyclin D1, were decreased in DU145 cells after
transfection with miR-1303 inhibitor (P<0.001). However,
co-transfection with siDKK3 and miR-1303 inhibitor markedly
reversed the decreased protein expressions of β-catenin, cyclin D1
and c-Myc induced by miR-1303 inhibitor (P<0.001). Furthermore,
the distribution and expression of β-catenin in DU145 cells were
measured by immunofluorescence staining (Fig. 5B). miR-1303 inhibitor resulted in a
reduction in the number of DU145 cells with β-catenin expression in
the nucleus compared with the control group (P<0.001), which was
reversed after siDKK3 and miR-1303 inhibitor co-transfection
(P<0.01). Accordingly, miR-1303 knockdown effectively suppressed
the activity of Wnt/β-catenin signaling pathway by regulating
DKK3.
Knockdown of miR-1303 suppresses tumor
growth in PCa-engrafted mice
To investigate the effects of miR-1303 on tumor
growth in vivo, DU145 ×enografts were performed to construct
a mouse PCa-engrafted model, and the tumor was subsequently treated
with miR-1303 inhibitor or negative control by local injection. As
shown in Fig. 6A, the growth of
tumors was suppressed by miR-1303 inhibitor, which was demonstrated
by the lower volume of the tumors treated with miR-1303 inhibitor
compared with the control group. In addition, IHC analysis of
tumors revealed that the expression of Ki67, an immunohistochemical
proliferation marker, was significantly reduced by miR-1303
inhibitor (P<0.001; Fig. 6B).
DKK3 expression levels were also increased in tumors treated with
miR-1303 inhibitor compared with control tumors (P<0.001).
Therefore, knockdown of miR-1303 may suppress tumor growth in
PCa-engrafted model.
Discussion
Results from the present study indicated that
miR-1303 expression was increased in PCa tissues and cell lines.
The level of miR-1303 was closely associated with higher Gleason
scores and a more advanced tumor stage. Patients with PCa expressed
higher levels of miR-1303 also displayed a reduced overall survival
rate. High expression of miR-1303 promoted the proliferation,
migration and invasion of PCa cells. In addition, inhibition of
miR-1303 effectively inhibited the growth of PCa-engrafted tumors
in mice. These results indicated that miR-1303 may act as an
oncogene in the progression and development of PCa. Thus, miR-1303
may be a potential biomarker for PCa treatment.
miRNAs have been reported to be closely related with
various biological processes, including cell proliferation,
metastasis and apoptosis (12).
Hence, the role of miRNAs in cancers has been widely studied.
Recently, miR-1303 was reported to function as an oncogene in
numerous types of tumors. For example, Li et al suggested
miR-1303 induced neuroblastoma proliferation via targeting glycogen
synthase kinase 3 β and secreted frizzled related protein 1
(15). Furthermore, miR-1303
overexpression may promote the metastasis and growth of gastric
cancer cells by inhibiting claudin 18 expression (16). Additionally, miR-1303 may be a new
target for microsatellite instability in colorectal cancer
(17). In the present study, it was
determined that DKK3 was negatively regulated and identified as a
direct target of miR-1303.
DKK3 is designated as a tumor suppressor since its
expression is downregulated in many types of tumor, including lung
cancer, pancreatic cancer and PCa (27–30). In
agreement with previous reports, data from the present study
confirmed that DKK3 was downregulated in PCa cells. Additionally,
DKK3 was negatively regulated by miR-1303. It is well known that
DKK3 can exert a tumor suppressive function via inhibiting the
activity of Wnt/β-catenin pathway (31). As such, the interaction between
miR-1303 and the Wnt/β-catenin pathway was studied in this present
study.
The Wnt/β-catenin pathway is crucial for cellular
biological processes such as cell growth and differentiation
(32). After activating the
Wnt/β-catenin pathway, β-catenin levels increase in the cytoplasm,
followed by translocation to the nucleus (33). β-catenin then activates transcription
of target genes mainly containing cyclin D1 and c-Myc (34). Aberrant activation of the
Wnt/β-catenin pathway is frequently observed in many types of
cancers, which contributes to carcinogenesis (35). Wang et al (36) demonstrated that DKK3 could regulate
the Wnt/β-catenin pathway to induce the apoptosis of
cisplatin-resistant lung adenocarcinoma cells. The present study
indicated that the raised expression of miR-1303 in PCa cells may
inhibit DKK3 expression and activate the Wnt/β-catenin pathway,
thus promoting cell proliferation, migration and invasion.
In conclusion, the present study indicated that
miR-1303 is upregulated in PCa tissues and cell lines, and that
miR-1303 inhibition may effectively suppress the growth of PCa
tumors in mice. Moreover, miR-1303 may promote the proliferation,
migration and invasion of PCa cells through activating the
Wnt/β-catenin pathway via regulating DDK3. Therefore, miR-1303 may
be regarded as a new biomarker for PCa treatment. A limitation of
this study is the lack of analysis in other cell types. Hence,
further studies should use more cell lines to further validate the
effects of miR-1303 on prostate cancer in the future. Further
studies should also utilize more comprehensive basic research to
verify any other mechanisms of action that are involved with the
effects of miR-1303 on PCa.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL and WDZ performed the experiments and wrote the
manuscript. HYJ collected and analyzed the experimental data. ZDX
analyzed the experimental data and revised the manuscript. LW
designed the experiments and approved the final version
manuscript.
Ethics approval and consent to
participate
The Human Research Ethics Committee of Tongji
Hospital approved this experiment. Informed consent was obtained
from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoter A, Rizk S and Naim HY: The multiple
roles and therapeutic potential of molecular chaperones in prostate
cancer. Cancers (Basel). 11(pii): E11942019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salinas CA, Tsodikov A, Ishak-Howard M and
Cooney KA: Prostate cancer in young men: An important clinical
entity. Nat Rev Urol. 11:317–323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhan F, Shen J, Wang R, Wang L, Dai Y,
Zhang Y and Huang X: Role of exosomal small RNA in prostate cancer
metastasis. Cancer Manag Res. 10:4029–4038. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Altwaijry N, Somani S and Dufes C:
Targeted nonviral gene therapy in prostate cancer. Int J
Nanomedicine. 13:5753–5767. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Di Nunno V, Gatto L, Santoni M, Cimadamore
A, Lopez-Beltran A, Cheng L, Scarpelli M, Montironi R and Massari
F: Recent advances in liquid biopsy in patients with castration
resistant prostate cancer. Front Oncol. 8:3972018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shore ND, Karsh L, Gomella LG, Keane TE,
Concepcion RS and Crawford ED: Avoiding obsolescence in advanced
prostate cancer management: A guide for urologists. BJU Int.
115:188–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Akram ON, Mushtaq G and Kamal MA: An
overview of current screening and management approaches for
prostate cancer. Curr Drug Metab. 16:713–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parikh RR, Byun J, Goyal S and Kim IY:
Local therapy improves overall survival in patients with newly
diagnosed metastatic prostate cancer. Prostate. 77:559–572. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bryzgunova OE, Konoshenko MY and Laktionov
PP: MicroRNA-guided gene expression in prostate cancer: Literature
and database overview. J Gene Med. 20:e30162018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gurtan AM and Sharp PA: The role of miRNAs
in regulating gene expression networks. J Mol Biol. 425:3582–3600.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharma N and Baruah MM: The microRNA
signatures: Aberrantly expressed miRNAs in prostate cancer. Clin
Transl Oncol. 21:126–144. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu Y, Gu L, Li Y, Lin X, Shen H, Cui K,
Chen L, Zhou F, Zhao Q, Zhang, et al: miR-148a inhibits colitis and
colitis-associated tumorigenesis in mice. Cell Death Differ.
24:2199–2209. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei H, Cui R, Bahr J, Zanesi N, Luo Z,
Meng W, Liang G and Croce CM: miR-130a deregulates PTEN and
stimulates tumor growth. Cancer Res. 77:6168–6178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Z, Xu Z, Xie Q, Gao W, Xie J and Zhou
L: miR-1303 promotes the proliferation of neuroblastoma cell
SH-SY5Y by targeting GSK3β and SFRP1. Biomed Pharmacother.
83:508–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang SJ, Feng JF, Wang L, Guo W, Du YW,
Ming L and Zhao GQ: miR-1303 targets claudin-18 gene to modulate
proliferation and invasion of gastric cancer cells. Dig Dis Sci.
59:1754–1763. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
El-Murr N, Abidi Z, Wanherdrick K, Svrcek
M, Gaub MP, Fléjou JF, Hamelin R, Duval A and Lesuffleur T: MiRNA
genes constitute new targets for microsatellite instability in
colorectal cancer. PLoS One. 7:e318622012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao L, Chen B, Li J, Yang F, Cen X, Liao Z
and Long X: Wnt/β-catenin signaling pathway inhibits the
proliferation and apoptosis of U87 glioma cells via different
mechanisms. PLoS One. 12:e01813462017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schneider JA and Logan SK: Revisiting the
role of Wnt/β-catenin signaling in prostate cancer. Mol Cell
Endocrinol. 462:(Pt A). 3–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kypta RM and Waxman J: Wnt/β-catenin
signalling in prostate cancer. Nat Rev Urol. 9:418–428. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Q, Ye L, Guo W, Wang M, Huang S and
Peng X: PHF21B overexpression promotes cancer stem cell-like traits
in prostate cancer cells by activating the Wnt/β-catenin signaling
pathway. J Exp Clin Cancer Res. 36:852017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Flores ML, Castilla C, Gasca J, Medina R,
Pérez-Valderrama B, Romero F, Japón MA and Sáez C: Loss of PKCδ
induces prostate cancer resistance to paclitaxel through activation
of Wnt/β-Catenin pathway and Mcl-1 accumulation. Mol Cancer Ther.
15:1713–1725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
24
|
Katase N, Nishimatsu SI, Yamauchi A,
Yamamura M and Fujita S: DKK3 overexpression increases the
malignant properties of head and neck squamous cell carcinoma
cells. Oncol Res. 26:45–58. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moskalev EA, Luckert K, Vorobjev IA,
Mastitsky SE, Gladkikh AA, Stephan A, Schrenk M, Kaplanov KD,
Kalashnikova OB, Pötz O, et al: Concurrent epigenetic silencing of
wnt/beta-catenin pathway inhibitor genes in B cell chronic
lymphocytic leukaemia. BMC Cancer. 12:2132012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Forsdahl S, Kiselev Y, Hogseth R, Mjelle
JE and Mikkola I: Pax6 regulates the expression of Dkk3 in murine
and human cell lines, and altered responses to Wnt signaling are
shown in FlpIn-3T3 cells stably expressing either the Pax6 or the
Pax6(5a) isoform. PLoS One. 9:e1025592014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shien K, Tanaka N, Watanabe M, Soh J,
Sakaguchi M, Matsuo K, Yamamoto H, Furukawa M, Asano H, Tsukuda K,
et al: Anti-cancer effects of REIC/Dkk-3-encoding adenoviral vector
for the treatment of non-small cell lung cancer. PLoS One.
9:e879002014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo Q and Qin W: DKK3 blocked
translocation of β-catenin/EMT induced by hypoxia and improved
gemcitabine therapeutic effect in pancreatic cancer Bxpc-3 cell. J
Cell Mol Med. 19:2832–2841. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Al Shareef Z, Kardooni H, Murillo-Garzon
V, Domenici G, Stylianakis E, Steel JH Rabano M, Gorroño-Etxebarria
I, Zabalza I, Vivanco MD, et al: Protective effect of stromal
Dickkopf-3 in prostate cancer: Opposing roles for TGFBI and ECM-1.
Oncogene. 37:5305–5324. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Veeck J, Bektas N, Hartmann A, Kristiansen
G, Heindrichs U, Knüchel R and Dahl E: Wnt signalling in human
breast cancer: Expression of the putative Wnt inhibitor Dickkopf-3
(DKK3) is frequently suppressed by promoter hypermethylation in
mammary tumours. Breast Cancer Res. 10:R822008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chiurillo MA: Role of the Wnt/beta-catenin
pathway in gastric cancer: An in-depth literature review. World J
Exp Med. 5:84–102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ashihara E, Takada T and Maekawa T:
Targeting the canonical Wnt/β-catenin pathway in hematological
malignancies. Cancer Sci. 106:665–671. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Arend RC, Londono-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Z, Ma LJ, Kang Y, Li X and Zhang XJ:
Dickkopf-3 (Dkk3) induces apoptosis in cisplatin-resistant lung
adenocarcinoma cells via the Wnt/β-catenin pathway. Oncol Rep.
33:1097–1106. 2015. View Article : Google Scholar : PubMed/NCBI
|