Introduction
Atherosclerosis is a chronic inflammatory and
vascular disease, which frequently causes heart disease and stroke
(1). Atherosclerosis is involved in
the accumulation of lipid-engorged macrophages in the
subendothelial layer of arterial vessels (2,3).
Endothelial dysfunction is one of the main contributors to
atherosclerosis (4) and results in a
phenotypic alteration that increases the expression of adhesion
molecules released by endothelial cells (ECs). Apoptosis of
endothelial cells plays an important role in the occurrence and
development of atherosclerosis (5).
MicroRNAs (miRNAs/miRs) are a group of small,
single-stranded, non-coding RNA molecules which influence the
synthesis of proteins via their interactions with target mRNAs
(6,7). It has been reported that miRNAs could
modulate a number of biological processes, including tumorigenesis,
development, invasion, metastasis and other biological
characteristics (8,9). Accumulating evidence suggests that
miRNAs serve as diagnostic and prognostic biomarkers of numerous
types of cancer (10,11). Some miRNAs have been reported to be
involved in atherosclerosis through regulating vascular cells
(12). Previous research indicated
that miRNAs were closely related to the pathogenesis of
atherosclerosis (13,14).
Previous studies reported that several miRNAs, such
as miR-34a, miR-217, and miR-146a, could regulate the proliferation
and differentiation of ECs and may also stimulate cellular
senescence, which can act as a trigger for endothelial dysfunction
(15–17). miR-144 is a conservative miRNA and
tumor suppressor gene that plays a role in tumor suppression in a
variety of cancers, such as bladder cancer (18), thyroid cancer (19), gastric cancer (20), colorectal cancer (21), ovarian cancer (22) and nasopharyngeal carcinoma (NPC)
(23). Moreover, previous studies
reported that miR-144 upregulation may inhibit the proliferation,
invasion and metastasis of cancer cells (21,24).
However, the specific function and mechanism of miR-144-5p in
atherosclerosis remain unclear. Apoptosis of endothelial cells
plays an important role in the occurrence and development of
atherosclerosis. Therefore, the present study aimed to investigate
the effects of miR-144-5p on human umbilical vein endothelial cells
(HUVECs) to explore the role of miR-144-5p in atherosclerosis.
Materials and methods
Cell culture
HUVECs were obtained from Shanghai Institute of Life
Sciences, Chinese Academy of Sciences. HUVECs were cultured in
medium 199 supplemented with 10% FBS, 1% penicillin and
streptomycin (Gibco; Thermo Fisher Scientific, Inc.), endothelial
cell growth supplement (Sigma-Aldrich; Merck KGaA) and epidermal
growth factor (10 ng/ml, Roche Diagnostics) in a humidified
atmosphere at 37°C with 5% CO2.
Cell transfection
Once cells reached 70–80% confluency, miR-144-5p
mimics (Guangzhou RiboBio Co., Ltd.) or the negative control of
miR-144-5p mimics (mimics-NC; Guangzhou RiboBio Co., Ltd.) were
transfected into HUVECs using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific Inc.) according to the
manufacturer's protocols. The transfection efficiency was detected
by reverse transcription-quantitative PCR (RT-qPCR), as described
below.
Flow cytometry analysis
After transfection, HUVECs were washed three times
with PBS and centrifuged at 1,000 × g for 5 min at 20°C. Harvested
cells were stained with 200 µl annexin V/FITC binding buffer
(Beyotime Institute of Biotechnology) and then incubated with 5 µl
propidium iodide (PI; Beyotime Institute of Biotechnology) for 25
min at room temperature without light. Cell apoptosis was analyzed
by flow cytometry (BD FACSCalibur™; Becton, Dickinson and Company)
and data were analyzed using FlowJo software (version 7.6.1; FlowJo
LLC). Each experiment was performed in triplicate.
Cell invasion assay
Cell invasion ability was detected using a Transwell
chamber (pore size, 8 µm; Costar; Corning Inc.) pre-coated with
Matrigel (1 mg/ml; BD Biosciences) at 4°C overnight. HUVECs
(2×104 cells/well) were briefly plated into the upper
chambers using human endothelial serum-free medium (cat. no.
11111044; Gibco; Thermo Fisher Scientific, Inc.) containing 0.5%
FBS. Medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS was added to the lower chamber as a
chemoattractant, stimulating cells to move to the lower chamber. At
24 h after incubation, the invasive cells on the lower chamber were
fixed with 4% paraformaldehyde at room temperature for 30 min.
Subsequently, the membrane was stained for 30 min with 0.1% crystal
violet at room temperature. Cells located on the top of membrane
were wiped with a cotton swab. The Transwell chamber was then
washed with 95% ethanol. A total of 10 fields of view were selected
randomly to count the number of cells under a light microscope
(magnification, ×200; Nikon Corporation). Each group was repeated
in triplicate and all experiments were repeated three times to
obtain average values.
Cell migration assay
A wound-healing assay was performed to detect cell
migration ability. HUVECs (3×105 cells/well) were seeded
in six-well plates. Cells were grown until 90% confluency before
each well was scraped with a 10 µl pipette tip to create a linear
region devoid of cells. Subsequently, the cells were washed with
PBS three times. Human endothelial serum-free culture medium (cat.
no. 11111044; Gibco; Thermo Fisher Scientific, Inc.) was then added
to cells and incubated at 37°C with 5% CO2. Wound
healing was monitored using a light microscope (magnification,
×100; Nikon Corporation) at 0 and 24 h after scraping. Percentage
of wound (%) = (Wound width at 24 h/Wound width at 0 h) ×100.
MTT assay
HUVECs were inoculated in a 96-well plate at a
density of 1×104 cells/well at 37°C with 5%
CO2 for 12, 24 and 48 h. Subsequently, MTT (20 µl; 5
mg/ml) was added to each well, and cells were incubated for 4 h. A
total of 150 µl DMSO was added to dissolve formazan crystals and
gently shaken for 10 min. The optical density of each sample was
determined with a microplate reader (Multiskan™ FC; Thermo Fisher
Scientific, Inc.) by measuring the absorbance at 490 nm. The
experiments were repeated at least three times.
Western blot analysis
Total protein was extracted from HUVECs using
radioimmunoprecipitation assay buffer containing 1 mM PMSF.
Bicinchoninic Acid Protein assay kit (Thermo Fisher Scientific,
Inc.) was then used to quantify protein concentration following
which the protein samples were separated by 10% SDS-PAGE (20
µg/lane) and transferred onto PVDF membranes. Subsequently, the
membrane was blocked with 5% skim milk for 1 h at room temperature
and incubated with primary antibodies against phosphorylated
(p)-AKT (cat. no. ab81283; 1:1,000; Abcam), AKT (cat. no. ab179463,
1:10,000, Abcam), p-PI3K (cat. no. ab182651, 1:1,000, Abcam), PI3K
(cat. no. ab86714, 1:1,000, Abcam), eNOS (cat. no. ab66127,
1:1,000, Abcam) or GAPDH (cat. no. ab181602, 1:10,000, Abcam) at
4°C overnight. The membrane was then incubated with horseradish
peroxidase-conjugated secondary antibody Goat Anti-Rabbit IgG
H&L (ab205718; 1:2,000; Abcam), Goat Anti-Mouse IgG H&L
(ab205719; 1:2,000; Abcam) and for 2 h at room temperature. The
GAPDH was used as internal reference. Afterwards, the protein bands
were detected and visualized using an enhanced chemiluminescence
method (RapidStep™ ECL Reagent; Merck KGaA). The densities of bands
were semi-quantitively analyzed using ImageJ (version 4.0; National
Institutes of Health).
RT-qPCR assay
Total RNA was extracted from HUVECs using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) following the manufacturer's protocols. Total RNA
concentration was detected by NanoDrop™ 2000 (Thermo Fisher
Scientific, Inc.) and RNA samples were subsequently stored at
−80°C. cDNA synthesis was performed using the RevertAid™ First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The
temperature protocol was as follows: 42°C for 30 min and 85°C for 5
min. RT-qPCR was performed using a QuantiFast SYBR Green PCR kit
(Qiagen GmbH) in accordance with the manufacturer's protocols. The
following primers were used: U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; miR-144-5p forward,
5′-GCGCGAATTCGAGATCTTAACAGACCCTAGCTC-3′ and reverse,
5′-GCGCGGATCCGTGCCCTGGCAGTCAGTAGG-3′. Relative expression levels
were calculated using the 2−ΔΔCq method (25) after normalization with reference to
U6 expression. All experiments were performed in triplicate.
Dual-luciferase reporter assay
TargetScan 7.2 (http://www.targetscan.org/vert_72/) was used to
predict the potential targets of miR-144-5p. The results showed the
binding sites between miR-144-5p and the RICTOR 3′-untranslated
region (UTR). Subsequently, a dual-luciferase reporter assay was
performed to confirm the predicted binding sites. According to the
bioinformatics analysis results, a fragment of the RICTOR 3′-UTR
containing the wild type (WT) or mutant
(5′-AGAAGAGGUUAGGAACACCCCGA-3′) seed regions of miR-144-5p was
chemically synthesized in vitro and cloned into BamHI
and AscI sites of the pMIR-RB-REPORT™ vector (Guangzhou
RiboBio Co., Ltd.). HUVECs were then co-transfected with miR-144-5p
mimics or mimics-NC and WT-RICTOR or Mutant-RICTOR with
Lipofectamine 2000 for 48 h. After a 48-h incubation, relative
luciferase activity was calculated by normalizing to Renilla
luciferase activity using the Dual Luciferase Reporter Assay System
(Promega Corporation) following the manufacturer's protocol.
Statistical analysis
All data were are presented as the mean ± standard
deviation. All experiments were performed at least three times.
SPSS 19.0 software (IBM Corp.) was used to perform data analysis.
Comparisons between groups were analyzed using Student's t-test and
one-way analysis of variance followed by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effect of miR-144-5p on cell
proliferation and apoptosis of HUVECs
An RT-qPCR assay was first performed to detect the
transfection efficiency of miR-144-5p. RT-qPCR results showed that
miR-144-5p mimics significantly increased the relative expression
of miR-144-5p in HUVECs compared with the mimic-NC (Fig. 1A). To investigate the effects of
miR-144-5p on HUVEC proliferation and apoptosis, MTT and flow
cytometry assays were performed. MTT assay results demonstrated
that miR-144-5p mimics significantly reduced cell proliferation
compared to the mimic-NC group (Fig.
1B). Flow cytometry results indicated that miR-144-5p mimics
significantly promoted cell apoptosis of HUVECs compared with the
mimic-NC group (Fig. 1C and D).
Effect of miR-144-5p on cell migration
and invasion of HUVECs
In order to investigate the migration and invasion
of cells in different groups, wound healing and transwell assays
were performed. Results from the wound healing assay revealed that
miR-144-5p mimics significantly inhibited the cell migration
ability of HUVECs compared with the mimic-NC group (Fig. 2A and B). The transwell assay results
showed that miR-144-5p mimics significantly reduced the invasive
capacity and decreased the number of invasive HUVECs (Fig. 2C and D).
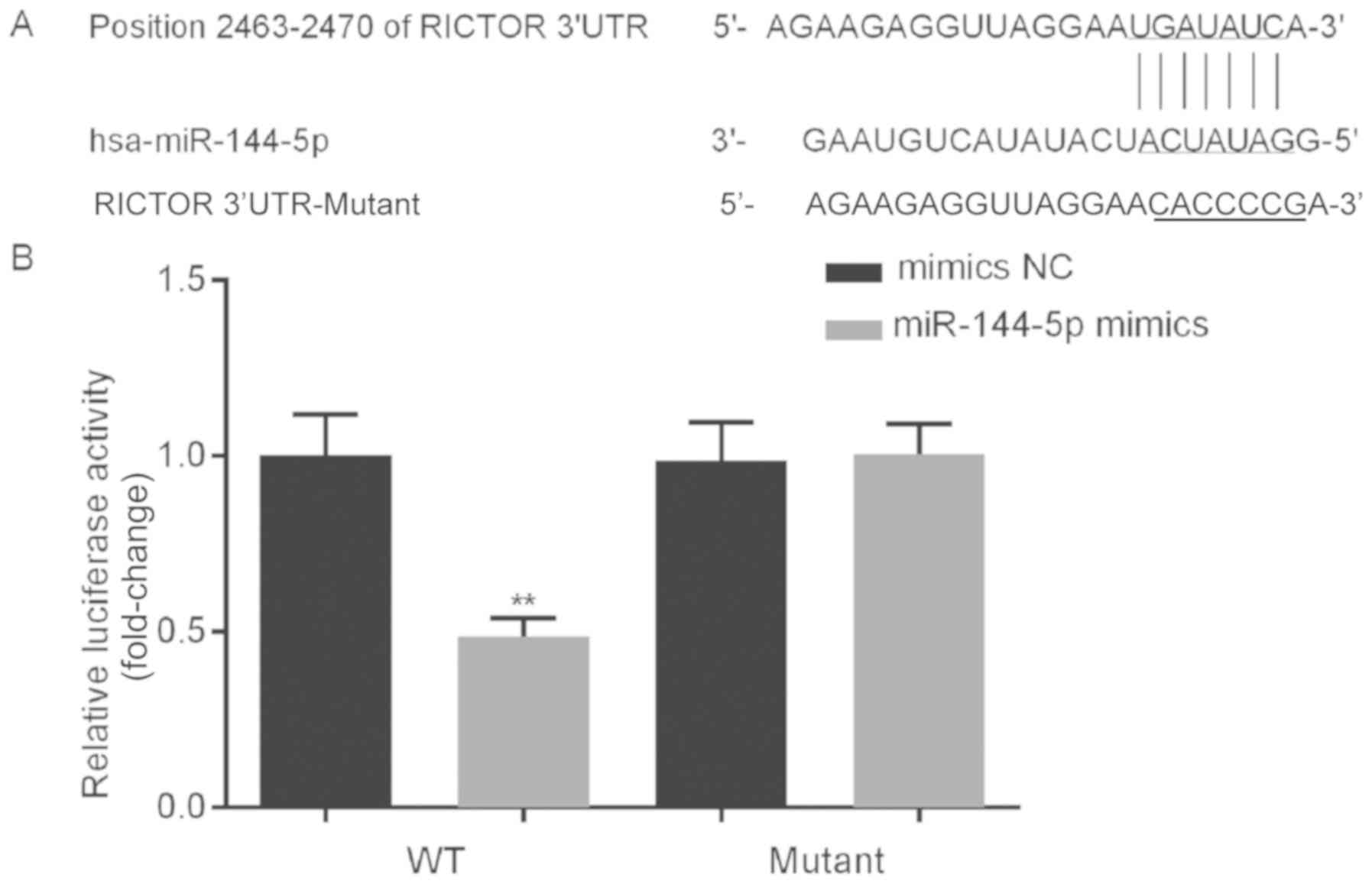
RICTOR is a direct target gene of
miR-144-5p
To investigate the potential role of miR-144-5p in
the growth of HUVECs, bioinformatics analysis was performed to
predict the potential targets of miR-144-5p. The results identified
binding sites between the 3′-UTR of RICTOR and miR-144-5p (Fig. 3A). A dual-luciferase reporter system
was used to determine if RICTOR was a direct target of miR-144-5p.
The results indicated that compared with the relative luciferase
activity of cells co-transfected with WT RICTOR 3′-UTR and
mimics-NC, the relative luciferase activity of cells co-transfected
with WT RICTOR 3′-UTR and miR-144-5p mimics significantly decreased
(Fig. 3B). Furthermore, when
comparing the relative luciferase activity of cells co-transfected
with mutant RICTOR 3′-UTR and mimics-NC, the relative luciferase
activity of cells co-transfected with mutant RICTOR 3′-UTR and
miR-144-5p mimics showed no significant changes (Fig. 3B). These data indicated that RICTOR
was a direct target gene of miR-144-5p.
miR-144-5p is associated with
PI3K-Akt-endothelial nitric oxide synthase (eNOS) pathway in
HUVECs
In order to further explore the specific mechanism
of the effect of miR-144-5p on HUVECs, the expression levels of
related proteins in the PI3K-Akt-eNOS pathway were detected.
Results from the western blot analysis indicated that miR-144-5p
mimics decreased the protein levels of phosphorylated (p)-PI3K,
p-Akt and eNOS in HUVECs (Fig. 4A and
B).
 | Figure 4.miR-144-5p mimics inhibit the protein
levels of p-PI3K, p-Akt and eNOS. (A) Western blot assay detected
the protein levels of p-PI3K, PI3K, p-Akt, Akt and eNOS, and GAPDH
was used as an internal control. (B) Ratios of p-PI3K/PI3K,
p-Akt/Akt, and eNOS/GAPDH were calculated. **P<0.01 vs. mimic-NC
eNOS, endothelial nitric oxide synthase; NC, negative control; miR,
microRNA. |
Discussion
Atherosclerosis is a relatively complex disease
affecting medium and large-sized arteries. Vascular injury plays
and important role in the pathogenesis of atherosclerosis, and
endothelial apoptosis plays a crucial role in endothelial
dysfunction and atherosclerosis lesion formation (26). Currently, atherosclerosis is being
extensively studied. Therefore, the current study aimed to explore
the role of miR-144-5p in atherosclerosis development through
investigating the effects of miR-144-5p on endothelial cells.
miR-144-5p mimics inhibited HUVEC proliferation and promoted cell
apoptosis. In addition, miR-144-5p mimics could decrease cell
migration and invasion of HUVECs. Moreover, RICTOR was a direct
target of miR-144-5p. Finally, miR-144-5p mimics could repress
PI3K-Akt-eNOS signaling pathway in HUVECs.
Previously, miRNAs have been confirmed to be
involved in numerous important physiological processes (27–30).
miRNAs are vital regulators for endothelial biology and
dysfunction. Deregulation of miRNA expression is involved in
endothelial angiogenesis, injury, inflammation and senescence
(31–33).
miR-144-5p has been studied in various cancers such
as renal cell carcinoma (34) and
miR-144-5p has also been studied in chronic periodontitis (35). A previous study demonstrated that
circulating miR-144-5p was associated with depressive disorders
(36). However, the role and
mechanism of miR-144-5p in atherosclerosis remains unclear.
Apoptosis of endothelial cells plays an important role in the
occurrence and development of atherosclerosis (26). The present study investigated the
effect of miR-144-5p on HUVECs to explore the role of miR-144-5p in
atherosclerosis.
The effect of miR-144-5p upregulation on the
biological behaviors of HUVECs was determined by performing MTT,
migration and invasion assays, and flow cytometry. The results
demonstrated that miR-144-5p mimics significantly inhibited HUVEC
proliferation, migration and invasion, and induced cell
apoptosis.
Subsequently, to explore the underlying mechanism of
miR-144-5p on HUVECs, bioinformatics analysis was used to predict
the potential targets of miR-144-5p. RICTOR was a direct target of
miR-144-5p in HUVECs. RICTOR is a crucial component of mTOR complex
2 (mTORC2), and the activation of mTORC2 depends on the presence of
the RICTOR protein. In addition, RICTOR is required for the
inactivating Ser473 phosphorylation on Akt (37). Furthermore, eNOS activity dysfunction
leads to a decrease in NO bioavailability, which results in
atherosclerosis (38). A previous
study revealed that the RICTOR/Akt/eNOS pathway played an important
role in regulating apoptosis and dysfunction in EC cells (39). Thus, the present study explored
whether the RICTOR/Akt/eNOS pathway was affected by miR-144-5p in
HUVECs. The results indicated that miR-144-5p mimics decreased the
protein levels of p-PI3K, p-Akt and eNOS in HUVECs. However, p-eNOS
levels were not detected in the current study, which is a
limitation.
In conclusion, miR-144-5p inhibited the
proliferation, invasion and migration of HUVECs, and induced
apoptosis through regulating the PI3K-Akt-eNOS signaling pathway,
at least partly by modulating RICTOR expression. Thus, miR-144-5p
might participate in the occurrence and development of
atherosclerosis through regulating the apoptosis of ECs. However,
the current study only included a preliminary analysis of
miR-144-5p in atherosclerosis. To completely elucidate the role of
miR-144-5p in atherosclerosis, further in-depth research is needed.
For example, the protective effects of RICTOR, PI3K, Akt, and eNOS
on HUVECs should be investigated. The role of miR-144-5p in
atherosclerosis should be studied in vivo. In addition, the
expression of miR-144-5p in patients with atherosclerosis and its
relationship with the clinicopathological features of patients can
be further explored.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study and revised the manuscript. WF
and JZ wrote the manuscript and collected the data. YS and RZ
searched the literature and interpreted the data. HZ collected the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lusis AJ: Atherosclerosis. Nature.
407:233–241. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hansson GK, Libby P and Tabas I:
Inflammation and plaque vulnerability. J Intern Med. 278:483–493.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Libby P and Hansson GK: Inflammation and
immunity in diseases of the arterial tree: Players and layers. Circ
Res. 116:307–311. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tabas I, García-Cardeña G and Owens GK:
Recent insights into the cellular biology of atherosclerosis. J
Cell Biol. 209:13–22. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mudau M, Genis A, Lochner A and Strijdom
H: Endothelial dysfunction: The early predictor of atherosclerosis.
Cardiovasc J Afr. 23:222–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thum T and Mayr M: Review focus on the
role of microRNA in cardiovascular biology and disease. Cardiovasc
Res. 93:543–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: MicroRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Natarelli L and Schober A: MicroRNAs and
the response to injury in atherosclerosis. Hamostaseologie.
35:142–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
96:776–780. 2006. View Article : Google Scholar
|
|
10
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song L, Peng L, Hua S, Li X, Ma L, Jie J,
Chen D, Wang Y and Li D: miR-144-5p enhances the radiosensitivity
of non-small-cell lung cancer cells via targeting ATF2. Biomed Res
Int. 2018:51094972018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji R, Cheng Y, Yue J, Yue J, Yang J, Liu
X, Chen H, Dean DB and Zhang C: MicroRNA expression signature and
antisense-mediated depletion reveal an essential role of microRNA
in vascular neointimal lesion formation. Circ Res. 100:1579–1588.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shan Z, Yao C, Li ZL, Teng Y, Li W, Wang
JS, Ye CS, Chang GQ, Huang XL, Li XX, et al: Differentially
expressed microRNAs at different stages of atherosclerosis in
ApoE-deficient mice. Chin Med J (Engl). 126:515–520.
2013.PubMed/NCBI
|
|
14
|
Hosin AA, Prasad A, Viiri LE, Davies AH
and Shalhoub J: MicroRNAs in atherosclerosis. J Vasc Res.
51:338–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alexandru N, Badila E, Weiss E, Cochior D,
Stępień E and Georgescu A: Vascular complications in diabetes:
Microparticles and microparticle associated microRNAs as active
players. Biochem Biophys Res Commun. 472:1–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kumar S, Kim CW, Simmons RD and Jo H: Role
of flow-sensitive microRNAs in endothelial dysfunction and
atherosclerosis: Mechanosensitive athero-miRs. Arterioscler Thromb
Vasc Biol. 34:2206–2216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun X, Belkin N and Feinberg MW:
Endothelial microRNAs and atherosclerosis. Curr Atheroscler Rep.
15:3722013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo Y, Ying L, Tian Y, Yang P, Zhu Y, Wang
Z, Qiu F and Lin J: miR-144 downregulation increases bladder cancer
cell proliferation by targeting EZH2 and regulating Wnt signaling.
FEBS J. 280:4531–4538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guan H, Liang W, Xie Z, Li H, Liu J, Liu
L, Xiu L and Li Y: Down-regulation of miR-144 promotes thyroid
cancer cell invasion by targeting ZEB1 and ZEB2. Endocrine.
48:566–574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu J, Xue H, Zhang J, Suo T, Xiang Y,
Zhang W, Ma J, Cai D and Gu X: MicroRNA-144 inhibits the metastasis
of gastric cancer by targeting MET expression. J Exp Clin Cancer
Res. 34:352015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iwaya T, Yokobori T, Nishidan N, Kogo R,
Sudo T, Tanaka F, Shibata K, Sawada G, Takahashi Y, Ishibashi M, et
al: Downregulation of miR-144 is associated with colorectal cancer
progression via activation of mTOR signaling pathway.
Carcinogenesis. 33:2391–2397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han S, Zhu J and Zhang Y: miR-144
potentially suppresses proliferation and migration of ovarian
cancer cells by targeting RUNX1. Med Sci Monit Basic Res. 24:46.
2018. View Article : Google Scholar
|
|
23
|
Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL,
Zhu YH, Dong SS, Kong KL, Chen J, Tsao SW, Guan XY and Fu L:
MicroRNA-144 promotes cell proliferation, migration and invasion in
nasopharyngeal carcinoma through repression of PTEN.
Carcinogenesis. 34:454–463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren K, Liu QQ, An ZF, Zhang DP and Chen
XH: miR-144 functions as tumor suppressor by targeting PIM1 in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:3028–3037.
2017.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alvarez RJ, Gips SJ, Moldovan N, Wilhide
CC, Milliken EE, Hoang AT, Hruban RH, Silverman HS, Dang CV and
Goldschmidt-Clermont PJ: 17beta-estradiol inhibits apoptosis of
endothelial cells. Biochem Biophys Res Commun. 237:372–381. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: miR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bou Kheir T, Futoma-Kazmierczak E,
Jacobsen A, Krogh A, Bardram L, Hother C, Grønbæk K, Federspiel B,
Lund AH and Friis-Hansen L: miR-449 inhibits cell proliferation and
is down-regulated in gastric cancer. Mol Cancer. 10:292011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical signifcance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–4284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai
CH, Kao HW, Fang WL, Huang KH, Chan WC and Lin WC: Epigenetic
regulation of miR-34b and miR-129 expression in gastric cancer. Int
J Cancer. 129:2600–2610. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bai M, Li J, Yang H, Zhang H, Zhou Z, Deng
T, Zhu K, Ning T, Fan Q, Ying G and Ba Y: miR-135b delivered by
gastric tumor exosomes inhibits FOXO1 expression in endothelial
cells and promotes angiogenesis. Mol Ther. 27:1772–1783. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng B, Yin WN, Suzuki T, Zhang XH, Zhang
Y, Song LL, Jin LS, Zhan H, Zhang H, Li JS and Wen JK:
Exosome-mediated miR-155 transfer from smooth muscle cells to
endothelial cells induces endothelial injury and promotes
atherosclerosis. Mol Ther. 25:1279–1294. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, Peng J, Ren Z, He NY, Li Q, Zhao
XS, Wang MM, Wen HY, Tang ZH, Jiang ZS, et al: Functional
regulatory roles of microRNAs in atherosclerosis. Clin Chim Acta.
460:164–171. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamada Y, Arai T, Kojima S, Sugawara S,
Kato M, Okato A, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of antitumor miR-144-5p targets oncogenes: Direct
regulation of syndecan-3 and its clinical significance. Cancer Sci.
109:2919–2936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, Wang R, Ge Y, Chen D, Wu B and Fang
F: Assessment of microRNA-144-5p and its putative targets in
inflamed gingiva from chronic periodontitis patients. J Periodontal
Res. 54:266–277. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang X, Sundquist K, Hedelius A, Palmér K,
Memon AA and Sundquist J: Circulating microRNA-144-5p is associated
with depressive disorders. Clin Epigenetics. 7:692015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zou Z, Chen J, Yang J and Bai X: Targeted
inhibition of rictor/mTORC2 in cancer treatment: A new era after
rapamycin. Curr Cancer Drug Targets. 16:288–304. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Heiss C, Rodriguez-Mateos A and Kelm M:
Central role of eNOS in the maintenance of endothelial homeostasis.
Antioxid Redox Signal. 22:1230–1242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qin B, Shu Y, Long L, Li H, Men X, Feng L,
Yang H and Lu Z: MicroRNA-142-3p induces atherosclerosis-associated
endothelial cell apoptosis by directly targeting rictor. Cell
Physiol Biochem. 47:1589–1603. 2018. View Article : Google Scholar : PubMed/NCBI
|