Introduction
Non-syndromic cleft lip or palate (NSCL/P) is the
most common congenital birth defect, with an incidence of 1–2%
(1). The interaction between
environmental risks and genetic factors is widely accepted as the
molecular pathological basis of congenital cleft palate (2). Palate fusion is regulated by a variety
of cytokines, signaling molecules and signaling pathways, including
transforming growth factor-β (TGF-β), fibroblast growth factor
(FGF), Wnt and sonic hedgehog (Shh) signaling pathways (3–6).
Previous studies have demonstrated that knocking out the
Tgfb3 gene can inhibit palatal plate fusion in normal mice
(7). In the palatal process
epithelium, Wnt/β-catenin signaling can control palatal fusion
through Tgfb3. It serves a bidirectional regulatory role
with TGF-β in regulating cell proliferation, differentiation and
apoptosis (7,8). In the early stages of embryonic
development, exogenous teratogenic agents can affect the normal
palate development by altering the expression of a number of
signaling factors, including Gli3, FGF, Tbx1 and TGF-β. If the
palatal process is not raised during development or the bilateral
palatal process cannot be contacted, it will lead to cleft palate
on a cellular level (9–12). Retinoic acid (RA) is a derivative of
vitamin A and is considered to be a common teratogenic factor
(13,14). The function of retinoic acid is
mediated through binding to its corresponding membrane receptor,
transporting retinoic acid binding protein into the nucleus through
the cytoplasm, where it regulates of gene transcription. It has
been previous demonstrated that Gli3, FGF, TBX1 and Hoxa2
expression are altered by retinoic acid (15–17)
which results in palatal mesenchymal cell apoptosis, resulting in
cleft palate.
Gene expression is spatiotemporal during palatal
formation (18). Illustrating the
temporal and spatial specificity of gene expression during palatal
formation, may help to establish the pathological mechanism of
cleft palate. Epidemiological studies have previously revealed that
platelet-derived growth factor receptor (PDGFR)-α is closely
related to growth and development, and its absence may lead to a
series of congenital malformations, including NSCL/P (19,20). The
PDGFR-α signaling pathway is one of a number of signaling pathways
involved in the regulation of craniofacial development, and PDGFR-α
plays an important role in human embryonic development, as well as
normal physiological activities (21–23). The
lack of PDGFR-α expression in neural crest cells can lead to
obstruction of the palate and the nasal septum in mouse models,
resulting in facial bone structure and cartilage abnormalities
(24). The platelet-derived growth
factor (PDGF) family consists of two receptor genes, PDGFR-α and
PDGFR-β. PDGFR-α is involved in cellular responses, including
migration, proliferation and division (24). Mice with disrupted PDGF-α signaling
display cheek fissures, bone deformities and cleft palate when they
are born, whilst those that do not survive typically die by day
10–15 during embryonic development (25).
During normal embryonic palatal development,
exposure to drugs or environmental chemicals may result in
alterations in palatal fusion that can cause cleft palate (26). There are several in vitro
models that can be used to assess the pathogenesis of cleft palate,
including mouse embryonic primary mesenchyme (MEPM) cell culture
(27,28), organ and tissue cultures of palatal
shelves (29–32) and whole mouse embryo cultures
(33,34). Each of these three models has their
own advantages and disadvantages, which should be carefully
considered when deciding which model to use to investigate specific
experimental questions. MEPM cell culture is commonly used to
investigate how MEPM cells proliferate and differentiate. Palatal
shelves are composed of MEPM and medial edge (MEE) cells. Organ
cultures of palatal shelves allow the study of the interactions
between MEPM and MEE cells during palatal fusion (28,35). The
whole mouse embryo culture allows the study of the mechanisms
underlying palatal development (36,37).
The present study reported a novel method of
cultivating palate cells from gestation day (GD) 13.5 mouse fetuses
in DMEM/F12 medium containing 10% fetal bovine serum. The palates
maintained in rotary organ culture displayed substantial growth and
fused similarly in vitro as in vivo mouse fetuses.
Furthermore, the expression of PDGFR-α was assessed and compared
between in vitro and in vivo palatal shelves to
further confirm the reliability of the rotary organ culture
method.
Materials and methods
Rotary organ culture of palatal
shelves
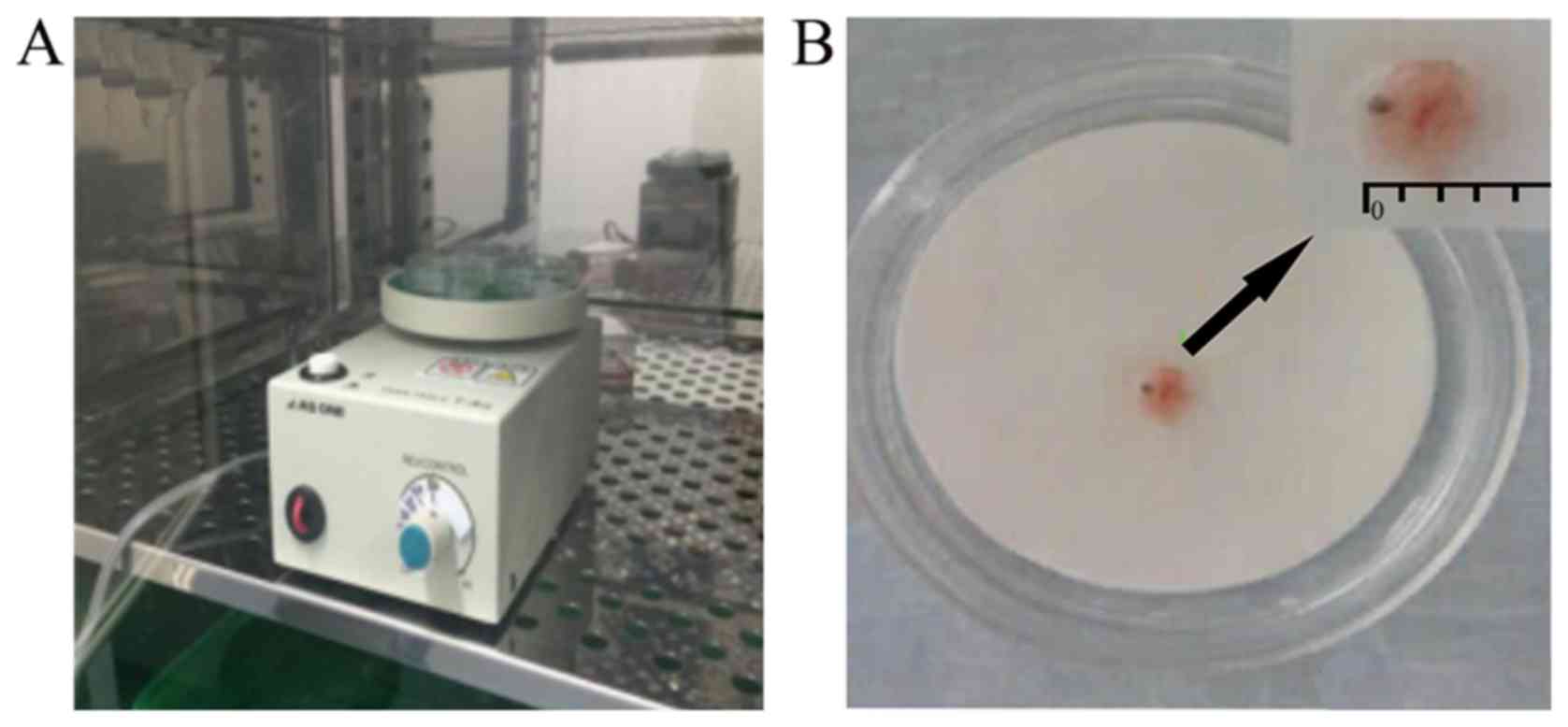
In the present study, electric rotary devices were
built based on the methods described by Shiota et al
(38). The organ culture instrument
used was designed based on the principle of the bacterial culture
instrument (Labstar 50; Shandong SCENKER Biotechnology Co., Ltd.)
used in the clinical laboratory of the Affiliated hospital of
Qingdao University (Shandong, China). The rotary table had an inner
diameter of 94 mm and culture dishes 3 cm in diameter were placed
onto the rotating table for the experiment. The rotary table was
able to accommodate a maximum of three dishes at any one time,
where the speed of rotation was adjusted to 20–25 rpm (Fig. 1).
In vivo and in vitro development of
the C57BL/6J mouse fetus palate
A total of 20 male and 32 female mice were used for
the present study (license no. HYXK20180116; Hua Fu Kang
Experimental Animal Center). The animals were housed at 22°C, 70%
humidity, with a 12-h light/dark cycle and were provided with
pellet food and tap water ad libitum. Specific pathogen-free
C57BL/6J male mice aged 7–8 weeks (weight, 22–25 g) and female mice
aged 6–7 weeks (weight, 17–20 g) were mated. The present study was
approved by the Qing Dao University Institutional Animal Care and
Use Committee (reference no. AHQU-MAL2018079). The mice were
subsequently housed at a male:female ratio of 1:2. The observation
of vaginal plugs the following morning was considered day 0 of the
embryo (GD 0.5). A total of 26 pregnant mice were euthanized on GD
13.5, and 2 were euthanized on each of GD 14.5, 15.0 and 15.5 by
cervical dislocation. Since they were utilized solely for breeding
purposes, none of the 20 male mice were harmed for the duration of
the present study and were given to researchers from other research
groups. Under sterile conditions, palatal explants were dissected
under a dissection microscope (magnification, ×2) using
micro-scissors. The jaw and tongue body were resected from the
horizontal left and right corners of the mouth, leaving the
bilateral palatal shelf exposed. The head was removed above the
eyes by horizontal incision. Briefly, a total of 195 palatal
explants were obtained. The procedure used for organ culture was
performed as previously described by Shiota et al (38). For in vivo experiments, 55
palatal explants at four different embryonic stages (GD 13.5, 14.5,
15.0 and 15.5) were collected. A total of six palatal explants from
each embryonic stage were fixed in 4% paraformaldehyde overnight at
4°C for immunohistochemistry (IHC). The remaining 31 palatal
explants were used for tissue protein extraction and western blot
analysis. On GD 13.5, the 140 palatal explant samples for in
vitro experimentation were removed and 6 palatal explant
samples were placed culture dishes 3 cm in diameter. These palatal
explant samples were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 1% penicillin-streptomycin
solution (Hyclone; GE Healthcare Life Sciences) and placed in
culture dishes on a rotary table (25 rpm) in an incubator at 37°C
with an atmosphere of 95% O2 and 5% CO2.
Palatal shelves were cultured in vitro for 0,
24, 36 or 48 h, similar to the four different embryonic stages (GD
13.5, 14.5, 15.0 and 15.5), respectively. After 0, 24, 36 or 48 h,
the palatal shelves were removed from culture and visualized under
a stereomicroscope (magnification, ×10; Leica M50; Leica
Microsystems GmbH) to image palatal development.
IHC staining
The fetal palatal shelves were fixed in 4%
paraformaldehyde for 48 h at 4°C. The samples were then embedded in
paraffin and were subsequently cut into 3-µm thick sections in the
coronal orientation. The tissue sections were then deparaffinized
with xylene at room temperature and rehydrated using a descending
ethanol series. Antigen retrieval was performed by incubating the
sections with citric acid buffer (pH 6.0; Beijing Solarbio Science
& Technology Co., Ltd.) for 30 min in a microwave at about
~60°C for antigen retrieval, followed by non-specific peroxidase
blocking using peroxidase blocking solution (cat no. ZLI-9310;
ZSGB-BIO; OriGene Technologies, Inc.) for 10 min at room
temperature. The slides were then incubated with primary rabbit
anti-mouse PDGFR-α monoclonal antibody (1:200; cat. no. ab32570;
Abcam) overnight at 4°C. Following the primary incubation, the
sections were washed with PBS and incubated with a secondary
horseradish peroxidase-conjugated goat anti-rabbit IgG antibody
(1:600; cat. no. ab205718; Abcam) at 37°C for 1 h. Immunoreactivity
was detected using an Histostain™-SP Kits (cat. no. SPN-9001;
ZSGB-BIO; OriGene Technologies, Inc.) and counterstained with
hematoxylin for 30 min at room temperature. Images were obtained
using an Olympus BX60 fluorescence microscope (magnification ×100;
Olympus Corporation).
Western blotting
Western blot analysis was performed to assess the
protein levels of PDGFR-α in palatal explants. Cells from the
palatal shelves were harvested at different time points (GD 13.5,
14.5, 15.0 and 15.5 in vivo or 0, 24, 36 and 48 h in
vitro) for protein extraction with mammalian protein extraction
reagent (Pierce; Thermo Fisher Scientific, Inc.). The total protein
content of the supernatant was determined using a bicinchoninic
acid protein assay kit (Beyotime Institute of Biotechnology). Equal
amounts of total protein (~50 µg) were electrophoresed on a 10%
SDS-PAGE gel and transferred onto a PVDF membrane (EMD Millipore)
in a Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad
Laboratories, Inc.) at 15 V for 30 min. The membrane was then
blocked for 2 h at room temperature with 5% skimmed milk in
Tris-buffered saline containing 0.05% Tween-20. Subsequently, the
membrane was incubated 4°C overnight with the following primary
antibodies: Anti-PDGFR-α (1:2,000; cat. no. ab32570; Abcam) and
monoclonal anti-GAPDH (1:5,000; cat. no. ab181602; Abcam).
Membranes were then incubated with a peroxidase-conjugated
secondary antibody (1:5,000; cat. no. ab205718; Abcam) for 1 h at
37°C. Protein bands were visualized using an ECL1 Detection kit
(cat. no. RPN2109; GE Healthcare) according to the manufacturer's
instructions. Protein expression was quantified using Gel-Pro
Analyzer software (version 3.1; Media Cybernetics, Inc.) with GAPDH
as the loading control. The grey level was analyzed using ImageJ
v1.8.0 software (National Institutes of Health).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 18.0; IBM Corp.). Comparisons were performed
using one-way ANOVA followed by Fisher's Least Significant
Difference post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of PDGFR-α protein was
observed by IHC
At GD 13.5 in vivo (Fig. 2A) and after 0 h cultivation in
vitro (Fig. 3A), PDGFR-α was
expressed in the vertical growth of the palatal shelf, primarily in
the epithelial region. The palatal mesenchyme was concentrated in
the central and lateral regions of the palatal shelf. The
expression of PDGFR-α in oral epithelium adjacent to palatal frame
was higher compared with that in nasal epithelium. The palatal
mesenchyme was concentrated in the central and lateral regions of
the palatal shelf (Figs. 2A and
3A). However, little or no PDGFR-α
expression was observed in the mesenchyme near the nasal cavity
epithelium. Interestingly, no notable expression of PDGFR-α was
observed in the apical palatal frame at GD 13.5 in vivo
(Fig. 2A), but a higher level of
expression was found in the apical part after 0 h cultivation in
vitro compared with at GD 13.5 in vivo (Figs. 2A and 3A).
It was observed that PDGFR-α was partially expressed
on the surface of palatal epithelium on GD 13.5 (Fig. 3A). With the elevation of palatal
frame, PDGFR-α was expressed in the palatal process, oral and nasal
palatal epithelial cells on GD 14.5 in vivo (Fig. 2B) and after 24 h cultivation in
vitro (Fig. 3B). The expression
of PDGFR-α in palatal mesenchymal tissue on GD 14.5 in vivo
(Fig. 2B) was more pronounced
compared with that in vitro (Fig.
3B).
At GD 15.0 in vivo (Fig. 2C) and in vitro for 36 h
(Fig. 3C), a marked reduction in the
expression of PDGFR-α was observed compared with GD14.5 in
vivo (Fig. 2B) or in
vitro for 24 h (Fig. 3B),
respectively. The expression of PDGFR-α in palatal frame epithelium
was limited to the middle palatal crest epithelium and its adjacent
palatal nasal epithelium. In the mesenchymal tissue, the expression
of PDGFR-α was mainly concentrated near the epithelium on both
sides of the palatal crest, but the expression level was low in the
mesenchyme near the oral epithelium near the palatal frame
(Figs. 2C and 3C). It was observed that the expression of
PDGFR-α in the triangular region composed of the middle palatal
crest epithelium and adjacent nasal palatal epithelium but not in
the oral epithelium near the palatal frame (Fig. 2C).
The expression of PDGFR-α in the epithelium and
mesenchyme of the palatal shelf was lower at GD 15.5 in vivo
(Fig. 2D) and after 48 h culture
in vitro (Fig. 3D) compared
with those on GD15.0 (Figs. 2C and
3C). The region of PDGFR-α
expression in the mesenchyme was primarily concentrated in the
middle of the palatal shelf and expression was weak. There was some
PDGFR-α expression in the nasal epithelium adjacent to the palatal
shelf, but no expression was observed in the oral epithelium
(Fig. 2D).
The expression of PDGFR-α displayed a continuous and
dynamic trend across the different stages of development of the
mouse embryonic palate. At 0 h of in vitro culture (GD 13.5
in vivo), PDGFR-α was primarily expressed in the palatal
shelf (Fig. 3A). After 24 h
cultivation in vitro (GD 14.5 in vivo), PDGFR-α
gradually covered the oral and nasal epithelium and mesenchymal
cells of the palate (Fig. 3B).
Starting with GD15.0, the expression of PDGFR-α decreased gradually
in the palatal frame, and only in the middle palatal crest
epithelium or adjacent nasal epithelium (Fig. 3C). With the fusion of palate in
GD15.5, the epithelium of middle palatal crest disappeared and the
expression of PDGFR-α was reduced in the palate (Fig. 3D).
Expression of PDGFR-α protein as
observed by western blotting
The expression of PDGFR-α protein was analyzed in
vivo and in vitro at different stages of palatal bone
development by western blot analysis (Fig. 4). The expression profiles of PDGFR-α
following in vivo and in vitro cultivation exhibited
comparable patterns, with peaks observed following 24 h culture
in vitro and GD 14.5 in vivo. The results of western
blot and immunohistochemistry exhibited consistent trends.
Observation of in vitro cultured
palatal shelves
On GD13.5, the nasal floor structure could be
observed in the oral cavity (Fig.
5A). Stereomicroscopy results revealed that after 24 h culture
in vitro, the bilateral palatal frames gradually gathered in
the middle, where the distance was markedly shortened compared with
that observed in GD13.5 (Fig. 5A and
B). After 36 h culture (Fig.
5C), all 36 pairs of palatal shelves were found to have made
contact. Histologically, 22 (61.1%) displayed mesenchymal fusion
and 14 (38.9%) displayed epithelial fusion without mesenchymal
confluence. After 48 h culture, contact between the palatal shelves
was observed in all 36 pairs of palatal shelves. Histologically, 36
(100%) displayed mesenchymal confluence and disappearance of the
midline epithelial seam (Fig.
5D).
Discussion
Palate development in mammals is a complex process
that involves the vertical formation, rotation and horizontal
proximity of the palatal shelf; the formation and degeneration of
epithelial ridges in the median palatine process; the adhesion of
epithelial cells and the fusion of mesenchymal cells (9,39). A
number of previous studies have reported that genetic and
environmental interactions contribute to the development of
congenital cleft lip and palate (18,40).
PDGFR-α is a cell surface receptor tyrosine kinase
for PDGFs (23). The human PDGFR-α
genes are located on chromosome 4q12 (41,42).
Under physiological conditions, PDGF interacts with its
corresponding receptor PDGFR-α to form and activate the
ligand-receptor complex. PDGFR-α signaling activation can activate
related genes, including Tbx1, SATB2 and Gli3
(15–17) and a number of signaling pathways,
including WNT, TGF, FGF and SHH, resulting in a variety of
physiological activities within the cells (23,43–45).
Disruption of PDGFR-α in zebra fish by microRNA 140
was found to cause craniofacial abnormalities, including cleft
palate (23,46). Previous studies in PDGFR-α knockout
mice have provided evidence supporting a significant role for
PDGFR-α in palatal development (47–49).
Qian et al (50) demonstrated
that the correct timing and level of PDGFR-α expression are crucial
for embryonic development. Furthermore, it was reported that
deletion of PDGFR-α caused developmental defects of multiple
endoderm- and mesoderm-derived structures, resulting in a complex
phenotype including orofacial cleft, spina bifida, rib deformities
and omphalocele in mice (51,52).
Results from studies utilizing animal models are not the only
evidence that indicates that PDGFR-α plays an important role in
cleft palate formation. In a previous study, 102 patients with
NSCL/P were examined by DNA genome and PCR sequencing analysis,
revealing seven mutations in PDGFR-α in nine patients, totaling to
an incidence of 8.8%. An incidence of 8.8% is significantly higher
than the mutation rate of the general population (1%; P<0.01)
(19,20). Therefore, the results obtained in the
present study further suggested that PDGFR-α plays an important
role in human embryonic palatal fusion.
PDGFR-α is an important regulator of embryonic
palatal development (23). However,
to the best of our knowledge, there have been no reports detailing
the spatiotemporal expression pattern of PDGFR-α in the development
of embryonic palatal shelves. In the present study, PDGFR-α was
detected in mouse palatal shelves at GD 13.5, 14.5, 15.0 and 15.5,
and the expression of PDGFR-α was highest at GD 14.5. These results
indicated that PDGFR-α displays a dynamic expression pattern in the
development of palatal shelves. The bilateral palatal process began
to fuse to form the middle palatal suture, where PDGFR-α expression
also began to decrease on GD15.0 and was mainly concentrated in the
middle palatal crest epithelium. On GD15.5, complete fusion was
observed in the specimens and PDGFR-α expression was largely
diminished. The expression patterns of PDGFR-α in the palatal
shelves were similar in vitro and in vivo, suggesting
that PDGFR-α serves a role in palatal fusion.
Current methods of cultivating palatal shelves in
vitro include metal fence culture (51), Trowell culture (32,53) and
other stationary culture methods (54,55). In
1990, the palatal suspension culture method was established by
Shiota et al (38). In this
method, the palatal shelves were cultured in a rotating culture
device, which displayed a number of advantages over the traditional
stationary culture method. Firstly, the method of scissoring the
palatal shelves was different. Palatal explants were dissected
under a dissection microscope using a microscopic shear and a
horizontal incision was made through the oral opening. The upper
part of the head was resected by making a second incision parallel
to the first incision, at the level of the eyeballs. Palatal
explants included palatal shelves and part of the maxillary
protrusion. The brain tissue, tongue and lower jaw were removed
with microscope forceps, following which the palatal shelves were
placed in a horizontal position in a culture dish (46,56).
This method described by Shiota et al (38) is easier to implement than other
methods, largely preserves the integrity of the palatal shelves and
ensures the survival rate of the palatal shelves in vitro
(57). The rotary culture device
rotates at a speed of 20–25 rpm, rotating the palatal shelves with
the filter membrane in a petri dish at the same rate as the
converted rotary culture device. Finally, palatal explants, petri
dishes and the rotary culture devices are placed in a cell
incubator for culture. The power line is drawn out through the edge
of the cell incubator door. The edge of the incubator door has
rubber strips, ensuring that the incubator is airtight when the
power line passes through (51,58). The
palatal explants remain almost suspended due to changing gravity
vectors and no single gravity factor was found to influence any
dominant direction of growth. This maintains the differentiation
and migration of palatal shelf cells, which establishes the
three-dimensional culture model in vitro of palate shelves
in the laboratory (50,58). In addition, the growth, elevation,
contact and fusion of palatal shelves can be observed in
vitro using this method. In the present study, the
morphological development and fusion process of palatal shelves in
rotary culture were similar to that observed in vivo. The
spatiotemporal expression of PDGFR-α also confirmed the feasibility
of the rotary culture method used in the present study, The level
of PDGFR-α protein expression after 24, 36 and 48 h cultivation
in vitro were comparable with that after GD 14.5, 15.0 and
15.5 in vivo.
The method used in the present study was modified
from the suspension organ culture originally described by Shiota
et al (38). Firstly, palatal
explants were placed in petri dishes rather than in culture
bottles. Secondly, palatal explants and rotary culture devices were
cultured in cell incubators instead of bottles, consisted of
oxygen, nitrogen and carbon dioxide. The palatal fusion rate of the
method used in the current study was 100%, which appeared to
completely simulate in vivo palatal development, suggesting
that the method used in the current study may be easier to
implement than the method originally described by Shiota et
al (38).
The experimental method used in the current study
reduced the number of steps required to complete the experiment,
but also provided a practical in vitro animal model for
studying normal palate development, the pathogenesis of cleft
palate and screening of teratogenic agents. Furthermore, the
present study suggested that PDGFR-α participates in palatal
development.
Acknowledgements
Not applicable.
Funding
The project is supported by the Natural Science Fund
of Shandong province. (grant. no. ZR2015HM022; Shandong,
China).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
WLX contributed to the conception of the study. WLX
and GY contributed significantly to analysis and manuscript
preparation. GY and NZ carried performed the experiment and
designed the figures. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All protocols were approved by the Ethics Committee
of the Affiliated Hospital of Qingdao University (IACUC Approval
No. QDU 93-016; Shandong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dixon MJ, Marazita ML, Beaty TH and Murray
JC: Cleft lip and palate: Understanding genetic and environmental
influences. Nat Rev Genet. 12:167–178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jugessur A and Murray JC: Orofacial
clefting: Recent insights into a complex trait. Curr Opin Genet
Dev. 15:270–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li C, Lan Y and Jiang R: Molecular and
cellular mechanisms of palate development. J Dent Res.
96:1184–1191. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vieira AR, Avila JR, Daack-Hirsch S,
Dragan E, Félix TM, Rahimov F, Harrington J, Schultz RR, Watanabe
Y, Johnson M, et al: Medical sequencing of candidate genes for
nonsyndromic cleft lip and palate. PLoS Genet. 1:e642005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Riley BM, Mansilla MA, Ma J, Daack-Hirsch
S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dodé C,
Mohammadi M, et al: Impaired FGF signaling contributes to cleft lip
and palate. Proc Natl Acad Sci USA. 104:4512–4517. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu D and Helms JA: The role of sonic
hedgehog in normal and abnormal craniofacial morphogenesis.
Development. 126:4873–4884. 1999.PubMed/NCBI
|
|
7
|
Nawshad A, LaGamba D and Hay ED:
Transforming growth factor beta (TGFbeta) signalling in palatal
growth, apoptosis and epithelial mesenchymal transformation (EMT).
Arch Oral Biol. 49:675–689. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reynolds K, Kumari P, Sepulveda Rincon L,
Gu R, Ji Y, Kumar S and Zhou CJ: Wnt signaling in orofacial clefts:
Crosstalk, pathogenesis and models. Dis Model Mech.
12:dmm0370512019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao L, Yin J and Wu W: Long non-coding RNA
H19-mediated mouse cleft palate induced by
2,3,7,8-tetrachlorodibenzo-p-dioxin. Exp Ther Med. 11:2355–2360.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bleyl SB, Saijoh Y, Bax NA,
Gittenberger-de Groot AC, Wisse LJ, Chapman SC, Hunter J, Shiratori
H, Hamada H, Yamada S, et al: Dysregulation of the PDGFRA gene
causes inflow tract anomalies including TAPVR: Integrating evidence
from human genetics and model organisms. Hum Mol Genet.
19:1286–1301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suzuki A, Sangani DR, Ansari A and Iwata
J: Molecular mechanisms of midfacial developmental defects. Dev
Dyn. 245:276–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoshioka W and Tohyama C: Mechanisms of
developmental toxicity of dioxins and related compounds. Int J Mol
Sci. 20:E6172019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duester G: Retinoic acid synthesis and
signaling during early organogenesis. Cell. 134:921–931. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakabe M, Kokubo H, Nakajima Y and Saga Y:
Ectopic retinoic acid signaling affects outflow tract cushion
development through suppression of the myocardial Tbx2-Tgfβ2
pathway. Development. 139:385–395. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gendron-Maguire M, Mallo M, Zhang M and
Gridley T: Hoxa-2 mutant mice exhibit homeotic transformation of
skeletal elements derived from cranial neural crest. Cell.
75:1317–1331. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao XY and Tang SJ: Effects of phenytoin
on Satb2 and Hoxa2 gene expressions in mouse embryonic craniofacial
tissue. Biochem Cell Biol. 88:731–735. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohbayashi N, Shibayama M, Kurotaki Y,
Imanishi M, Fujimori T, Itoh N and Takada S: FGF18 is required for
normal cell proliferation and differentiation during osteogenesis
and chondrogenesis. Genes Dev. 16:870–879. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Setó-Salvia N and Stanier P: Genetics of
cleft lip and/or cleft palate: Association with other common
anomalies. Eur J Med Genet. 57:381–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ding H, Wu X, Boström H, Kim I, Wong N,
Tsoi B, O'Rourke M, Koh GY, Soriano P, Betsholtz C, et al: A
specific requirement for PDGF-C in palate formation and PDGFR-α
signaling. Nat Genet. 36:1111–1116. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Choi SJ, Marazita ML, Hart PS, Sulima PP,
Field LL, McHenry TG, Govil M, Cooper ME, Letra A, Menezes R, et
al: The PDGF-C regulatory region SNP rs28999109 decreases promoter
transcriptional activity and is associated with CL/P. Eur J Hum
Genet. 17:774–784. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoch RV and Soriano P: Roles of PDGF in
animal development. Development. 130:4769–4784. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Niederreither K and Dollé P: Retinoic acid
in development: Towards an integrated view. Nat Rev Genet.
9:541–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eberhart JK, He X, Swartz ME, Yan YL, Song
H, Boling TC, Kunerth AK, Walker MB, Kimmel CB and Postlethwait JH:
MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat
Genet. 40:290–298. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Smith CL and Tallquist MD: PDGF function
in diverse neural crest cell populations. Cell Adh Migr. 4:561–566.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stephenson DA, Mercola M, Anderson E, Wang
CY, Stiles CD, Bowen-Pope DF and Chapman VM: Platelet-derived
growth factor receptor alpha-subunit gene (Pdgfra) is deleted in
the mouse patch (Ph) mutation. Proc Natl Acad Sci USA. 88:6–10.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Higashihori N, Song Y and Richman JM:
Expression and regulation of the decoy bone morphogenetic protein
receptor BAMBI in the developing avian face. Dev Dyn.
237:1500–1508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Okello DO, Iyyanar PPR, Kulyk WM, Smith
TM, Lozanoff S, Ji S and Nazarali AJ: Six2 plays an intrinsic role
in regulating proliferation of mesenchymal cells in the developing
palate. Front Physiol. 8:9552017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iyyanar PPR and Nazarali AJ: Hoxa2
inhibits bone morphogenetic protein signaling during osteogenic
differentiation of the palatal mesenchyme. Front Physiol.
8:9292017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abbott BD: Embryonic midfacial palatal
organ culture methods in developmental toxicology. Methods Mol
Biol. 1965:93–105. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shin JO, Lee JM, Bok J and Jung HS:
Inhibition of the Zeb family prevents murine palatogenesis through
regulation of apoptosis and the cell cycle. Biochem Biophys Res
Commun. 506:223–230. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abbott BD and Pratt RM: Human embryonic
palatal epithelial differentiation is altered by retinoic acid and
epidermal growth factor in organ culture. J Craniofac Genet Dev
Biol. 7:241–265. 1987.PubMed/NCBI
|
|
32
|
Koch WE and Smiley GR: In-vivo and
in-vitro studies of the development of the avian secondary palate.
Arch Oral Biol. 26:181–187. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Wang J, Dai X, Ding Y and Li Y:
Prevention of retinoic acid-induced early craniofacial
abnormalities by vitamin B12 in mice. Cleft Palate Craniofac J.
48:355–362. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamada M, Yamamoto N, Ohgami S, Kanazawa
M, Harada J, Ohno N and Natsume N: The effect of sevoflurane on
developing A/J strain mouse embryos using a whole-embryo culture
system-the incidence of cleft lip in culture embryos. In Vitro Cell
Dev Biol Anim. 50:237–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dudas M, Li WY, Kim J, Yang A and
Kaartinen V: Palatal fusion-where do the midline cells go? A review
on cleft palate, a major human birth defect. Acta Histochem.
109:1–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Harris C and Hansen JM: In vitro methods
for the study of mechanisms of developmental toxicology.
Developmental and Reproductive Toxicology-a Practical Approach.
Hood RD: Taylor and Francis; Boca Raton: pp. 647–695. 2006
|
|
37
|
Harris C: Overview of in vitro models in
developmental toxicology. Methods Mol Biol. 889:105–113. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shiota K, Kosazuma T, Klug S and Neubert
D: Development of the fetal mouse palate in suspension organ
culture. Acta Anat (Basel). 137:59–64. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Takahara S, Takigawa T and Shiota K:
Programmed cell death is not a necessary prerequisite for fusion of
the fetal mouse palate. Int J Dev Biol. 48:39–46. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Watanabe T, Dakshinamurti K and Persaud
TV: Biotin influences palatal development of mouse embryos in organ
culture. J Nutr. 125:2114–221. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lan Y, Xu J and Jiang R: Cellular and
molecular mechanisms of palatogenesis. Curr Top Dev Biol.
115:59–84. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fredriksson L, Li H and Eriksson U: The
PDGF family: Four gene products form five dimeric isoforms.
Cytokine Growth Factor Rev. 15:197–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tallquist M and Kazlauskas A: PDGF
signaling in cells and mice. Cytokine Growth Factor Rev.
15:205–213. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu E, Palmer N, Tian Z, Moseman AP,
Galdzicki M, Wang X, Berger B, Zhang H and Kohane IS: Comprehensive
dissection of PDGF-PDGFR signaling pathways in PDGFR genetically
defined cells. PLoS One. 3:e37942008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jones AV and Cross NC: Oncogenic
derivatives of platelet-derived growth factor receptors. Cell Mol
Life Sci. 61:2912–2923. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Eberhart JK, He X, Swartz ME, Yan YL, Song
H, Boling TC, Kunerth AK, Walker MB, Kimmel CB, et al: MicroRNA
Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet.
40:290–298. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
47
|
Soriano P: The PDGF alpha receptor is
required for neural crest cell development and for normal
patterning of the somites. Development. 124:2691–2700.
1997.PubMed/NCBI
|
|
48
|
Ding H, Wu X, Boström H, Kim I, Wong N,
Tsoi B, O'Rourke M, Koh GY, Soriano P, Betsholtz C, et al: A
specific requirement for PDGF-C in palate formation and PDGFR-alpha
signaling. Nat Genet. 36:1111–1116. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu X, Bringas P Jr, Soriano P and Chai Y:
PDGFR-alpha signaling is critical for tooth cusp and palate
morphogenesis. Dev Dyn. 232:75–84. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qian C, Wong CWY, Wu Z, He Q, Xia H, Tam
PKH, Wong KKY and Lui VCH: Stage specific requirement of
platelet-derived growth factor receptor-α in embryonic development.
PLoS One. 12:e01844732017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rattanasopha S, Tongkobpetch S,
Srichomthong C, Siriwan P, Suphapeetiporn K and Shotelersuk V:
PDGFRa mutations in humans with isolated cleft palate. Eur J Hum
Genet. 20:1058–1062. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu SJ, He W, Shi B, Meng T, Li XY and Liu
YR: A preliminary study on the teratogenesis of dexamethasone and
the preventive effect of vitamin B12 on murine embryonic palatal
shelf fusion in vitro. J Zhejiang Univ Sci B. 9:306–312. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Brunet CL, Sharpe PM and Ferguson MW: The
distribution of epidermal growth factor binding sites in the
developing mouse palate. Int J Dev Biol. 37:451–458.
1993.PubMed/NCBI
|
|
54
|
Chen TC, Holick MF, Lokeshwar BL,
Burnstein KL and Schwartz GG: Evaluation of vitamin D analogs as
therapeutic agents for prostate cancer. Recent Results Cancer Res.
164:273–288. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu JW, Lin KH, Weber C, Bhalla S, Kelso
S, Wang K and Tang SY: An in vitro organ culture model of the
murine intervertebral disc. J Vis Exp. Apr 11–2017.(Epub ahead of
print). doi: 10.3791/55437. doi: 10.3791/55437.
|
|
56
|
Ke CY, Xiao WL, Chen CM, Lo LJ and Wong
FH: IRF6 is the mediator of TGFβ3 during regulation of the
epithelial mesenchymal transition and palatal fusion. Sci Rep.
5:127912015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Thomas AJ, Eberly LE, Davey Smith G and
Neaton JD; Multiple Risk Factor Intervention Trial (MRFIT) research
group, : ZIP-code-based versus tract-based income measures as
long-term risk-adjusted mortality predictors. Am J Epidemiol.
164:586–590. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Abbott BD, Diliberto JJ and Birnbaum LS:
2,3,7,8-Tetrachlorodibenzo-p-dioxin alters embryonic palatal medial
epithelial cell differentiation in vitro. Toxicol Appl Pharmacol.
100:119–131. 1989. View Article : Google Scholar : PubMed/NCBI
|